Abstract
Y chromosomes are different from other chromosomes because of a lack of recombination. Until now, complete sequence information of Y chromosomes has been available only for some primates, although considerable information is available for other organisms, e.g., several species of Drosophila. Here, we report the gene organization of the Y chromosome in the dioecious liverwort Marchantia polymorpha and provide a detailed view of a Y chromosome in a haploid organism. On the 10-Mb Y chromosome, 64 genes are identified, 14 of which are detected only in the male genome and are expressed in reproductive organs but not in vegetative thalli, suggesting their participation in male reproductive functions. Another 40 genes on the Y chromosome are expressed in thalli and male sexual organs. At least six of these genes have diverged X-linked counterparts that are in turn expressed in thalli and sexual organs in female plants, suggesting that these X- and Y-linked genes have essential cellular functions. These findings indicate that the Y and X chromosomes share the same ancestral autosome and support the prediction that in a haploid organism essential genes on sex chromosomes are more likely to persist than in a diploid organism.
Keywords: bryophyte, dioecy, Marchantia polymorpha
In many sexually dimorphic organisms, sex chromosomes play key roles in sex determination and sexual development. In mammals, for example, a female has two X chromosomes, whereas a male has one X and one Y chromosome (XX/XY system). The mammalian Y chromosome carries a gene that induces testis development (e.g., mouse SRY; refs. 1 and 2) and thus determines male sex. In humans, the Y chromosome is smaller than most of the other chromosomes and harbors <200 genes, whereas the X chromosome contains 1,098 genes (3). The Y chromosome has accumulated mutations and lost many genes, because of suppressed recombination with the X chromosome as reviewed and discussed by Charlesworth and Charlesworth (4). The canonical Drosophila Y chromosome is recombinationally isolated from the X and carries few genes, which appear to be autosomally rather than X-chromosomally derived. It is required for male fertility but plays no role in sex determination. This system has been drastically modified in a subset of Drosophila species (D. affinis and D. pseudoobscura groups; ref. 5). Intriguing is the observation and analysis of recently established sex chromosomes in Drosophila species such as the neo-Y and neo-X in D. miranda (6, 7). Here, the loss of recombination has been found to lead to drastic changes in gene content and structures (8, 9).
Sex determination systems in dioecious plants are thought to have evolved many times from hermaphroditic ancestors (10). Analogous to the mammalian XX/XY system, in the dioecious angiosperm white campion (Silene latifolia) males are X/Y and females XX. The Y chromosome induces male development (11). In papaya (Carica papaya), a dominant male-determining locus resides in a chromosomal region where recombination is suppressed and the DNA sequence has extensively diverged from its homologous region on the X chromosome (12). Therefore, this papaya chromosome is considered an immature Y chromosome in an emerging XX/XY system. In dioecious sorrel, Rumex acetosa, a male individual has one X and two Y chromosomes, whereas a female has two X chromosomes. Here, the presence of Y chromosomes has no influence on triggering male development, but as is the case in Drosophila, the X/autosomal balance determines the sex (13, 14).
In general, Y chromosomes preserve their genetic identity by suppressing recombination with X chromosomes, thus placing themselves on a unique evolutionary path. Investigations in various diploid organisms revealed that Y chromosomes tend to accumulate repeats and lose genes (4). However, comprehensive sequence data of Y chromosomes are available only for humans and chimpanzees (15–17).
Y chromosomes in haploid dioecious organisms also appear different from other chromosomes, but have been poorly studied. Under haploidy, both X and Y chromosomes should evolve in the same way (18), which will be discussed in detail later. The liverwort Marchantia polymorpha, an extant species of the earliest land plants (19), is dioecious, and the dominant forms in its life cycle are female and male haploid thalli. These are phenotypically identical until the female or male sexual organs differentiate. After fertilization by flagellated sperm, a diploid zygote develops into a sporophyte by mitosis, followed by meiosis to produce haploid spores, which germinate and develop into the next-generation thalli. The haploid thalli of both sexes can also propagate asexually by so-called gemmae.
Several bryophyte species have been reported to possess sex chromosomes (20); in M. polymorpha the haploid set of chromosomes consists of eight autosomes and a single sex chromosome, an X chromosome in females (n = 8+X) and a Y chromosome in males (n = 8+Y). Therefore, unlike in the XX/XY system, the X and Y chromosomes in M. polymorpha are separated from each other during most of the life cycle. In M. polymorpha, the X and Y chromosomes align during meiotic metaphase but keep some distance from each other and are in the diakinesis stage separated earlier than the autosomes (21, 22), suggesting that no recombination between the X and Y chromosomes is possible. The extensive sequence analysis of the M. polymorpha Y chromosome reported here not only provides an in-depth view into the gene content and structure of a plant sex chromosome but also yields insights into the evolution of recombination-suppressed sex chromosomes in a haploid genome.
Results and Discussion
Sequencing of the Marchantia Y Chromosome.
We had previously found that the M. polymorpha Y chromosome (10 Mb) contains a family of unique repeats that are represented by a 2.4-kb BamHI repeat confined to a 4-Mb segment of the Y chromosome (23) designated YR1 (Y chromosome region 1). We now obtained the sequence of YR1 by sequencing 28 P1-derived artificial chromosome (PAC) clones that collectively cover YR1, because the accumulation of the 2.4-kb BamHI repeat family made the construction of contigs impractical [see supporting information (SI) Text for details]. The total sequence obtained amounts to 3,200,899 bp (SI Table 3, SI Table 4, and SI Fig. 4).
For sequence analysis of the other 6-Mb segment of the Y chromosome region 2 (YR2), 59 tiled PAC clones were selected from two contigs of aligned PAC clones, contig A and contig B, that cover YR2 (see SI Text for details and SI Fig. 4). Three PCR-amplified DNA fragments filled mapping and tiling gaps in contig B. The total lengths of the sequences obtained were 3,467,261 bp for contig A and 2,530,874 bp for contig B, which account for >95% of their size as estimated from the physical mapping.
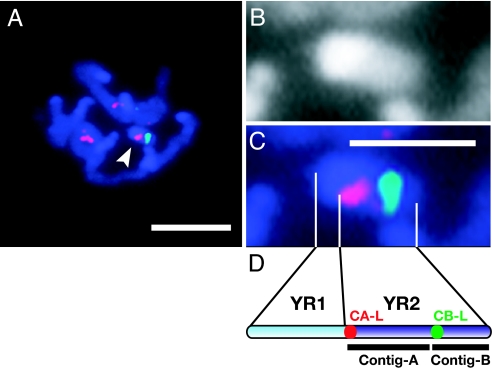
Contig A and contig B were cytologically mapped on the Y chromosome by FISH. One of the signals for the end of contig A terminated by clone pMM23–431A8 (CA-L; red signals in Fig. 1A and C) was detected in the immediate vicinity of the more condensed YR1 (Fig. 1B). The other signals for CA-L indicate the presence of sequences similar to CA-L on autosomes. The signal for the end of contig B terminated by clone pMM23–359F1 (CB-L; green signals in Fig. 1 A and C) was detected in the central region of YR2. This result aligns YR1, contig A, and contig B on the Y chromosome in the order illustrated in Fig. 1D.
Fig. 1.
Ordering of contig A and contig B on the M. polymorpha Y chromosome. (A) Detection of the left terminal regions of contig A (CA-L, red) and contig B (CB-L, green) on DAPI-stained prometaphase chromosomes (blue). The Y chromosome is indicated by an arrowhead. (B and C) Magnification of DAPI-stained and FISH images of the Y chromosome, respectively. (D) Schematic alignment of contig A and contig B on YR2. The positions of CA-L and CB-L are indicated by red and green dots, respectively. (Scale bars: 5 μm, A; 2 μm, C.)
Genes on the Marchantia Y Chromosome.
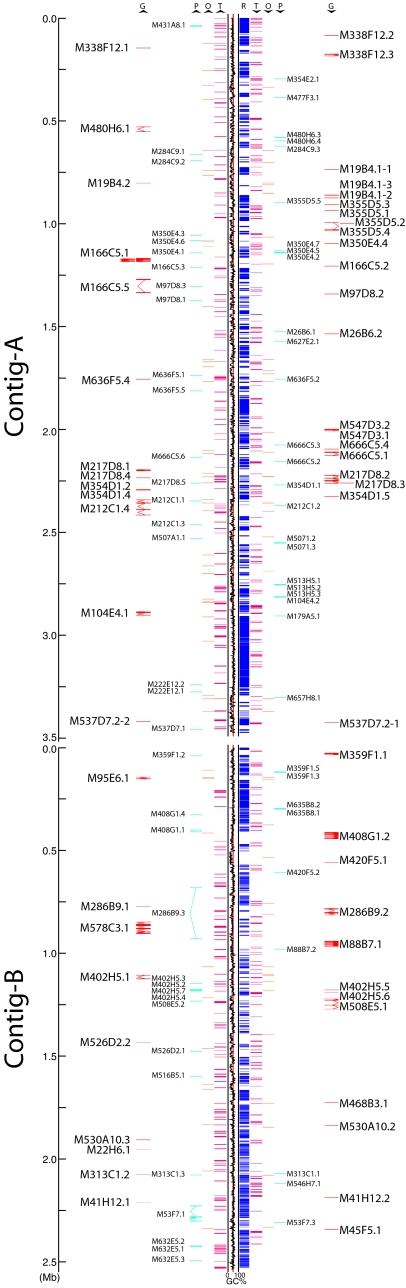
Similarity searches against the public sequence databases and M. polymorpha ESTs (see SI Text for details; SI Dataset 1) detected 64 genes, 9 in YR1 and 55 in YR2 (Table 1, Fig. 2, SI Table 5, and SI Table 6). Genomic PCR and/or Southern blotting shows sequences similar to 25 of the Y-chromosome genes are also in female DNA, whereas the remaining 39 genes appear to be present only in the male DNA (Table 1). However, six of the apparently male-specific genes turned out to have putative X-linked homologs (Table 2). Fourteen genes, one (M2D3.5) in YR1 and 13 in YR2, are unique to the male genome and in addition show sexual organ-specific expression (Table 1). These genes are thus candidates for male reproductive functions.
Table 1.
Genes identified on the M. polymorpha Y chromosome
| Chromosome region | Name | Presence in female | Expression |
|
|---|---|---|---|---|
| Sexual organ | Thallus | |||
| YR1 | M2D3.1 | + | + | + |
| M2D3.2 | + | + | + | |
| M2D3.3 | + | - | - | |
| M2D3.4 | - | + | + | |
| M2D3.5 (ORF162) | - | + | - | |
| M2D3.6 | + | + | + | |
| M8H2.3 | + | + | + | |
| M205B1.4 | + | + | - | |
| M123D8.8 | + | + | + | |
| YR2, Contig A | M338F12.2 | - | + | + |
| M338F12.1 | - | + | + | |
| M338F12.3 | + | + | + | |
| M480H6.1 | - | + | + | |
| M19B4.1-1 | - | - | - | |
| M19B4.1-2 | - | - | - | |
| M19B4.1-3 | - | + | - | |
| M19B4.2 | - | + | - | |
| M355D5.3 | + | + | + | |
| M355D5.1 | + | + | - | |
| M355D5.4 | - | + | - | |
| M355D5.2 | - | + | - | |
| M350E4.4 | - | + | - | |
| M166C5.1 | - | + | + | |
| M166C5.2 | - | + | + | |
| M166C5.5 | - | + | - | |
| M97D8.2 | + | + | + | |
| M26B6.2 | - | + | + | |
| M363F5.4 | + | + | + | |
| M547D3.2 | + | + | + | |
| M547D3.1 | - | + | + | |
| M666C5.4 | + | + | + | |
| M666C5.1 | - | + | - | |
| M217D8.1 | - | + | + | |
| M217D8.2 | - | + | + | |
| M217D8.4 | - | + | + | |
| M354D1.5 | + | + | - | |
| M212C1.4 | - | + | - | |
| M104E4.1 | - | + | + | |
| M537D7.2-1 | + | + | + | |
| M537D7.2-2 | + | + | - | |
| YR2, Contig B | M359F1.1 | - | + | + |
| M95E6.1 | - | + | + | |
| M408G1.2 | - | + | + | |
| M286B9.1 | - | + | + | |
| M420F5.1 | - | + | + | |
| M286B9.2 | - | + | + | |
| M578C3.1 | - | + | + | |
| M88B7.1 | - | + | + | |
| M402H5.1 | - | + | + | |
| M402H5.5 | + | + | + | |
| M402H5.6 | + | + | + | |
| M508E5.1 | - | + | - | |
| M526D2.2 | - | + | - | |
| M468B3.1 | - | + | - | |
| M530A10.2 | + | + | + | |
| M530A10.3 | - | + | - | |
| M22H6.1 | + | + | + | |
| M323C1.2 | + | + | + | |
| M41H12.2 | + | + | - | |
| M41H12.1 | - | - | - | |
| M45F5.1 | + | + | + | |
Genes whose homologs are found in animals but not in angiosperms are in bold.
Fig. 2.
Overview of YR2. The linear arrangements of contig A and contig B are shown with pertinent features indicated by the following letters: G, putative gene; P, pseudogene or EST homolog; O, organellar DNA insertion; T, transposable element; R, repeat. Arrowheads under the letters indicate their orientation.
Table 2.
Y chromosomal genes with X-linked putative homologs
| Name | Length, bp* Y/X-linked | Identity, % | ps† | pn† | ds† | dn† | Homolog (UniProt accession no.) | Accession X-linked |
|---|---|---|---|---|---|---|---|---|
| YR2, Contig A | ||||||||
| M338F12.1 | 709/709 | 80.1 | 0.86 | 0.02 | NC | 0.02 | Calcium-dependent protein kinase (Q94G52) | AB288007 |
| M547D3.1 | 142/142 | 93.6 | 0.31 | 0.00 | 0.39 | 0.00 | GTP-binding protein, ras-like (Q9FJH0) | AB288008 |
| M104E4.1 | 2,157/2,139 | 62.6 | 0.74 | 0.09 | 3.35 | 0.10 | Nonphototrophic hypocotyl 1a (Q9ST26) | AB272580 |
| YR2, Contig B | ||||||||
| M408G1.2 | 1,182/1,135 | 74.4 | 0.84 | 0.04 | NC | 0.04 | Serine/threonine protein phosphatase (BSL2_ARATH) | AB288009 |
| M88B7.1 | 430/430 | 85.6 | 0.50 | 0.03 | 0.82 | 0.03 | ZIM-like 1 protein (Q94B66) | AB288010 |
| M402H5.1 | 374/374 | 70.6 | 0.84 | 0.12 | NC | 0.13 | Hypothetical protein (Q8RWB1) | AB288011 |
A paternal BC2 population and primers listed in SI Table 8 were used for testing linkage to the X chromosome.
*Length of X-linked sequences amplified by degenerate PCR and that of corresponding regions of Y chromosomal genes. Note that the entire coding sequence of the M104E4.1 gene (AB272579) was compared with that of the F62B12.1 gene (AB272580) carried by the X-linked PAC clone, pMF28-62B12 (AB272581).
†ps(pn), the proportion of observed synonymous (nonsynonymous) substitutions; ds(dn), the Jukes-Cantor correction for multiple hits of ps(pn); NC, not calculated due to ps >0.75.
Although no closely similar homologs of the M. polymorpha Y-linked genes were found in searches of other species' Y chromosomes, we identified some putative genes from ORFs whose translated sequences resemble some animal male-fertility proteins. Among the 14 putative male reproductive genes, six encode proteins whose homologs are found in animals but not in angiosperms (genes in bold in Table 1, SI Table 5, SI Table 7, and SI Fig. 5). Because in bryophytes male gametes are flagellated sperm, spermatogenesis in M. polymorpha and animals could very well share some proteins of common evolutionary descent, and these six genes may be involved in analogous functions in spermatogenesis.
Another 40 genes on the Y chromosome are expressed in vegetative thalli and male sexual organs and thus may code for functions not related to male sexual differentiation. At least five genes in YR1 have homologs on autosomes and thus may not be essential (24).
Gene Density in the Y Chromosome.
In total, 64 genes are detected in >9 Mb of the Y chromosome by our similarity probing with genes identified in other organisms and/or the M. polymorpha ESTs. The current set of M. polymorpha EST sequences contains ≈10,000 nonredundant sequences, but is not saturated. Conservatively assuming 10,000 genes in the genome of 280 Mb, the 10-Mb Y chromosome would thus be predicted to carry 10,000 × 10/280 = 350 genes, or more. The observed lower gene content may be explained by successive degeneration of the Y chromosome, although the accumulation of repeats cannot be excluded as a possible cause. Further data on gene content and density in the other chromosomes of this plant will allow a more detailed evaluation of the observed gene density in the future.
Repeats and Transposable Elements.
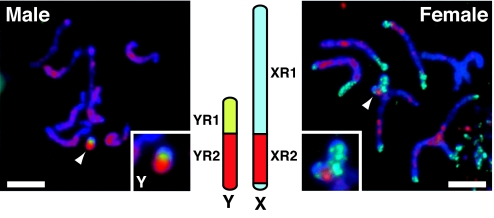
Both YR1 and YR2 are rich in repeats, but the origins of these repeats are strikingly different (see also SI Fig. 6). The YR1 domain consists of unique small repeat sequences of only several hundred nucleotides, which are assembled in various stoichiometries to form different arrangements of the 2.4-kb BamHI repeat family. Structures and sequences of these elements have been analyzed (23). When DNA of clone pMM23-104E4 (contig A of YR2), which is one of the first clones isolated from YR2 (25), and a fragment of the 2.4-kb BamHI repeat were simultaneously applied to prometaphase chromosomes of male plants in FISH experiments, the signal of the 2.4-kb BamHI repeat (yellowish green signal in Fig. 3Left) identifies only YR1, and the signal of pMM23-104E4 lights up only YR2 in its entirety (red signals in Fig. 3 Left).
Fig. 3.
Visualization of segments in the M. polymorpha sex chromosomes. Detection of pMM23-104E4 (red) on Giemsa-stained prometaphase chromosomes (blue). Signals of the 2.4-kb BamHI DNA fragment (yellowish green) were merged on the male chromosomes (Left). Similarly, signals of the 45S rDNA fragment (17S rDNA, light blue) were merged on the female chromosomes (Right). The male chromosomes were not probed with the 45S rDNA fragment. Arrowheads indicate the Y and the X chromosomes; the insets show magnifications. Schematic diagrams of the Y and X chromosomes displayed in the center indicate the color coding of the FISH images. (Scale bars: 5 μm.)
The M. polymorpha X chromosome (20 Mb) carries a large cluster of ribosomal DNA (45S rDNA) designated XR1 (X chromosome region 1), whereas no rDNA is found on the Y chromosome (26). When DNA of pMM23-104E4 and a fragment of the rDNA sequence on the X chromosome (27) were used in a FISH analysis of female chromosomes, the signal of pMM23-104E4 (red signals in Fig. 3 Right) was detected on a segment of the X chromosome (designated X chromosome region 2; XR2) distinct from the rDNA signals (light blue signals in Fig. 3 Right) and on autosomes. The contig-A region in YR2 of the Y chromosome thus contains sequences similar to motifs on a specific segment of the X chromosome and on autosomes.
Several repeats in YR2 are related to transposable elements. Although the total complexity of common interchromosomal repeats remains unknown without sequence information on the entire genome, intrasegmental repeats of at least 200 bp and at least 90% identity alone account for 43% of YR2 (Fig. 2). An interesting similarity to YR2 is seen in humans, where common repeats such as LINE1 (long interspersed nuclear element 1), Alu, and retroviral elements account for 47% of the euchromatic MSY (male-specific region of the Y chromosome; ref. 15).
Evolution of the M. polymorpha Sex Chromosomes.
Like X and Y chromosomes of other organisms, the M. polymorpha sex chromosomes probably originated from a regular autosome. A widely accepted scheme for the evolution of Y chromosomes in the XX/XY system consists of three major events: acquisition of the sex-determining loci, suppression of recombination, and genetic degeneration driven by evolutionary processes such as Muller's ratchet (4, 28). However, Bull (18) predicted that the evolution of sex chromosomes in a haploid system is different from that in a diploid system. In a haploid organism, degeneration should not occur, because it would impair essential genes. Essential genes on the Y chromosome should thus also be present on the X, because females will likewise require these genes. To test this hypothesis, we investigated five of the putative genes for general functions from YR2 by degenerate PCR based on their conserved amino acid sequences for similar coding sequences in the X chromosome (Table 2). We indeed detected female homologs for all five genes tested and confirmed their X-linkage by genomic PCR using a paternal BC2 population and primers specific to the female sequences (data not shown). The X- and Y-linked coding sequences of each pair have diverged to some extent (70.6–93.6% nucleotide identity; Table 2). Only one X-linked sequence homolog was recovered for each of the YR2 genes tested, suggesting that the X-linked sequences do not represent copies of gene families that incidentally reside on the X chromosome. Three of the X–Y gene pairs, M338F12.1, M408G1.2, and M402H5.1, appear to have reached saturation of synonymous substitutions, suggesting that recombination between the X and Y chromosomes has been suppressed for a long time. The relatively low synonymous site divergence of the M547D3.1 pair could be explained by gene conversion that occurred after the establishment of the sex chromosomes. The results above also suggest that the putative genes for general functions in YR2 are essential for the plant. In addition, one X chromosome-linked PAC clone carries a homolog of the Y chromosomal gene M104E4.1 (contig A of YR2 in Table 1, SI Fig. 7, and SI Text for details), and the M104E4.1 pair also shows a very high synonymous substitution rate (Table 2). These results suggest that YR2 and XR2 evolved from the same ancestral autosome, as predicted by Bull (18).
Although the high synonymous substitution rate observed among the X–Y gene pairs suggests that the M. polymorpha Y chromosome has been long established, the 14 putative male reproductive genes are far fewer than the 40 putative general function genes expressed in thalli and sexual organs. The higher proportion of putative general function genes on the M. polymorpha Y chromosome is consistent with the prediction that in a predominant haploid lifestyle degeneration must not impair nonredundant genes whose expression is essential to survival of the organism (18).
According to another prediction by Bull (18), the X and Y chromosomes in a haploid organism should show similar characteristics, including the extent of their degeneration. In a haploid organism, both X and Y chromosomes lack partners for recombination or genetic complementation, which places the two sex chromosomes under identical evolutionary pressures. Although we have shown that XR1, like YR1, consists of specific repetitive sequences in the form of an rDNA cluster with distinct intergenic sequences (27), it is necessary to examine XR2 in detail to verify the above prediction.
Materials and Methods
Plant Materials.
Thalli and sexual organs of M. polymorpha (E lines) were obtained as described (29).
DNA Sequencing.
Sequencing of PAC clones was performed by a shotgun method as described (23). Some of the shotgun reads were produced by Shimadzu, Kyoto, Japan. Paired-end sequence reads with a coverage of six to eight times were generated for each PAC clone. Sequence reads were then assembled and edited by using the computer programs phred (30, 31), Paracel GenomeAssembler (Paracel, Pasadena, CA), and consed version 12.0 (32). Some sequence gaps were filled by primer walking.
Sequence Analysis.
Searches for protein coding regions were performed against the nonredundant protein sequence database of the European Bioinformatics Institute by using the BLASTX program (33) and against M. polymorpha ESTs (34, 35) with the BLASTN program (33). Amino acid sequences were aligned by using the CLUSTALW program (36). The rate of synonymous substitutions per potential synonymous site and the rate of nonsynonymous substitutions per potential nonsynonymous site were calculated by using the SNAP package (37).
Genomic PCR.
PCR for determining sex specificity of putative genes and linkage analysis was performed basically as described (23). Template genomic DNAs were isolated as described (29), and 10 ng each was used per assay.
cDNA Library Construction and EST Analysis.
Poly(A)+ RNAs were isolated from WT male thalli and sexual organs by using Concert Plant RNA Reagent (Invitrogen, Carlsbad, CA) and PolyATract System 1000 (Promega, Madison, WI), and cDNA libraries were constructed by using the poly(A)+ RNAs and SUPERSCRIPT Plasmid System with GATEWAY Technology for cDNA Synthesis and Cloning (Invitrogen) according to the manufacturers' instructions. Complementary DNAs were cloned directionally and sequenced to generate 5′ ESTs. Sequence reads were first processed with the base-caller program phred. Low-quality sequences, vector sequences, and poly(A) tails at the end of each sequence were removed, and cleaned sequences <90 bp in length were rejected by using the sequence processing software Paracel TranscriptAssembler (Paracel). Subsequently, sequences originating from Escherichia coli, organelles and structural RNAs were removed. ESTs were clustered by using Paracel TranscriptAssembler, and each cluster was visually inspected and edited for consistency by using consed.
RT-PCR Analysis.
First-strand cDNA was synthesized from DNase-treated poly(A)+ RNA by using SuperScript II reverse transcriptase (Invitrogen) at 42°C with XhoSseEcoR-dT primer (5′-GAGAATTCCTGCAGGCTCGAGTTTTTTTTTTTTTTTTTT) for 60 min. A 20-μl reaction mixture was diluted to 400 μl with Tris–EDTA, and 1 μl of the diluted mixture was used as template in a 20-μl PCR amplification mix containing 10 pmol of the same primers used for the genomic PCR. Reactions without reverse transcriptase were performed to check genomic DNA contaminations.
Degenerate RT-PCR.
Total RNA was isolated from male and female thalli with a RNeasy Plant Mini kit (Qiagen, Hilden, Germany) and reverse-transcribed with ReverTra Ace (Toyobo, Osaka, Japan) and oligo(dT) (20) primer. Degenerate primers were designed based on conserved amino acid sequences (SI Table 8). Amplified DNA fragments of the predicted sizes were gel-purified, cloned into the HincII site of pUC18 or pUC19 vectors, and sequenced.
FISH.
Preparation of chromosome spreads from young thalli of M. polymorpha and FISH was performed as described (23, 26, 38). For ordering contig A and contig B (Fig. 1), an ≈400-kb sequence at one end of each contig was masked with Y chromosomal sequences other than itself by using RepeatMasker (www.repeatmasker.org) to remove dispersed repetitive sequences. Using the resulting unmasked stretches of sequences, 9 and 10 primer pairs were designed for contig A and contig B, respectively, by using Primer3 (39) (SI Table 9). DNA fragments amplified with the primer pairs of each contig were pooled and designated CA-L for contig A and CB-L for contig B. The pooled DNAs of CA-L and CB-L were labeled with digoxigenin (DIG)-11-dUTP and biotin-16-dUTP (both Roche Diagnostics, Basel, Switzerland), respectively. Biotin-labeled probes were detected by FITC-avidin DCS (Vector Laboratories, Burlingame, CA) with one cycle of avidin-biotin-conjugated amplification. DIG-labeled probe was detected with anti-DIG-rhodamine (Roche Diagnostics) and Alexa-546 donkey anti-sheep IgG (Invitrogen). For visualization of segments of the Y and X chromosomes (Fig. 3), DNA of pMM23–104E4 (25) was labeled with biotin-16-dUTP and detected with Cy5-streptavidin (GE Healthcare, Piscataway, NJ) after tyramide signal amplification (PerkinElmer, Wellesley, MA). The entire length of the 2.4-kb BamHI fragment for male chromosomes, and the 17S rDNA sequence, part of 45S rDNA, for female were labeled with Spectrum Green-dUTP (Abbott Molecular, Des Plaines, IL).
Supplementary Material
Acknowledgments
We thank C. J. Leaver and M. Takemura for discussion and comments on the manuscript; Y. Nakamura, S. Tabata, and K. Nakai for technical advice in sequence analyses; K. Ando, H. Shiba, S. Takayama, A. Isogai, and M. Ishiura for the use of sequencing equipment; K. J. Newton and F. Sato for discussions and encouragement; K. Fukui, N. Ohmido, and Y. Akiyama for suggestions in cytological studies; and N. Iwabe for advice in evolutionary studies. This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences, Ministry of Education, Culture, Sports, Science, and Technology Grants 16011257, 15011224, 14011222, 13460150, 10760188, 09306006, 07281101, 07281102, 08273211, and 07281208, and Research for the Future Program Grant JSPS-RFTF00L01607 from the Japan Society for the Promotion of Science. H.B., K.Y., T.B., A.H., T. Nishio, R.S., A.Y., M.K., C.K., Y.K. and H.N. were supported by the 21st Century Centers of Excellence Program of the Ministry of Education, Culture, Sports, Science, and Technology. K.I. and S.Y. were recipients of the Predoctoral Fellowship from the Japan Society for the Promotion of Science. S.-H.C. was supported by The Brain Korea 21 Program from the Korea Research Foundation.
Abbreviations
- PAC
P1-derived artificial chromosome
- rDNA
ribosomal DNA
- YR2
Y chromosome region 2.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. BJ840459–BJ873180).
This article contains supporting information online at www.pnas.org/cgi/content/full/0609054104/DC1.
References
- 1.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 2.Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R. Nature. 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 3.Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlesworth B, Charlesworth D. Philos Trans R Soc London B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho AB, Clark AG. Science. 2005;307:108–110. doi: 10.1126/science.1101675. [DOI] [PubMed] [Google Scholar]
- 6.Bachtrog D, Charlesworth B. Nature. 2002;416:323–326. doi: 10.1038/416323a. [DOI] [PubMed] [Google Scholar]
- 7.Bachtrog D. Genome Res. 2005;15:1393–1401. doi: 10.1101/gr.3543605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachtrog D. Nat Genet. 2004;36:518–522. doi: 10.1038/ng1347. [DOI] [PubMed] [Google Scholar]
- 9.Bachtrog D. Nat Genet. 2003;34:215–219. doi: 10.1038/ng1164. [DOI] [PubMed] [Google Scholar]
- 10.Charlesworth D. Heredity. 2002;88:94–101. doi: 10.1038/sj.hdy.6800016. [DOI] [PubMed] [Google Scholar]
- 11.Westergaard M. Adv Genet. 1958;9:217–281. doi: 10.1016/s0065-2660(08)60163-7. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu Q, Pearl HM, Kim MS, Charlton JW, Stiles JI, et al. Nature. 2004;427:348–352. doi: 10.1038/nature02228. [DOI] [PubMed] [Google Scholar]
- 13.Ainsworth C, Parker J, Buchanan-Wollaston V. Curr Top Dev Biol. 1998;38:167–223. doi: 10.1016/s0070-2153(08)60247-1. [DOI] [PubMed] [Google Scholar]
- 14.Juarez C, Banks JA. Curr Opin Plant Biol. 1998;1:68–72. doi: 10.1016/s1369-5266(98)80130-1. [DOI] [PubMed] [Google Scholar]
- 15.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 16.Hughes JF, Skaletsky H, Pyntikova T, Minx PJ, Graves T, Rozen S, Wilson RK, Page DC. Nature. 2005;437:100–103. doi: 10.1038/nature04101. [DOI] [PubMed] [Google Scholar]
- 17.Kuroki Y, Toyoda A, Noguchi H, Taylor TD, Itoh T, Kim DS, Kim DW, Choi SH, Kim IC, Choi HH, et al. Nat Genet. 2006;38:158–167. doi: 10.1038/ng1729. [DOI] [PubMed] [Google Scholar]
- 18.Bull JJ. Evolution of Sex Determining Mechanisms. Menlo Park, CA: Benjamin-Cummings; 1983. [Google Scholar]
- 19.Qiu YL, Palmer JD. Trends Plant Sci. 1999;4:26–30. doi: 10.1016/s1360-1385(98)01361-2. [DOI] [PubMed] [Google Scholar]
- 20.Newton ME. In: The Experimental Biology of Bryophytes. Dyer AF, Duckett JG, editors. London: Academic; 1984. pp. 65–96. [Google Scholar]
- 21.Haupt G. Z Indukt Abstamm Vererbungsl. 1932;62:367–428. [Google Scholar]
- 22.Lobeer G. Jahrb Wiss Bot. 1934;80:567–817. [Google Scholar]
- 23.Okada S, Sone T, Fujisawa M, Nakayama S, Takenaka M, Ishizaki K, Kono K, Shimizu-Ueda Y, Hanajiri T, Yamato KT, et al. Proc Natl Acad Sci USA. 2001;98:9454–9459. doi: 10.1073/pnas.171304798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishizaki K, Shimizu-Ueda Y, Okada S, Yamamoto M, Fujisawa M, Yamato KT, Fukuzawa H, Ohyama K. Nucleic Acids Res. 2002;30:4675–4681. doi: 10.1093/nar/gkf604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujisawa M, Hayashi K, Nishio T, Bando T, Okada S, Yamato KT, Fukuzawa H, Ohyama K. Genetics. 2001;159:981–985. doi: 10.1093/genetics/159.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama S, Fujishita M, Sone T, Ohyama K. Chromosome Res. 2001;9:469–473. doi: 10.1023/a:1011676328165. [DOI] [PubMed] [Google Scholar]
- 27.Fujisawa M, Nakayama S, Nishio T, Fujishita M, Hayashi K, Ishizaki K, Kajikawa M, Yamato KT, Fukuzawa H, Ohyama K. Chromosome Res. 2003;11:695–703. doi: 10.1023/a:1025941206391. [DOI] [PubMed] [Google Scholar]
- 28.Charlesworth B. Proc Natl Acad Sci USA. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takenaka M, Yamaoka S, Hanajiri T, Shimizu-Ueda Y, Yamato KT, Fukuzawa H, Ohyama K. Transgenic Res. 2000;9:179–185. doi: 10.1023/a:1008963410465. [DOI] [PubMed] [Google Scholar]
- 30.Ewing B, Hillier L, Wendl MC, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 31.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 32.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiyama R, Yamato KT, Miura K, Sakaida M, Okada S, Kono K, Takahama M, Sone T, Takenaka M, Fukuzawa H, et al. DNA Res. 2000;7:165–174. doi: 10.1093/dnares/7.3.165. [DOI] [PubMed] [Google Scholar]
- 35.Nagai J, Yamato KT, Sakaida M, Yoda H, Fukuzawa H, Ohyama K. DNA Res. 1999;6:1–11. doi: 10.1093/dnares/6.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama S, Fujishita M, Ohyama K. In: Plant Genome: Biodiversity and Evolution: Lower Groups. Sharma AK, Sharma A, editors. Vol 2A. Enfield, NH: Science Publishers; 2004. pp. 235–246. [Google Scholar]
- 39.Rozen S, Skaletsky H. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editors. Totowa, NJ: Humana; 2000. pp. 365–386. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.