Abstract
In regenerating muscle cells, muscle regulatory factor (MRF) 4 is normally the last of the four MRFs to be expressed. To analyze how the timing of MRF4 expression affects muscle regeneration, we compared regeneration after local freeze injury of muscles from wild-type mice with muscles from transgenic mice in which MRF4 expression was under control of an ∼1.6-kb fragment of the myogenin promoter. Three days after injury, masseter and tibialis anterior (TA) muscles in wild-type mice expressed little or no MRF4 mRNA; whereas these muscles in transgenic mice expressed abundant MRF4 mRNA from both the transgene and the endogenous gene. Thus, MRF4 up-regulation was accelerated in transgenic compared to wild-type regenerating muscles, and expression of the transgene appeared to activate, perhaps indirectly, expression of the endogenous MRF4 gene. At 11 days after injury, regeneration, as measured by cross-sectional area and density of regenerated fibers, was significantly impaired in transgenic TA compared to wild-type TA, whereas at 19 days after injury both transgenic and TA muscle fibers had fully recovered to preinjury values. Regeneration of masseter muscles, which normally regenerate much less completely than TA muscles, was unaffected by the transgene. Thus, the timing of MRF4 up-regulation, as well as additional muscle-specific factors, can determine the progress of muscle regeneration.
Regeneration of skeletal muscles after injury and formation of skeletal muscles during development are both regulated by a family of muscle-specific, basic helix-loop-helix transcription factors, including muscle regulatory factor (MRF) 4, myogenin, MyoD, and Myf-5. 1-3 After muscle injury, Myf-5 and MyoD are typically the first of the MRFs to be expressed in the regenerating muscle cells, followed by myogenin, and finally MRF4. 4,5 Each of these MRFs also has a unique spatial and temporal pattern of expression during development. 1-3,6 These expression patterns raise the possibility that normal muscle regeneration requires a precise sequence of expression of the four MRFs. In this study we examine this possibility by determining how muscle regeneration is affected when the timing of MRF4 expression is altered.
Muscle regeneration after injury is performed by a population of mononucleate cells, termed satellite cells. Satellite cells reside in uninjured muscle as quiescent cells, but are activated in injured muscle to first divide and then differentiate to repair the injury. 7,8 Quiescent satellite cells respond within a day to injury by beginning to express MyoD and/or Myf-5, 9 although the complete process of regeneration to reform normal tissue architecture typically requires 3 to 4 weeks. 10 After injury, different muscles regenerate to different extents and muscles with the capacity for complete regeneration contain more activated satellite cells than muscles that do not regenerate completely. 10,11 For example, the tibialis anterior (TA) of the mouse can regenerate completely in less than 3 weeks and the injured TA contains two to three times more activated satellite cells than the masseter muscle that shows incomplete regeneration after 3 to 4 weeks. 10
Previous work has used knockout mice to examine the role of individual MRFs and other muscle transcription factors in muscle regeneration. Because regeneration is severely impaired in MyoD (−/−) mice, 12 it is clear that MyoD is a key regulator of regeneration. Because MRF4 expression is greatly reduced in regenerating MyoD(−/−) compared to wild-type muscles, it seems that one role of MyoD in activated satellite cells is to directly or indirectly activate the high level of MRF4 expression found in the later stages of regeneration. Similar experiments have not been reported for MRF4 (−/−) mice and cannot be performed for myogenin (−/−) or Myf-5 (−/−) mice, because these die at birth. 2,3 The myocyte nuclear factor is also required for successful muscle regeneration after injury. 13
In this work we use transgenic mice to examine how an alteration in the timing of MRF expression affects muscle regeneration after injury. In particular, we examined regeneration in mice carrying a transgene in which expression of MRF4 is under control of a myogenin promoter fragment. 14,15 We show that MRF4 is expressed sooner after injury in transgenic muscles than in wild-type muscles. In addition, premature expression of MRF4 from the transgene was accompanied by a similarly premature expression of the endogenous MRF4 gene, suggesting that MRF4 acts in a positive feedback loop to regulate its own expression in regenerating muscle. Furthermore, in transgenic TA, but not masseter, there was a significant, although transient, attenuation of regeneration at intermediate stages of recovery. Thus, muscles with different intrinsic capacities for regeneration showed different responses to an altered timing of MRF4 expression.
Experimental Procedures
Transgenic Mice, Genotyping, mRNA Analyses
Transgenic mice carrying a myogenin promoter fragment-MRF4 cDNA fusion gene, termed myo1565-MRF4 (locus 43), were generated previously. 14 The myo1565-MRF4 fusion gene includes the rat MRF4 cDNA 16 under control of nucleotides −1565 to +18, relative to the transcription start site, of the mouse myogenin promoter region. 17 A polymerase chain reaction assay of tail genomic DNA was used to identify mice that carried the myo1565-MRF4 transgene. 14 To distinguish the shorter mRNA produced by the endogenous MRF4 gene from the longer mRNA produced by the transgene, 10-μg samples of total RNA isolated from skinned and deboned hindlimb muscles were separated by electrophoresis, transferred to nylon, and hybridized with a 32P-labeled MRF4 probe. 14,15 Probes for myogenin, MyoD, and Myf-5 mRNAs were also used as previously described. 14,15,18 Consistent results were found when RNAs were obtained from multiple muscles (for replicates, from two to four muscles were analyzed; representative blots are shown).
Induced Regeneration of Skeletal Muscle
Mice (2 to 5 months old) were anesthetized by an intraperitoneal injection of 87 mg/kg ketamine and 13 mg/kg xylazine, and an incision ∼3 mm was made overlying the TA and masseter muscles. Muscles were subject to a standardized local freeze injury as previously described. 10 The incisions were sutured shut. At different times after injury, animals were euthanized using CO2 inhalation and the muscles were removed using standardized dissection methods. Time points were chosen based on the previously determined time course of regeneration in wild-type TA and masseter muscles. 10
Histological Analyses
The isolated muscles were embedded in OCT mounting medium and frozen in isopentane cooled in liquid nitrogen. For histological analyses, 10 to 12 14-μm cross sections were collected along the entire length of the muscles at 400- to 500-μm intervals and stained with hematoxylin and eosin (H&E). All analyses and photography were performed on a Zeiss Axioplan microscope equipped with a video camera and Scion Image software (Scion Corp., Frederick, MD).
In experiments analyzing regenerating muscles, standardized methods were used as previously described. 19-22 Briefly, sections containing the largest area of damage were selected for measurements. The core of the damaged area, as defined by the region that was least regenerated, was visualized using a ×10 objective, and the image was captured to a computer screen. The cross-sectional area (CSA) and total number of individual central nucleate, regenerating myofibers within these 0.15- to 0.3-mm2 fields were determined at specified times after injury. This method of analysis is reproducible as multiple trained observers (n = 4) choose the same sections for analysis. For example, in one examination of 79 samples, results were identical for 76 (96.2%). Because multiple samples were used to determine averages at each time point and for each genotype, such small differences among observers did not affect the results. No difference was observed either in the size of the injured area or in the percentage of the injured area that was analyzed between wild-type and transgenic mice. Myofibers with CSA smaller than 100 μm2 were not included in the analysis so as not to mistake regenerating myofibers with mononucleated cells in the damaged area.
For analyses of myofiber CSA in undamaged muscles, anatomical markers were used to find the same region in different samples and these sections were subsequently used for analysis. The CSA of individual myofibers was determined by capturing an image in the center of each section and analyzing 100 to 250 myofibers within this 0.3-mm2 field. Both the unpaired Student’s t-test and the nonparametric Mann-Whitney test were used to evaluate the significance of differences in the mean myofiber CSA or mean number of fibers per unit area between control and transgenic mice.
Results
This study was designed to determine how muscle regeneration is affected by premature expression of MRF4 in injured muscles. To this end, we compared regeneration in muscles from myo1565-MRF4 transgenic mice with that in their nontransgenic littermates. We expected that MRF4 would be expressed sooner after injury in transgenic muscles than in nontransgenic muscles. One group of our analyses, therefore, was used to confirm that the transgenic mice showed the predicted patterns of MRF4 expression.
First, however, we showed that the myo1565-MRF4 transgene did not alter the outcome of normal muscle development. TA and masseter muscles were prepared from transgenic and nontransgenic littermates at 2 to 5 months of age, sectioned, and H&E stained. Muscles in the transgenic and nontransgenic mice were of similar weight, had similar numbers of myofibers, and had similar fiber type percentages (J. B. Miller, unpublished). 14,15 In addition, both the mean CSAs of myofibers and the mean number of myofibers per unit area (myofiber density) were the same in transgenic and nontransgenic muscles (Table 1) ▶ . These results were consistent with previous analyses showing that adult myo1565-MRF4 mice have normal fiber type composition and muscle morphology. 14,15 Thus, by these measures, the uninjured TA and masseter muscles in myo1565-MRF4 transgenic mice were histologically identical to the same muscles in their nontransgenic littermates.
Table 1.
Effect of Genotype on Regeneration in Tibialis Anterior and Masseter Muscles
| Muscle | Days after injury | Genotype | Myofiber cross-sectional area (μm2), mean ± SE (no. mice)* | Number myofibers, mean ± SE (no. mice)* |
|---|---|---|---|---|
| Tibialis anterior | Uninjured | myo1565-MRF4 | 2227 ± 50 (4) | 112.0 ± 3.0 (4) |
| wild-type | 2203 ± 125 (3) | 118.3 ± 6.4 (3) | ||
| 6 | myo1565-MRF4 | 672 ± 35 (4) | 186.8 ± 19.7 (4) | |
| wild-type | 784 ± 166 (3) | 150.0 ± 20.7 (3) | ||
| 11 | myo1565-MRF4 | 639 ± 70 (6)‡ | 280.3 ± 27.3 (6)† | |
| wild-type | 1050 ± 60 (8)‡ | 201.9 ± 9.5 (8)† | ||
| 19 | myo1565-MRF4 | 2420 ± 144 (4) | 105.3 ± 7.5 (4) | |
| wild-type | 2477 ± 250 (4) | 106.3 ± 2.7 (4) | ||
| Masseter | Uninjured | myo1565-MRF4 | 1401 ± 39 (4) | 149.5 ± 8.8 (4) |
| wild-type | 1571 ± 73 (4) | 155.3 ± 9.3 (4) | ||
| 11 | myo1565-MRF4 | 548 ± 84 (4) | 314.3 ± 19.1 (4) | |
| wild-type | 567 ± 74 (3) | 306.0 ± 8.7 (3) | ||
| 19 | myo1565-MRF4 | 924 ± 34 (3) | 247.0 ± 25.0 (3) | |
| wild-type | 1009 ± 31 (5) | 243.2 ± 16.1 (5) | ||
| 26 | myo1565-MRF4 | 997 ± 106 (4) | 269.3 ± 31.8 (4) | |
| wild-type | 1046 ± 85 (5) | 238.0 ± 29.8 (5) |
*In uninjured muscles, the cross-sectional area and number of normal, peripherally nucleate myofibers/0.3 mm2 were determined. In injured muscles, the cross-sectional area and number of central nucleate, regenerating myofibers were determined.
†Transgenic and wild-type myofiber densities are significantly different (P < 0.01 by Student’s unpaired, two-tailed t-test; and P = 0.029 by Mann-Whitney nonparametric test).
‡Transgenic and wild-type myofiber areas are significantly different (P < 0.001 by Student’s unpaired, two-tailed t-test; and P = 0.0027 by Mann-Whitney nonparametric test).
We next used Northern blotting to determine how muscle injury and expression of the transgene affected expression of endogenous MRF mRNAs. At 3 or 11 days after surgery, we dissected both the injured and the contralateral uninjured TA and masseter muscles from each of the transgenic and nontransgenic littermates. Northern blot analysis was then used to examine expression of the transgene MRF4 mRNA, as well as the endogenous MRF4, myogenin, and MyoD mRNAs in individual muscles.
As expected, 16 uninjured muscles in both transgenic and wild-type mice had abundant MRF4 mRNA produced from the endogenous gene (Figure 1) ▶ . (The endogenous MRF4 mRNA is smaller and thus distinguishable on Northern blots from the transgene MRF4 mRNA 15 .) Thus, expression of the myo1565-MRF4 transgene did not alter expression of the endogenous MRF4 mRNA in uninjured muscles. Unexpectedly, the transgene MRF4 mRNA was also expressed in uninjured transgenic muscles (Figure 1 ▶ , see Discussion).
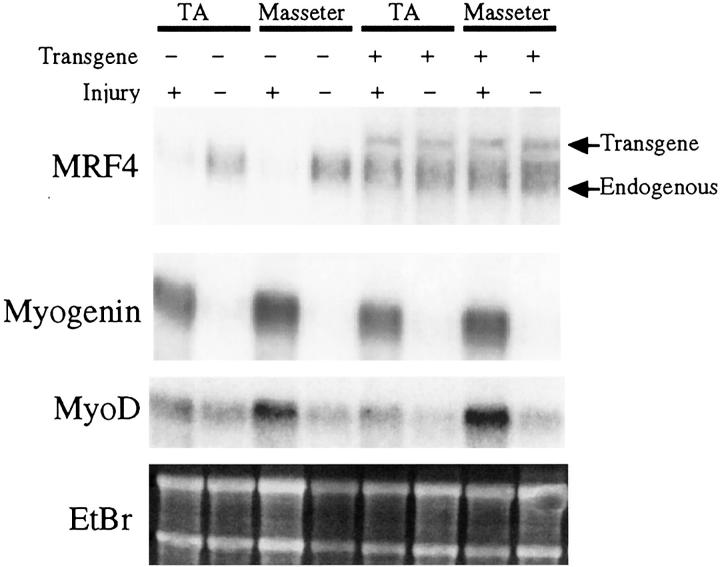
Figure 1.
Effect of myo1565-MRF4 expression on MRF expression at 3 days after injury. RNAs were prepared from the TA and masseter muscles of injured (3 days after injury) and uninjured wild-type and transgenic mice as indicated. Ethidium bromide-stained ribosomal RNAs (EtBr) show equal amounts of RNA in each lane. Northern blots analyzed the myo1565-MRF4 mRNA (transgene), the endogenous MRF4 mRNA (which, as indicated, is shorter than the transgene mRNA), the endogenous myogenin mRNA, and the endogenous MyoD mRNA. Injured transgenic, but not wild-type, muscles had abundant MRF4 mRNA (both endogenous and transgenic), indicating that up-regulation of MRF4 was accelerated in transgenic muscle cells after injury. In contrast, myogenin and MyoD mRNA levels were unaffected by the transgene.
At 3 days after injury, regenerating wild-type and transgenic muscles showed a clear difference in expression of the endogenous MRF4 mRNA. The endogenous MRF4 mRNA was nearly undetectable at 3 days after injury in regenerating wild-type muscles, but it was abundant in regenerating transgenic muscles at the same time (Figure 1) ▶ . The dramatic decrease in endogenous MRF4 mRNA levels on injury of wild-type muscles reflects the loss of the mature myofibers in which MRF4 is highly expressed. The transgene MRF4 mRNA was also expressed at 3 days after injury in the transgenic muscles. The level of MRF4 mRNAs in the transgenic muscles was similar to that in uninjured muscles, suggesting that the transgene produced a physiologically significant level of MRF4, as expected from previous analyses of this transgene during development in utero. 14,15 In contrast to the difference at 3 days after injury, the transgenic and wild-type muscles at 11 days after injury no longer differed in expression of the endogenous MRF4 mRNA (Figure 2) ▶ . The transgene MRF4 mRNA was expressed at 11 days after injury, as it was in both uninjured muscle and at 3 days after injury. These results showed that expression of MRF4 (from both the endogenous gene and transgene) was higher at 3 days after injury in transgenic compared to wild-type muscles.
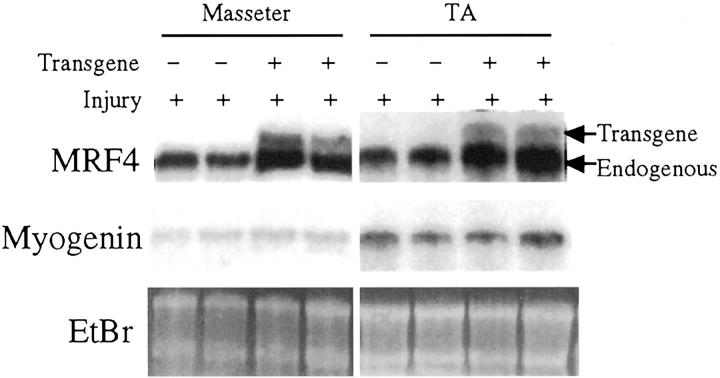
Figure 2.
MRF4 and myogenin mRNA levels in injured muscles at 11 days after injury were unaffected by expression of myo1565-MRF4. In contrast to 3 days after injury in which MRF4 mRNA was much more abundant in injured transgenic than wild-type muscles (see Figure 1 ▶ ), the MRF4mRNA levels at 11 days after injury were similar in transgenic and wild-type TA and masseter muscles. Myogenin mRNA values were also unaffected by the transgene at 11 days, as at 3 days (Figure 1) ▶ , after injury. Ethidium bromide-stained ribosomal RNAs (EtBr) show equal amounts of RNA in each lane.
In contrast to the effect on endogenous MRF4 mRNA levels at 3 days after injury, expression of the transgene had no effect on the levels of the endogenous MyoD or myogenin mRNAs (Figure 1) ▶ . In particular, uninjured wild-type and transgenic muscles both had a nearly undetectable level of myogenin mRNA, whereas injured wild-type and transgenic muscles both had abundant myogenin mRNA by 3 days after injury. At 11 days after injury, the myogenin mRNA remained readily detectable in both transgenic and nontransgenic muscles (Figure 2) ▶ . For all conditions, the TA and masseter muscles showed similar patterns of expression. At both 3 days and 11 days after injury, MyoD mRNA levels also appeared unaffected by the transgene, as levels were similar in transgenic and nontransgenic muscles (Figure 1 ▶ and not shown).
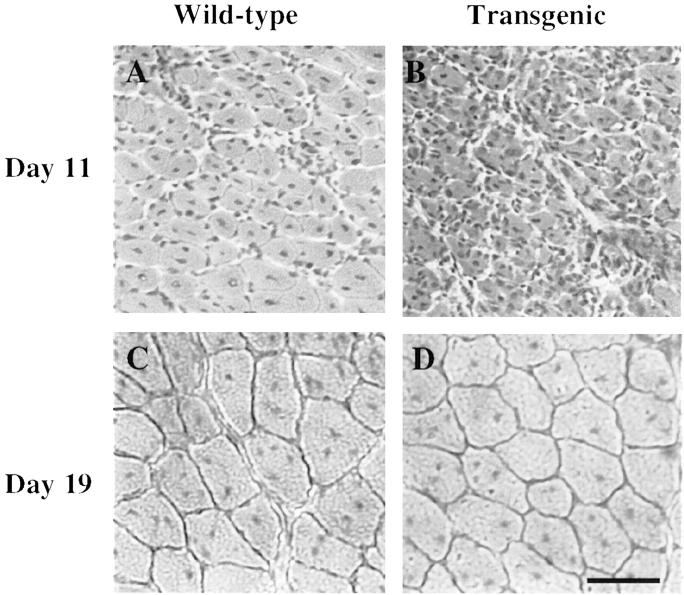
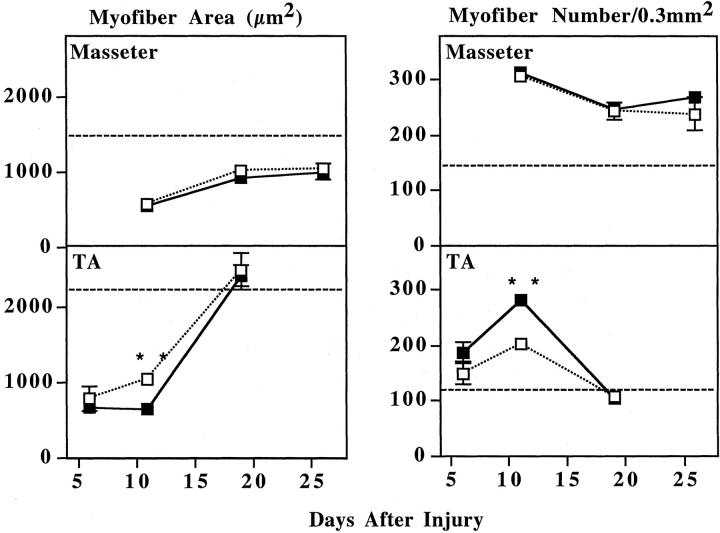
By histology, regeneration in transgenic TA appeared to be attenuated compared to wild-type TA at 11 days after injury. When sections from regenerating TA muscles were examined by H&E staining at 11 days after injury, the transgenic TA muscles appeared to contain many more small-diameter fibers than the nontransgenic muscles (Figure 3, A and B) ▶ . By 19 days after injury, however, the transgenic and wild-type TA muscles showed similar extents of regeneration (Figure 3, C and D) ▶ . In contrast to the TA results, the transgenic and nontransgenic masseter muscles appeared very similar to each other at 11 days, as well as at 19 and 26 days, after injury (not shown). Quantitative analyses of myofiber CSAs at multiple times after injury confirmed that, at the intermediate stage of recovery, the regenerated myofibers were smaller in transgenic than in nontransgenic TA muscles. In particular, the average myofiber CSA in the transgenic TA at 11 days after injury was only approximately two-thirds of that in the wild-type TA, which was a significant difference (P < 0.001 by Student’s unpaired two-tailed t-test and P = 0.0027 by Mann-Whitney nonparametric test) (Figure 4 ▶ , Table 1 ▶ ). The transgenic TA myofibers were also smaller than wild-type in CSA at day 6 after injury, but this difference did not reach significance. By day 19 after injury the myofibers in transgenic and nontransgenic TA muscles no longer differed in CSA and both had recovered to their values before injury (Figure 4 ▶ , Table 1 ▶ ).
Figure 3.
Attenuated regeneration in transgenic TA at 11 days, but not 19 days, after injury. H&E-stained sections were prepared from regenerating wild-type (A, C) and transgenic (B, D) TA muscles at 11 days (A, B) and 19 days (C, D) after injury as indicated. Regeneration appeared attenuated in the transgenic compared to the wild-type TA at 11 days after injury, but not at 19 days after injury. See Figure 4 ▶ and Table 1 ▶ for quantitation. Scale bar, 60 μm.
Figure 4.
Transgenic TA, but not masseter, muscles showed transient attenuation of regeneration. Masseter and TA muscles from wild-type (dotted line, open squares) and transgenic (solid line, filled squares) mice were injured and collected at 6 (TA only), 11, 19, or 26 (masseter only) days after injury. As described in Experimental Procedures, both the mean CSA and the density (number/0.3 mm2) of regenerating, central nucleate myofibers was measured. At 11 days after injury, the transgenic TA muscles showed a significantly smaller mean CSA and a significantly higher fiber density when compared to similarly injured wild-type TA muscles. In contrast, transgenic and wild-type values did not differ significantly at day 6 or day 19 in the TA or at any day in the masseter. The TA muscles regenerated much more rapidly and completely than the masseter muscles. Data are mean ± SE; see Table 1 ▶ for values and number of mice examined. Dashed horizontal lines indicate the approximate myofiber area or density of uninjured muscles (see Table 1 ▶ ). Asterisks indicates that the two values are significantly different (by both Student’s two-tailed, unpaired t-test and Mann-Whitney nonparametric test; see text for values).
In addition to the difference in CSAs, transgenic and nontransgenic TA muscles also differed in the density of central nucleate myofibers at 11 days after injury. The mean density of central nucleate fibers in the transgenic TA at 11 days after injury was ∼1.4 times greater than in the wild-type TA, which was a significant difference (P < 0.01 by Student’s t-test and P = 0.029 by Mann-Whitney test) (Figure 4 ▶ , Table 1 ▶ ). In contrast to this fiber density difference, we found that wild-type and transgenic TAs at day 11 had statistically identical total CSAs (average ± SE = 6.9 ± 0.5 mm2, n = 8, for wild-type; and 6.5 ± 0.2 mm2, n = 6, for transgenic; P = 0.49 by Mann-Whitney test), showing that neither muscle size nor injury-induced edema differed between transgenic and wild-type TA at 11 days after injury. Furthermore, the wild-type and transgenic TAs at day 11 were statistically identical in the total muscle area that was damaged by the freeze injury (average ± SE was 1.30 ± 0.10 mm2, n = 8 for wild-type and 1.47 ± 0.19 mm2, n = 6 for transgenic; t-test, P = 0.41) and in the area of damage that we analyzed (average ± SE was 0.164 ± 0.025 mm2, n = 8 for wild-type and 0.195 ± 0.019 mm2, n = 6 for transgenic; t-test, P = 0.37), suggesting that the significant differences in wild-type and transgenic TA fiber densities at day 11 were not because of variability in injury induction or measurement bias.
The density of central nucleate myofibers was also higher in the transgenic than in the wild-type TA at day 6 after injury, but this difference did not reach significance. By day 19 after injury, the density of central nucleate myofibers was the same in transgenic and nontransgenic TA muscles.
In contrast to the TA, no significant differences were detected between the transgenic and nontransgenic masseter muscles at 11, 19, or 26 days after injury (Figure 4 ▶ , Table 1 ▶ ). Both the CSAs and the densities of central nucleate myofibers were similar at all stages of regeneration in transgenic and nontransgenic masseters. Even 26 days was not sufficient for the masseter muscles to return to preinjury values, whereas the TA returned to preinjury values after only 19 days. Regeneration in masseter muscles, whether transgenic or not, was thus less extensive than in TA muscles, as was also found in an earlier study on wild-type masseter regeneration. 10
Discussion
At 3 days after injury, transgenic myo1565-MRF4, but not wild-type, mice expressed MRF4 in regenerating TA and masseter muscles. Thus, MRF4 mRNA was abundant in transgenic muscles at an earlier time after injury than in nontransgenic muscles. In transgenic muscles, MRF4 was expressed at this early time after injury from both the endogenous gene and the transgene, indicating that expression of the transgene also activated expression of the endogenous gene. In transgenic TA muscles, there was also a transient delay in regeneration compared to nontransgenic TA muscles. In contrast, masseter muscles regenerated at the same rate irrespective of the presence of the transgene. We discuss these findings below.
Because the endogenous MRF4 mRNA (as well as the transgene mRNA) was abundant in transgenic, but not nontransgenic, muscles at 3 days after injury, we conclude that MRF4 participates in a positive autoregulatory loop to activate its own promoter in regenerating muscle. Previous studies of myo1565-MRF4 mice showed that MRF4 participates in a positive autoregulatory loop during embryonic and fetal development. 14,15 In regenerating adult muscle and in activated satellite cells, MRF4 appears after myogenin 5,23 and myogenin appears to be required for activation of MRF4 expression in embryos. 15,24 The delay between myogenin and MRF4 expression in normal muscle regeneration suggests that factors in addition to myogenin may be required to activate MRF4 expression in wild-type muscles. Our results are consistent with this idea, because at 3 days after injury, myogenin was abundant in both wild-type and transgenic muscles, whereas MRF4 was abundant only in transgenic muscles. Positive autoregulation by MRF4 of its own promoter, if it indeed occurs in normal myofibers, could maintain the high level of MRF4 found in adult myofibers that ordinarily have little or no myogenin. 16,25
Expression of the myo1565-MRF4 transgene in uninjured adult muscle was unexpected. Because the endogenous myogenin mRNA is rare in adult muscle, we expected that the myo1565 fragment of the myogenin promoter would not be highly expressed in adult muscle. It may be that regulatory elements that down-regulate myogenin expression in some or all adult myofibers are missing from the myo1565 fragment of the myogenin promoter. 14,17 Additionally, a small amount of myogenin promoter activity could result in abundant transgene mRNA, if the transgene mRNA is much more stable than the endogenous mRNA. Despite this unexpected expression in uninjured muscle, the important finding for the current study was that MRF4 mRNA was abundant shortly after injury in the transgenic, but not wild-type, muscles, thus allowing us to compare regeneration in muscles with different patterns of MRF4 expression.
From day 6 to day 11 after injury, the transgenic and wild-type TA muscles showed different patterns of myofiber regeneration. In particular, the mean CSA of central nucleate myofibers remained constant in transgenic TA, but increased by approximately one-third in wild-type TA, from day 6 to day 11. As a result, the mean CSA of regenerating myofibers in transgenic TA was significantly smaller than that in wild-type TA at day 11 after injury. The density of regenerating myofibers in transgenic and wild-type TAs also differed significantly at day 11 after injury. Although myofiber density increased in both transgenic and wild-type muscles from day 6 to day 11 after injury, the increase was greater in transgenic TA (∼1.5×) than in wild-type TA (∼1.3×). These results suggest that expression of transgene in the TA appeared to attenuate regeneration at day 11 by inhibiting the growth of previously regenerated myotubes (myofiber area did not increase as in wild-type) and/or by enhancing the formation (or growth to larger than our 100-μm2 minimum size) of new myofibers.
The transgenic TA muscles eventually recovered to the same extent as the nontransgenic TA muscles. By 19 days after injury, the central nucleate myofibers in transgenic and nontransgenic TA muscles had similar CSAs and densities. Thus, from 11 to 19 days after injury, the transgenic TA myofibers appeared to grow more rapidly than wild-type myofibers so that transgenic myofibers caught up to the wild-type myofibers in CSA. As TA regeneration progressed, the density of central nucleate myofibers decreased and the CSAs increased to reach levels similar to those of uninjured TA muscles by day 19 after injury. 10
Taken together, these results suggest that expression of the myo1565-MRF4 transgene produces a biphasic alteration in the balance of myofiber formation and myofiber growth during recovery of the TA from injury. From day 6 to day 11 after injury, transgenic TA muscles showed less myofiber growth and a faster increase in myofiber density than wild-type muscles, whereas from day 11 to day 19 the transgenic TA muscles showed more myofiber growth and a faster decrease in density than wild-type muscles.
The possibility that transgene expression promotes myotube formation at the intermediate stage of transgenic TA regeneration is consistent with previous studies suggesting that increased MRF4 expression promotes myotube formation. 9,14,26-29 In myo1565-MRF4 embryos, in particular, myofiber formation occurs ∼0.5 to 1 day earlier than in nontransgenic embryos, although the muscles in transgenic and nontransgenic fetuses become indistinguishable as development proceeds in utero and into adulthood (this study). 14 High level expression of MRF4 also promotes early differentiation and myotube formation in a myogenic cell line. 26 Conversely, in MRF4-null embryos, some somitic myocytes fail to form. 27 Furthermore, MRF4 is not up-regulated and myotube formation is decreased in MyoD-null and myocyte nuclear factor-null satellite cells, although it is not known if addition of MRF4 would normalize myogenesis in these cells. 9,13,28,30
The changes in myofiber formation and growth produced by the myo1565-MRF4 transgene are different from those produced by other treatments that affect muscle regeneration. As examples, inactivation of desmin or MyoD or treatment with hepatocyte growth factor produce a transient attenuation of regeneration similar to that seen here in the transgenic TA. 19,30,31 Under those conditions, however, myofiber density was initially decreased compared to untreated or wild-type cells, rather than increased as seen here with the MRF4 transgene. On the other hand, both the formation and the growth of regenerating myofibers are accelerated by inhibitors of nuclear factor-κB such as curcumin; 20 and leukemia inhibitory factor treatment speeds the growth of regenerated myofibers. 32,33 None of those changes in regeneration matches the change seen in the myo1565-MRF4 TA muscles, suggesting that MRF4 acts via a distinct mechanism to affect TA regeneration.
It is possible that the transgene altered TA regeneration by increasing the total MRF (or basic helix-loop-helix protein) concentration, thus, perhaps, titrating out intracellular inhibitors of muscle gene activation. An alternative possibility is that MRF4 plays a unique role in myocyte formation, consistent with many studies showing that each MRF has some unique functions. 2,3 One way to distinguish between these possibilities might be to analyze regeneration in mice that express MRFs other than MRF4 under control of the myo1565 promoter fragment.
Why did expression of myo1565-MRF4 alter regeneration of the TA, but not the masseter? This difference was unlikely to be because of differences in transgene expression, because the transgene appeared to be expressed to the same extent and at the same early time after injury in both TA and masseter. In addition, both TA and masseter showed accelerated up-regulation of the endogenous MRF4 gene on transgene expression. TA muscles do, however, produce more satellite cells and regenerate to a greater extent than masseter muscles. 10 In the present study, TA myofiber CSAs were fully recovered by 19 days after injury, whereas masseter myofiber CSAs reached only approximately two-thirds recovery after 26 days. Perhaps the extensive production of new myofibers in the TA was sensitive to accelerated up-regulation of MRF4, whereas the already limited production in the masseter was not.
A number of studies have uncovered extracellular and intracellular factors that either promote or inhibit muscle regeneration. Among the extracellular agents that improve the outcome or speed of muscle regeneration in vivo are glucocorticoids, the nuclear factor-κB inhibitor curcumin, leukemia inhibitory factor, basic fibroblast growth factor, and insulin-like growth factor 1. 20,32-35 On the other hand, hepatocyte growth factor inhibits muscle regeneration after injury, 19 as does inactivation of the muscle transcription factors MyoD and myocyte nuclear factor. 12,13,28 Different muscles also have different capacities for regeneration 10 and as shown here, have different responses to accelerated up-regulation of MRF4 different gene expression. Physiologically distinct muscles, such as the extraocular muscles, also express distinct profiles of regulatory proteins that likely affect the extent and timing of satellite cell proliferation and subsequent muscle regeneration. 36-38 Understanding muscle-specific mechanisms of regeneration could lead to improved treatments for injuries to specific muscles.
Footnotes
Address reprint requests to Jeffrey B. Miller, Boston Biomedical Research Institute, 64 Grove St., Watertown, MA 02472. E-mail: miller@bbri.org.
Supported by the National Institutes of Health (grants DE11987, DE13040, and AR47314 to G. K. P; grant AR45840 to J. A. D.; and grant AR43565, ES11384, and HL64641 to J. B. M.), the United States Department of Agriculture (grant 99-35206-7951), and the Muscular Dystrophy Association (to J. B. M.).
References
- 1.Brand-Saberi B, Christ B: Genetic and epigenetic control of muscle development in vertebrates. Cell Tissue Res 1999, 296:199-212 [DOI] [PubMed] [Google Scholar]
- 2.Perry RL, Rudnicki MA: Molecular mechanisms regulating myogenic determination and differentiation. Front Biosci 2000, 5:D750-D767 [DOI] [PubMed] [Google Scholar]
- 3.Miller JB: Developmental biology of skeletal muscle. Hilton-Jones D Griggs RC Karpati G eds. Disorders of Voluntary Muscle ed 6 2001:pp 26-41 Cambridge University Press, Cambridge
- 4.Smith TH, Block NE, Rhodes SJ, Konieczny SF, Miller JB: A unique pattern of expression of the four muscle regulatory factors distinguishes somitic from embryonic, fetal, and newborn mouse myogenic cells. Development 1993, 117:1125-1133 [DOI] [PubMed] [Google Scholar]
- 5.Zhou Z, Bornemann A: MRF4 protein expression in regenerating rat muscle. J Muscle Res Cell Motil 2001, 22:311-316 [DOI] [PubMed] [Google Scholar]
- 6.Smith TH, Kachinsky AM, Miller JB: Somite subdomains, muscle cell origins, and the four muscle regulatory factor proteins. J Cell Biol 1994, 127:95-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawke TJ, Garry DJ: Myogenic satellite cells: physiology to molecular biology. J Appl Physiol 2001, 91:534-551 [DOI] [PubMed] [Google Scholar]
- 8.Zammit P, Beauchamp J: The skeletal muscle satellite cell: stem cell or son of stem cell? Differentiation 2001, 68:193-204 [DOI] [PubMed] [Google Scholar]
- 9.Cornelison DD, Olwin BB, Rudnicki MA, Wold BJ: MyoD(−/−) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev Biol 2000, 224:122-137 [DOI] [PubMed] [Google Scholar]
- 10.Pavlath GK, Thaloor D, Rando TA, Cheong M, English AW, Zheng B: Heterogeneity among muscle precursor cells in adult skeletal muscles with differing regenerative capacities. Dev Dyn 1998, 212:495-508 [DOI] [PubMed] [Google Scholar]
- 11.Mitchell CA, McGeachie JK, Grounds MD: Cellular differences in the regeneration of murine skeletal muscle: a quantitative histological study in SJL/J and BALB/c mice. Cell Tissue Res 1992, 269:159-166 [DOI] [PubMed] [Google Scholar]
- 12.Megeney L, Kablar B, Garrett K, Anderson JE, Rudnicki MA: MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev 1996, 10:1173-1183 [DOI] [PubMed] [Google Scholar]
- 13.Garry DJ, Meeson A, Elterman J, Zhao Y, Yang P, Bassel-Duby R, Williams RS: Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc Natl Acad Sci USA 2000, 97:5416-5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Block NE, Zhu Z, Kachinsky AM, Dominov JA, Miller JB: Acceleration of somitic myogenesis in embryos of myogenin promoter-MRF4 transgenic mice. Dev Dyn 1996, 207:384-392 [DOI] [PubMed] [Google Scholar]
- 15.Zhu Z, Miller JB: MRF4 can substitute for myogenin during early myogenesis. Dev Dyn 1997, 209:233-241 [DOI] [PubMed] [Google Scholar]
- 16.Rhodes SJ, Konieczny SF: Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev 1989, 3:2050-2061 [DOI] [PubMed] [Google Scholar]
- 17.Cheng TC, Hanley TA, Mudd J, Merlie JP, Olson EN: Mapping of myogenin transcription during embryogenesis using transgenes linked to myogenin control region. J Cell Biol 1992, 119:1649-1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominov JA, Miller JB: POU homeodomain genes and myogenes. Dev Genet 1996, 19:108-118 [DOI] [PubMed] [Google Scholar]
- 19.Miller KJ, Thaloor D, Matteson S, Pavlath GK: Hepatocyte growth factor affects satellite cell activation and differentiation in regenerating skeletal muscle. Am J Physiol 2000, 278:C174-C181 [DOI] [PubMed] [Google Scholar]
- 20.Thaloor D, Miller KJ, Gephart J, Mitchell PO, Pavlath GK: Systemic administration of the NF-kappaB inhibitor curcumin stimulates muscle regeneration after traumatic injury. Am J Physiol 1999, 277:C320-C329 [DOI] [PubMed] [Google Scholar]
- 21.Horsley V, Friday BB, Matteson S, Kegley KM, Gephart J, Pavlath GK: Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J Cell Biol 2001, 153:329-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavlath GK: Regulation of class I MHC expression in skeletal muscle: deleterious effect of aberrant expression on myogenesis. J Neuroimmunol 2002, 125:42-50 [DOI] [PubMed] [Google Scholar]
- 23.Cornelison DD, Wold BJ: Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol 1997, 191:270-283 [DOI] [PubMed] [Google Scholar]
- 24.Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH: Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 1993, 364:501-506 [DOI] [PubMed] [Google Scholar]
- 25.Musaro A, Deangelis MGC, Germani A, Ciccarelli C, Molinaro M, Zani BM: Enhanced expression of myogenic regulatory genes in aging skeletal muscle. Exp Cell Res 1995, 221:241-248 [DOI] [PubMed] [Google Scholar]
- 26.Block NE, Miller JB: Expression of MRF4, a myogenic helix-loop-helix protein, produces multiple changes in the myogenic program of BC3H-1 cells. Mol Cell Biol 1992, 12:2484-2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patapoutian A, Yoon JK, Miner JH, Wang S, Stark K, Wold B: Disruption of the mouse MRF4 gene identifies multiple waves of myogenesis in the myotome. Development 1995, 121:3347-3358 [DOI] [PubMed] [Google Scholar]
- 28.Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA: Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J Cell Biol 1999, 144:631-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sumariwalla VM, Klein WH: Similar myogenic functions for myogenin and MRF4 but not MyoD in differentiated murine embryonic stem cells. Genesis 2001, 30:239-249 [DOI] [PubMed] [Google Scholar]
- 30.White JD, Scaffidi A, Davies M, McGeachie J, Rudnicki MA, Grounds MD: Myotube formation is delayed but not prevented in MyoD-deficient skeletal muscle: studies in regenerating whole muscle grafts of adult mice. J Histochem Cytochem 2001, 48:1531-1544 [DOI] [PubMed] [Google Scholar]
- 31.Smythe GM, Davies MJ, Paulin D, Grounds MD: Absence of desmin slightly prolongs myoblast proliferation and delays fusion in vivo in regenerating grafts of skeletal muscle. Cell Tissue Res 2001, 304:287-294 [DOI] [PubMed] [Google Scholar]
- 32.Bernard W, Bower J, Brown MA, Murphy M, Austin L: Leukemia inhibitory factor (LIF) infusion stimulates skeletal muscle regeneration after injury: injured muscle expresses LIF mRNA. J Neurol Sci 1994, 123:108-113 [DOI] [PubMed] [Google Scholar]
- 33.Kurek JB, Bower JJ, Romanella M, Koentgen F, Murphy M, Austin L: The role of leukemia inhibitory factor in skeletal muscle regeneration. Muscle Nerve 1997, 20:815-822 [DOI] [PubMed] [Google Scholar]
- 34.Anderson JE, Weber M, Vargas C: Deflazacort increases laminin expression and myogenic repair, and induces early persistent functional gain in mdx mouse muscular dystrophy. Cell Transplant 2000, 9:551-564 [DOI] [PubMed] [Google Scholar]
- 35.Menetrey J, Kasemkijwattana C, Day CS, Bosch P, Vogt M, Fu FH, Moreland MS, Huard J: Growth factors improve muscle healing in vivo. J Bone Joint Surg Br 2000, 82:131-137 [DOI] [PubMed] [Google Scholar]
- 36.Fischer MD, Gorospe JR, Felder E, Bogdanovich S, Pedrosa-Domellof F, Ahima RS, Rubinstein NA, Hoffman EP, Khurana TS: Expression profiling reveals metabolic and structural components of extraocular muscles. Physiol Genomics 2002, 9:71-84 [DOI] [PubMed] [Google Scholar]
- 37.Cheng G, Porter JD: Transcriptional profile of rat extraocular muscle by serial analysis of gene expression. Invest Ophthalmol Vis Sci 2002, 43:1048-1058 [PubMed] [Google Scholar]
- 38.Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, Leahy P, Li J, Andrade FH: Extraocular muscle is defined by a fundamentally distinct gene expression profile. Proc Natl Acad Sci USA 2001, 98:12062-12067 [DOI] [PMC free article] [PubMed] [Google Scholar]