Abstract
Pancreatic cancer is the fifth leading cause of cancer death in the United States. We used cDNA microarrays to analyze global gene expression patterns in 14 pancreatic cancer cell lines, 17 resected infiltrating pancreatic cancer tissues, and 5 samples of normal pancreas to identify genes that are differentially expressed in pancreatic cancer. We found more than 400 cDNAs corresponding to genes that were differentially expressed in the pancreatic cancer tissues and cell lines as compared to normal pancreas. These genes that tended to be expressed at higher levels in pancreatic cancers were associated with a variety of processes, including cell-cell and cell-matrix interactions, cytoskeletal remodeling, proteolytic activity, and Ca++ homeostasis. Two prominent clusters of genes were related to the high rates of cellular proliferation in pancreatic cancer cell lines and the host desmoplastic response in the resected pancreatic cancer tissues. Of 149 genes identified as more highly expressed in the pancreatic cancers compared with normal pancreas, 103 genes have not been previously reported in association with pancreatic cancer. The expression patterns of 14 of these highly expressed genes were validated by either immunohistochemistry or reverse transcriptase-polymerase chain reaction as being expressed in pancreatic cancer. The overexpression of one gene in particular, 14-3-3ς, was found to be associated with aberrant hypomethylation in the majority of pancreatic cancers analyzed. The genes and expressed sequence tags presented in this study provide clues to the pathobiology of pancreatic cancer and implicate a large number of potentially new molecular markers for the detection and treatment of pancreatic cancer.
Most pancreatic adenocarcinomas share similar genetic alterations and the clinical course of the disease is relatively homogenous. 1 The majority of pancreatic adenocarcinomas contain activating point mutations in the K-ras gene (>90% in most studies), and a significant number of these neoplasms also exhibit genetic inactivation of the p16 gene. A sizable group of genes has been found that are mutated less frequently in pancreatic cancers. 2,3 Other molecular mechanisms that may contribute to carcinogenesis of the pancreas include overexpression of growth factors or their receptors, or changes in activity of signal transduction pathways. 4,5 Despite significant progress, much remains to be learned regarding the fundamental changes that occur in the development and progression of adenocarcinomas of the pancreas.
Gene expression methodologies have shown promising utility in identifying novel markers or genes of interest in solid tumors, particularly in the study of pancreatic ductal adenocarcinoma. Compared to only 5 years ago, we are now aware of hundreds of genes with potential importance in the biology of pancreatic cancer. 6-11 Strategies for identifying differentially expressed genes in pancreatic cancer have progressed from initial studies using gridded cDNA libraries, 12 and later representational difference analysis of cDNAs, 7,9,13 to the newer strategies of serial analysis of gene expression, 14,15 oligonucleotide microarrays, 11 and cDNA microarrays. 8 To differentiate gene expression patterns arising from the primary cancer from those arising in the surrounding stroma investigators have used several strategies including limiting their analysis to comparison of pancreatic cancer cell lines with normal pancreas, 8 comparing pancreatic cancer cell lines with normal pancreatic ductal epithelium, 15 whereas other investigators have used laser capture microdissection. 10 Overexpressed genes now recognized as potentially important in pancreatic cancer include, but are not limited to, mesothelin, 16 prostate stem cell antigen, 17 claudin-4, 18 biglycan, 19 S100A4, 20 TMPRSS3, 21 transglutaminase II, 22 fascin, and hsp47. 23 The identification of these genes provides new opportunities for drug and therapeutic development aimed at targeting pancreatic cancers. 18,24
In an effort to further our efforts to identify novel genes highly expressed in pancreatic cancers with the potential for development into serological markers or therapeutic targets, we analyzed a large set of surgically resected pancreatic cancer tissues, pancreatic cancer cell lines, and normal pancreas tissues using a 45,000 cDNA microarray. The data presented not only confirm other earlier reports of highly expressed genes in pancreas cancer identified through a variety of approaches, but also provides new information regarding the genes and cellular pathways that play a role in this tumor type.
Materials and Methods
Tissues
Samples of primary invasive pancreatic ductal adenocarcinoma from pancreaticoduodenectomy specimens were collected from patients undergoing Whipple resections at the Johns Hopkins Hospital or the Stanford University School of Medicine. In each case, specimens of bulk tumor were harvested within 10 minutes of resection and snap-frozen in liquid nitrogen before storage at −80°C. Hematoxylin and eosin-stained sections of the adjacent tissue were prepared before snap-freezing to confirm the presence of infiltrating adenocarcinoma within the section.
Cell Lines
Human pancreatic cancer cell lines AsPc1, BxPc3,CAPAN1, CAPAN2, CFPAC1, Hs766T, MiaPaca2, Panc-1, and Su86.86 cell lines were obtained from the American Type Culture Collection, Rockville, MD. COLO357 was obtained from the European Collection of Animal Cell Cultures, Salisbury, UK. The pancreas cancer line (PL) cell lines used are low-passage pancreatic carcinoma cell lines established in our laboratories. 25,26 Cell lines were grown in their recommended media. Use of different media minimized the variance in growth rates but presumably introduced other variations in gene expression patterns.
mRNA Extractions
Total RNA was obtained from homogenized frozen tissues and cell lines grown at 70 to 90% confluence were using TRIzol reagent (Life Technologies, Inc., Grand Island, NY). Polyadenylated mRNAs were purified from total RNA using the Fast Track 2.0 mRNA isolation kit (InVitrogen, Carlsbad, CA).
cDNA Microarrays and Statistical Analysis of Data
cDNA microarrays were printed and used as previously described in detail 27 (detailed protocols are available at http://cmgm.Stanford.EDU/pbrown/). Briefly, mRNA from 11 different normal cultured cell lines were pooled and used as a reference control sample to prepare cDNA labeled with Cy3-deoxyuridine triphosphate (dUTP), and mRNA harvested from the 14 individual pancreatic cancer cell lines or 22 resected pancreas tissues (5 normal pancreas, 12 ductal adenocarcinomas, 2 ampullary carcinomas, 1 islet cell tumor, and 2 carcinomas arising in intraductal papillary mucinous neoplasms of the pancreas) was used to prepare cDNA labeled with Cy5-dUTP. The two differentially labeled cDNA probes were mixed and simultaneously hybridized to cDNA microarrays. Microarrays were scanned with a Genepix 4000B microarray scanner (Axon Instruments) using Genepix 5.0 software, and analyzed using the Cluster and TreeView programs (http://www.microarrays.org/software.html). 28 The complete data for each sample described in this report are available through the Stanford Microarray Database site (http://genome-www4.stanford.edu/MicroArray/SMD/).
Significance analysis of microarrays (SAM) v1.13 (http://www-stat.stanford.edu/∼tibs/SAM/) 29 was used to perform the two-class comparison for differentially expressed genes between the 31 samples with pancreatic cancer (cell lines and resected cancer tissues) and the 5 samples of normal pancreas. The log-transformed and filtered gene expression data used for the original hierarchical cluster analysis described above was exported into an Excel 5.0 spreadsheet, and reformatted according to specifications outlined by the SAM v1.13 program. The K-nearest neighbor imputation was used to account for missing data within the dataset, and output criteria selected for SAM included at least threefold greater expression in the pancreatic cancers as compared to normal tissues, and a significance threshold expected to produce fewer than five false-positive genes. Complete data are available at http://genome-www.stanford.edu/pancreatic 1.
Immunohistochemistry
Immunohistochemical analysis was performed to validate the translation and differential expression of selected genes in archival tissue sections of infiltrating pancreatic ductal adenocarcinomas. Adjacent sections of the infiltrating primary adenocarcinoma and normal nonneoplastic pancreatic tissue were formalin-fixed and paraffin-embedded. The proteins analyzed were S100A10, RON, Trop-2, cytokeratin 19, transglutaminase II, cdc-2, gamma synuclein, 14-3-3ς, and fibronectin. A detailed description of the methods involved in the immunolabeling of these proteins is available from the authors. Staining was evaluated by two of the authors (AM and RHH).
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated from 20 pancreatic cancer cell lines and an aliquot of 1 μg of total RNA from each sample was reverse-transcribed to cDNA using the SuperScript II kit (Life Technologies, Inc.) with oligo(dT)12-18 primer. Gene expression was compared against the simultaneous PCR of glyceraldehyde-3-phosphate dehydrogenase cDNA.
Methylation-Specific PCR (MSP)
Methylation status of the 5′ region of 14-3-3ς was determined by MSP as described previously. 30 PCR primers and conditions are provided in supplemental data.
Treatment with 5-Aza-2′-Deoxycitidine (5-Aza-dC)
MiaPaCa2 cells were treated with a demethylating agent, 5-aza-2′-deoxycitidine (5-aza-dC; Sigma Chemical Co., St. Louis, MO) at a final concentration of 1 μmol/L for 5 days. Total RNA was isolated from the untreated and treated cells using Trizol and was subjected to RT-PCR for 14-3-3ς expression.
Results
Data Filtering and Hierarchical Clustering
Human cDNA microarrays containing 45,000 individual cDNAs were hybridized with cDNAs prepared from 14 pancreatic cancer cell lines, 5 samples of normal pancreatic tissue, and 17 samples of primary pancreatic cancer tumor tissue. The complete dataset is freely available at http://genome-www.stanford.edu/pancreatic 1. Samples of normal pancreas were analyzed to provide a basis for assessing the contributions of acinar and islet cells to the gene expression profiles detected in primary tumors. Pancreas cancer cell lines were similarly analyzed to identify the gene expression patterns in the neoplastic cells. Those cDNAs with the greatest variation in expression among these samples were retained for the analysis. For each sample, the R/G ratio was normalized to the mean across all samples for each cDNA and log2-transformed. A filter was applied to remove those cDNAs whose expression did not vary by at least two standard deviations from the mean in this sample set in at least two of the samples. As a result, 1492 cDNAs were selected for use in the analyses described below.
Global Gene Expression Profiles in Pancreatic Cancer
We analyzed the global gene expression patterns of pancreatic cancer to search for features that might provide insights into the biology of this tumor type. We first organized the data using hierarchical clustering of the cDNAs, the cell lines, and the tissue samples based on their global gene expression profiles (Figure 1, A and B) ▶ . As expected, normal and tumor tissue samples clustered separately from the cell lines, primarily on the basis of differential expression of proliferation-related genes, which were much more highly expressed in the rapidly dividing cell lines, or the presence of nonneoplastic stromal and inflammatory cell gene expression within the tissue specimens. Among the tissues, normal pancreas was distinguished from invasive carcinomas, predominantly because of the presence of acinar and islet cell gene expression in the former (Figure 1A) ▶ .
Figure 1.

Profiles of gene expression in pancreatic samples. A: Dendrogram representing the results of hierarchical cluster analysis of the gene expression patterns in 36 pancreatic samples, based on data for 1492 genes. Sample names highlighted in green are normal pancreatic tissue (n = 5), samples in red are pancreatic primary tumor tissues (n = 17), and samples listed in black are pancreatic cancer cell lines (n = 14). B: Hierarchical cluster analysis of 1492 genes whose expression varied by at least two standard deviations from the mean in at least two samples. Five clusters of gene expression with similar patterns of variation in expression are highlighted, corresponding to acinar or islet gene expression in normal pancreas (white vertical bar), desmoplasia-associated gene expression (dark gray vertical bar), cell line-specific expression (black vertical bar), and genes differentially expressed in pancreas cancers (light gray vertical bars). The ratio of the abundance of transcripts of each gene in a given sample to its median abundance across all cell lines or tissue samples is represented by the color of the corresponding cell in the Tree View-generated diagram. Green cells are those with transcript levels below the median, black cells are equal to the median, and red cells are greater than the median. Gray cells represent technically inadequate or missing data. Color saturation of each cell reflects the magnitude of the ratio relative to the mean for each gene. The complete dataset is freely available at http://genome-www.stanford.edu/pancreatic1.
Hierarchical clustering of the 1492 cDNAs based on the similarity in the patterns of gene expression revealed systematic features of the gene expression programs in these samples, which could be related to biological or histological features of the pancreatic samples (Figure 1B) ▶ . For example, gene expression patterns related to acinar and islet cells were identified in normal pancreas, a gene expression pattern that appeared to be related to the desmoplastic response was noted in pancreas cancer tissues, and genes related to rates of cellular proliferation in cancer cell lines could be recognized.
Specific Gene Clusters Identified by cDNA Microarray
Two major clusters of genes were differentially expressed in the pancreatic cancer cell lines and primary pancreatic cancer tissues as compared to normal pancreas tissues. These pancreas cancer-specific clusters together contained 424 cDNAs. A detailed account of all of the cDNAs included within these various clusters are presented on our web page (http://pathology.jhu.edu/pancreas/microarray). The genes represented in the pancreas cancer-specific clusters appeared to reflect a diversity of functions, including cell-cell junctions (annexins A4 and A11; claudins 3, 4, and 7), cell/matrix interactions (integrin-α3 and -α6), cytoskeletal assembly (cytokeratins 7, 17, and 19; pleckstrin), cell-cycle regulation (Cdc42 effector protein 3), transcription factors (TCF7), tissue invasion (S100A4, S100P, S100A10, and S100A11), proteolytic processing (urokinase plasminogen activator; matrix metalloproteinases 7, 14, and 24), and interferon- or retinoic acid-induced functions (interferon gamma-induced protein 16; interferon α-induced protein 27; interferon-induced transmembrane proteins 1, 2, and 3; retinoic acid receptor responder 3; retinoic acid receptor gamma).
To confirm the expression of genes identified by hierarchical clustering, the expression patterns of four of the genes represented in the pancreas cancer-specific clusters (S100A10, Trop-2, RON, and cytokeratin 19) 15,31 were analyzed by immunohistochemical labeling of paraffin-embedded sections of 6 of the 17 pancreas ductal adenocarcinomas analyzed by cDNA microarray. Strong labeling of the neoplastic epithelium was seen with each marker, in contrast to the weak to negative labeling of the normal duct epithelium present in the same tissue sections. However, three of the four markers (S100A10, Trop-2, and RON) also showed variable amounts of labeling of the surrounding stromal tissues, whereas cytokeratin 19 labeled the neoplastic epithelium of the infiltrating carcinomas only (see supplementary figure at http://pathology.jhu.edu/pancreas/microarray/supplementfigure1.cfm).
Two additional and informative clusters were found among the 1492 cDNAs analyzed by hierarchical cluster analysis. The first cluster associated with cellular proliferation has been well described. This proliferation cluster included chromosome-remodeling genes (ie, SMC4-like 1), cell cycle-regulating genes (ie, cyclin A2), and genes associated with cytoskeletal remodeling (ie, myosin heavy polypeptide 1). Proliferating cell nuclear antigen was also present in this cluster. Pancreatic cancer cell lines showed high levels of expression of these genes, in contrast to primary tumor tissues that had low levels of expression.
Pancreatic cancers are well recognized for their exuberant host stromal response to invasive carcinoma, known as desmoplasia, which often accounts for the majority of the cellularity of the actual mass produced by the carcinoma. 14,32,33 A cluster of genes that appeared to be related to this prominent desmoplastic response was also identified by hierarchical cluster analysis. This cluster of several genes highly expressed in the invasive pancreatic tumor tissues as compared to pancreas cancer cell lines or normal pancreas included collagen 1α1 and 1α2, matrix metalloproteinases and their inhibitors, apolipoprotein C-1 and C-II, hevin, osteonectin and biglycan. Some of these genes (biglycan, MMPs, TIMP1) have been previously identified as overexpressed in pancreatic cancer. 10,19,34
Identification of Novel Tumor Markers of Pancreatic Cancer
To determine those genes with statistically significant differences in expression in pancreatic cancer cell lines and pancreatic cancer tissues compared to normal pancreas, we used Significance Analysis of Microarray (SAM) as described in the Materials and Methods section. Using a threefold differential cutoff, we found 216 cDNAs expressed at higher levels and 236 cDNAs expressed at lower levels in pancreatic cancers compared to normal pancreatic tissues at a rate of five false-positives. Many of the genes that were expressed at lower levels in pancreatic cancer tissues or cell lines appeared to reflect the loss of acinar and islet cell-associated gene expression in pancreatic cancers because of the atrophy and/or destruction of these cell types within the infiltrative mass. We were primarily interested in the 216 up-regulated cDNAs, which included 149 different known genes whose average expression level in pancreatic cancers was at least threefold higher than in the normal pancreas samples we analyzed.
A Pub Med search of each of these 149 known genes revealed that 46 were previously reported as highly expressed in pancreatic cancer by ourselves and others, in strong support of the ability of SAM to identify significantly overexpressed cDNAs in pancreatic cancer cell lines or tissues (Table 1) ▶ . Of the remaining 103 known genes identified, 40 have been reported as playing a role in other tumor types, whereas 63 of these known genes have not been reported in reference to any tumor type ▶ (Table 2) ▶ .
Table 1.
Highly Expressed Genes Previously Reported in Pancreatic Cancer and Confirmed by Significance Analysis of Microarray*
| Gene symbol | Known gene name |
|---|---|
| AGR2 | Anterior gradient 2 homolog |
| ANXA1 | Annexin A1 |
| ARL7 | ADP-ribosylation factor-like 7 |
| CAPG | Capping protein (actin filament), gelsolin-like |
| CAV2 | Caveolin 2 |
| CD9 | CD9 antigen (p24) |
| CDC2 | Cell division cycle 2, G1 to S and G2 to M |
| CEACAM5 | Carcinoembryonic antigen-related cell adhesion molecule 5 |
| CLDN 4 | Claudin 4 |
| COTL 1 | Coactosin-like 1 |
| CTSD | Cathepsin D |
| CTSE | Cathepsin E |
| DAF | Decay accelerating factor for complement (CD55, Cromer blood Group system) |
| ERBB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) |
| FER1L3 | fer-1-like 3, myoferlin (C. elegans) |
| FN 1 | Fibronectin 1 |
| FTL | Ferritin, light polypeptide |
| FXYD3 | FXYD domain-containing ion transport regulator 3 |
| HMGIY | High-mobility group (nonhistone chromosomal) protein isoforms I and Y |
| IGFBP3 | Insulin-like growth factor binding protein 3 |
| INHBB | Inhibin, beta B (activin AB beta polypeptide) |
| ITGA1 | Integrin, alpha 1 |
| ITGA2 | Integrin, alpha 2 |
| ITGA3 | Integrin, alpha 3 |
| KRT17 | Keratin 17 |
| KRT19 | Keratin 19 |
| LAMC2 | Laminin, gamma 2 (nicein (100 kd), kalinin (105 kd), BM600 (100 kd), Herlitz junctional epidermolysis bullosa)) |
| LCN2 | Lipocalin 2 (oncogene 24p3) |
| LGALS3 | Lectin, galactoside-binding, soluble, 3 (galectin 3) |
| MMP14 | Matrix metalloproteinase 14 (membrane-inserted) |
| PLAUR | Plasminogen activator, urokinase receptor |
| PSMA7 | Proteasome (prosome, macropain) subunit, alpha type, 7 |
| RAI3 | Retinoic acid induced 3 |
| S100A10 | S100 calcium-binding protein A10 (annexin II ligand, calpactin I, light polypeptide (p11)) |
| S100A11 | S100 calcium-binding protein A11 (calgizzarin) |
| S100P | S100 calcium-binding protein P |
| SDC1 | Syndecan 1 |
| SFN | Stratifin |
| SLPI | Secretory leukocyte protease inhibitor (antileukoproteinase) |
| SNRPB | Small nuclear ribonucleoprotein polypeptides B and B1 |
| SPUVE | Protease, serine, 23 |
| TGFBI | Transforming growth factor, beta-induced,68 kd |
| TGM2 | Transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) |
| TIMP1 | Tissue inhibitor of metalloproteinase 1 (erythroid potentiating activity, collagenase inhibitor) |
| TSPAN-1 | Tetraspan 1 |
| TXNL2 | Thioredoxin-like 2 |
*Information compiled 8/02 from the PubMed and LocusLink databases available on the NCBI website (http://www.ncbi.nlm.gov).
Table 2.
Novel Highly Expressed Genes in Pancreatic Cancer Identified by Significance Analysis of Microarray (SAM)*
| Unigene ID | Symbol | Known gene name | Function |
|---|---|---|---|
| 2442 | ADAM9 | A disintegrin and metalloproteinase domain 9 (meltrin gamma) | Integral membrane metalloproteinase |
| 323342 | ARPC4 | Actin-related protein 2/3 complex, subunit 4 (20 kd) | Cytoskeletal protein; actin polymerization |
| 82425 | ARPC5 | Actin-related protein 2/3 complex, subunit 5 (16 kd) | Cytoskeletal protein; actin polymerization |
| 78045 | ACTG2 | Actin, gamma 2, smooth muscle, enteric | Cell motility, intestinal smooth muscle |
| 41644 | ASP | AKAP-associated sperm protein | Sperm protein; anchoring protein |
| 75746 | ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 | Ethanol detoxification |
| 164960 | BARX1 | BarH-like homeobox 1 | Homeobox gene; CNS development |
| 40323 | BUB3 | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | Mitotic checkpoint protein |
| 356181 | CAPN1 | Calpain 1, (mu/I) large subunit | Subunit of calcium regulated neutral protease |
| 74034 | CAV1 | Caveolin 1, caveolae protein, 22 kD | Caveolar protein/endocytosis |
| 348669 | CKS1B | CDC28 protein kinase 1B | Cell-cycle protein (p27 inhibition) |
| 1600 | CCT5 | Chaperonin-containing TCP1, subunit 5 (epsilon) | Chaperon protein, protein folding |
| 75724 | COPB2 | Coatomer protein complex, subunit beta 2 (beta prime) | Golgi transport protein |
| 117938 | COL17A1 | Collagen, type XVII, alpha 1 | Cell-cell matrix adhesion |
| na† | COX5BL3 | Cytochrome c oxidase subunit Vb-like 3 | Mitochondrial protein |
| 12802 | DDEF2 | Development and differentiation enhancing factor 2 | GTPase activating protein |
| 274464 | DIA1 | Diaphorase (NADH/NADPH) (cytochrome b-5 reductase) | Quinone reductase |
| 254105 | ENO1 | Enolase 1, (alpha) | Enzyme; may bind c-myc promoter element |
| 116651 | EVA1 | Epithelial V-like antigen 1 | Cell adhesion; immunoglobulin superfamily |
| 11638 | FACL5 | Fatty-acid-Coenzyme A ligase, long-chain 5 | Fatty acid metabolism |
| 4756 | FEN1 | Flap structure-specific endonuclease 1 | DNA damage repair |
| 239 | FOXM1 | Forkhead box M1 | Forkhead protein; Transcription factor; targets Cyclin D1/B1 |
| 13040 | GPR87 | G protein-coupled receptor 87 | G-protein coupled orphan receptor |
| 132942 | GRAF | GTPase regulator associated with the focal adhesion kinase pp125 | Signal transduction, may be a tumor suppressor in some leukemias |
| 147097 | H2AFX | H2A histone family, member X | DNA binding and compaction protein |
| 40968 | HS3ST1 | Heparan sulfate (glucosamine) 3-O-sulfotransferase | Golgi membrane protein |
| 1690 | HBP17 | Heparin-binding growth factor-binding protein | Mobilizes and activates FGF, “angiogenic switch” in cancers |
| 22554 | HOXB5 | Homeobox B5 | Homeobox gene |
| 98428 | HOXB6 | Homeobox B6 | Homeobox gene |
| 8004 | HAPIP | Huntingtin-associated protein interacting protein (duo) | Vescicular transport; contains a PLHD |
| 85962 | HAS3 | Hyaluronan synthase 3 | Hyaluron synthesis |
| 77348 | HPGD | Hydroxyprostaglandin dehydrogenase 15-(NAD) | Prostaglandin inactivation |
| 348553 | IMUP | Immortalization-upregulated protein | Putative role in immortalization |
| 833 | ISG15 | Interferon-stimulated protein, 15 kDa | Cytokine; augments IFN-gamma and lymphokines |
| 75117 | ILF2 | Interleukin enhancer binding factor 2, 45kD | DNA-binding transcription factor |
| 159557 | KPNA2 | Karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | Nuclear transport protein |
| 80342 | KRT15 | Keratin 15 | Intermediate filament |
| 335952 | KRT6B | Keratin 6B | Intermediate filament |
| 242463 | KRT8 | Keratin 8 | Intermediate filament |
| 85226 | LIPA | Lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | Lysosomal hydrolase |
| 20815 | MAEA | Macrophage erythroblast attacher | Erythrocyte maturation |
| 110309 | HLA-F | Major histocompatibility complex, class I, F | HLA Class I |
| 198253 | HLA-DQA1 | Major histocompatibility complex, class II, DQ alpha 1 | HLA Class II |
| 76550 | MAL2 | Mal, T-cell differentiation protein 2 | Vescicular transport; myelin biogenesis |
| 343521 | MDH2 | Malate dehydrogenase 2, NAD (mitochondrial) | Metabolic enzyme |
| 26638 | MS4A8B | Membrane-spanning 4-domains, subfamily A, member 8B | Immunoglobulin receptor |
| 154443 | MCM4 | Minichromosome maintenance deficient (S. cerevisiae) 4 | DNA replication and chromosomal stability |
| 333823 | MRPL13 | Mitochondrial ribosomal protein L13 | Ribosomal protein |
| (Table continues) |
Table 2A.
Continued
| Unigene ID | Symbol | Known gene name | Function |
|---|---|---|---|
| 10724 | MRPS35 | Mitochondrial ribosomal protein S35 | Ribosomal protein |
| 76307 | NBL1 | Neuroblastoma, suppression of tumorigenicity 1 | Putative tumor suppressor gene in neuroblastoma |
| 302649 | NAP1L1 | Nucleosome assembly protein 1-like 1 | Nucleosome assembly protein |
| 178761 | POH1 | 26S proteasome-associated pad 1 homolog | Protein degradation, Drug resistance |
| 180909 | PRDX1 | Peroxiredoxin 1 | Oxidative stress protein; increased in proliferating cells |
| 78771 | PGK1 | Phosphoglycerate kinase 1 | Glycolytic enzyme |
| 252587 | PTTG1 | Pituitary tumor-transforming 1 | Anaphase-promoting complex substrate; oncoprotein |
| 375567 | PSG11 | Pregnancy specific beta-1-glycoprotein 11 | CEA family member |
| 238527 | RAP2B | RAP2B, member of RAS oncogene family | GTP-binding protein |
| 203559 | MRPL44 | Mitochondrial ribosomal protein L44 | Ribosomal protein |
| 272822 | RUVBL1 | RuvB (E coli homolog)-like 1 | ATPase/helicase; transcriptional activation cofactor |
| 115166 | SCEL | Sciellin | Keratinocyte protein |
| 250822 | STK15 | Serine/threonine kinase 15 | TGFbeta family, causes centrosome destabilization |
| 168913 | STK24 | Serine/threonine kinase 24 (Ste20, yeast homolog) | Signal transduction (activates MAPK) |
| 50758 | SMC4L1 | SMC4 (structural maintenance of chromosomes 4, yeast)-like 1 | Mitotic assembly protein |
| 75231 | SLC16A1 | Solute carrier family 16 (monocarboxylic acid transporters), member 1 | Monocarboxylate transporter |
| 169902 | SLC2A1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | Glucose transporter |
| 284291 | SNX6 | Sorting nexin 6 | Intracellular trafficking |
| 8215 | STRBP | Spermatid perinuclear RNA-binding protein | RNA binding and repair |
| 349470 | SNCG | Synuclein, gamma (breast cancer-specific protein 1) | Neurofilament protein |
| 83848 | TPI1 | Triosephosphate isomerase 1 | Glycolytic enzyme |
| 22826 | TMOD3 | Tropomodulin 3 (ubiquitous) | Cytoskeletal protein; actin filament binding |
| 250641 | TPM4 | Tropomyosin 4 | Cytoskeletal protein; actin filament binding |
| 75318 | TUBA1 | Tubulin, alpha 1 (testis specific) | Cytoskeletal protein; microtubule formation |
| 349695 | TUBA2 | Tubulin, alpha 2 | Cytoskeletal protein; microtubule formation |
| 272897 | TUBA3 | Tubulin, alpha 3 | Cytoskeletal protein; microtubule formation |
| 93002 | UBE2L6 | Ubiquitin-conjugating enzyme E2L 6 | Ubiquiting conjugating protein |
| 69009 | B3GNT3 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 | Enzyme; involved in L-selectin ligand biosynthesis |
| 198307 | VBP1 | von Hippel-Lindau binding protein 1 | Chaperone protein, binds VHL |
| 150930 | XRCC4 | X-ray repair complementing defective repair in Chinese hamster cells 4 | DNA double strand break repair |
*Information compiled 8/02 from the PubMed and LocusLink databases available on the NCBI website (http://www.ncbi.nlm.gov).
†na, not available.
Because normal pancreas tissue contains a predominance of acinar cells and islets relative to normal duct epithelium, we realize that the contribution of pancreatic duct epithelium to the gene expression detected of the normal pancreas cannot be easily recognized. 10 Therefore, the 103 candidate cDNAs identified by SAM were screened using the data available on the SAGEmap database (http://www.ncbi.nlm.nih.gov/SAGE/) of normal pancreas duct epithelium libraries HX and H126. For each of the 103 candidate cDNAs identified as differentially expressed in the present study, the corresponding SAGE tags were identified, and the total number of SAGE tags present in the SAGE map database (http://www.ncbi.nlm.nih.gov/SAGE/) of normal pancreas duct epithelium libraries HX and H126 was determined. Twenty-four genes were found to have five or more tags present in at least one of the two normal duct libraries and were excluded from further study. The remaining 79 known genes identified by SAM did not show high levels of expression in the two normal duct epithelium SAGE libraries and appear to be newly recognized differentially expressed genes in pancreatic cancers.
Immunohistochemistry
Despite their identification as highly expressed genes in pancreatic cancer, not all genes previously reported have been validated in pancreas cancer tissue specimens. Therefore, immunohistochemical labeling patterns were determined for five of the cDNAs identified as significantly overexpressed in pancreatic cancers by SAM (14-3-3ς, transglutaminase II, cdc2, fibronectin, and gamma synuclein) (Figure 2) ▶ .
Figure 2.
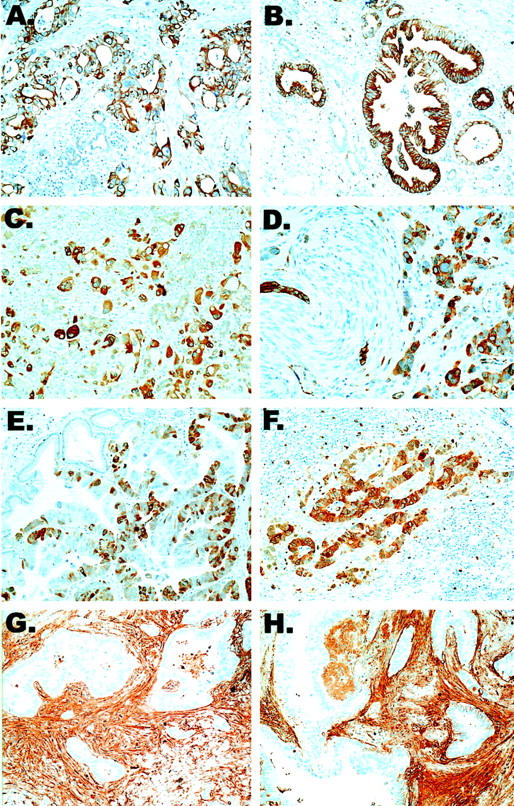
Immunohistochemical validation of selected highly expressed gene products detected by SAM. Shown are 14-3-3ς (A and B), transglutaminase II (C and D), cdc2 (E and F), and fibronectin (G and H). Strong positive immunohistochemical labeling of 14-3-3ς, transglutaminase II, and cdc2 proteins are detected in the neoplastic epithelium of infiltrating pancreatic ductal adenocarcinomas, but not in normal duct epithelium within the same tissue sections. In contrast, fibronectin is expressed predominantly by the host stromal response. Original magnifications, ×400.
Moderate to intense 14-3-3ς expression was demonstrable in seven of eight (90%) infiltrating adenocarcinomas (Figure 2A) ▶ . 14-3-3ς was expressed primarily in the cytoplasm, with membranous accentuation. Adjacent nonneoplastic pancreas was essentially nonreactive or demonstrated weak labeling (Figure 2B) ▶ . Similarly, eight of eight (100%) infiltrating pancreatic ductal adenocarcinomas demonstrated cytoplasmic transglutaminase II labeling within the neoplastic epithelium, whereas nonneoplastic ductal and acinar structures did not label (Figure 2, C and D) ▶ .
A nascent pool of cdc2 protein exists in the cytoplasm, but nuclear translocation of cdc2 is required for its role in promoting G2-M progression. In accordance, cdc2 was localized in both nucleus and cytoplasm in eight of eight (100%) infiltrating pancreatic ductal adenocarcinomas, and expression was limited to the neoplastic cells only (Figure 2, E and F) ▶ . Contrary to the epithelial-specific overexpression of these proteins in pancreatic adenocarcinomas, intense fibronectin expression was localized primarily to the tumor-associated stromal desmoplasia, with only focal weak labeling of the neoplastic epithelium, consistent with the role of fibronectin as a collagen-binding protein (Figure 2, G and H) ▶ . Focal gamma synuclein expression was seen in only two of eight (25%) adenocarcinomas, albeit no labeling was present in the nonneoplastic ductal and acinar structures in any instance (not shown).
RT-PCR
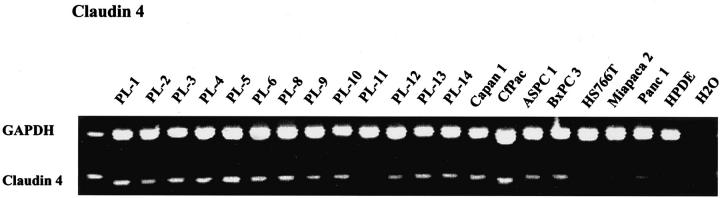
Ten genes [forkhead box M1 (FOXM1), claudin 4, S100 calcium-binding protein P (S100P), myoferlin (fer-1), XRCC4, caveolin-2, transforming growth factor β-induced 68-kd protein (TGFBI), secretory leukocyte proteinase inhibitor (SLP-1), ADAM9, and 14-3-3ς] were selected for validation by RT-PCR in 20 pancreatic cancer cell lines and the immortal human pancreatic ductal epithelial cell line (HPDE6) (Figure 3) ▶ . FOXM1 was expressed in all 20 cell lines, 14-3-3ς, fer-1, claudin 4, and S100P in 19 of the 20 cell lines, XRCC4 and caveolin-2 in 17 of the 20 cell lines, TGFBI in 16 of the 20 cell lines, SLP1 in 15 of the 20 cell lines, and ADAM9 in 13 of the 20 cell lines analyzed. Five of the 10 genes were expressed in the immortalized HPDE6 cell line including FOXM1, fer-1, caveolin-2, TGFB1, and SLP-1. Interestingly, three of these five genes had low-level expression in normal pancreatic epithelium by SAGE (fer-1, FOXxM1, and SLP-1).
Figure 3.
RT-PCR validation of highly expressed genes identified by SAM. Shown are 20 pancreatic cancer cell lines, an immortal human pancreatic ductal epithelial cell line (HPDE6), and a water control. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) serves as an RNA control. Claudin 4 is expressed in 19 of 20 pancreatic cancer cell lines.
Promoter Methylation Analyses of 14-3-3ς in Pancreatic Cancer
The 14-3-3ς gene was further investigated to determine the mechanism of overexpression in pancreatic cancers, previously reported as highly expressed in one pancreatic cancer cell line by two-dimensional gel electrophoresis. 35 In contrast, the epigenetic silencing of 14-3-3ς by CpG island hypermethylation is among the most common molecular abnormalities in breast carcinoma. 36 We therefore used MSP to analyze the methylation status of the 5′ region of 14-3-3ς in a panel of 15 normal pancreatic tissues and 20 pancreatic cancer cell lines to determine whether altered methylation of this gene might be associated with its increased expression in pancreatic cancer (Figure 4) ▶ .
Figure 4.
Methylation of 14-3-3ς promoter region in pancreatic cancer cell lines. A: DNA extracted from normal pancreatic epithelium (NP) and pancreatic cancer cell lines were amplified with primers specific to the unmethylated (U) or the methylated (M) templates after modification with sodium bisulfite. All normal pancreas samples permitted amplification of both methylated and unmethylated alleles, in contrast to 17 of 20 (85%) pancreatic cancer cell lines in which there was amplification of only unmethylated templates. One cell line, MiaPaCa2, shows complete methylation at 14-3-3ς. B: 14-3-3ς mRNA expression is detected in 19 cell lines with at least one unmethylated 14-3-3ς allele. Treatment of MiaPaCa2 cells with 5-aza-dC resulted in re-expression of 14-3-3ς.
All normal pancreas samples permitted amplification of both methylated and unmethylated alleles, suggesting that variable methylation of this gene occurs in normal pancreas cells. On the other hand, in 17 of 20 (85%) pancreatic cancer cell lines there was amplification of only unmethylated templates, suggesting that aberrant hypomethylation of 14-3-3ς occurs during pancreatic cancer evolution (Figure 4A) ▶ . Two cell lines (Hs766T and PL14) recapitulated the partial methylation pattern seen in normal pancreas, and one cell line, MiaPaCa2, had complete methylation at 14-3-3ς.
All of 19 cell lines with at least one unmethylated 14-3-3ς allele expressed the transcript for 14-3-3ς by RT-PCR (Figure 4B) ▶ . With the current assays, we were unable to distinguish between levels of mono- and biallelic expression of 14-3-3ς. Notably, 14-3-3ς mRNA was not expressed in MiaPaCa2, which had complete methylation at this gene, but treatment of MiaPaCa2 cells with the demethylating agent 5-aza-dC resulted in re-expression of 14-3-3ς (Figure 4B) ▶ .
Discussion
In this study we have characterized a large number of genes differentially expressed in pancreatic cancer some of which are promising markers of the disease. Although several global gene expression studies of pancreatic cancer have recently been reported, 6,8,10-12,15,37-39 in this study we have identified numerous additional genes overexpressed in pancreatic cancer not previously reported. One notable feature of the genes reported as differentially expressed in pancreatic cancers to date is that while many genes are repeatedly found in all studies, some genes identified as overexpressed in one study are not always identified in another. These differences probably occur for several reasons including the number of genes available for study on an array, the type of biostatistical analysis used, and the number and type of tumor specimens characterized. Our current study complements previous pancreatic cancer gene expression studies from ours and other groups in that we used a microarray of 45,000 cDNAs from the Stanford University’s microarray facility, one several times more comprehensive than cDNA microarrays used in previous studies. 8,10,38 In addition, in this study we have analyzed gene expression patterns by both hierarchical cluster analysis and SAM analysis to identify overexpressed genes. Finally, we confirmed and characterized the expression patterns of a large number of overexpressed genes in both primary pancreatic cancers and in pancreatic cancer cell lines. Further characterization of the gene expression patterns in these tumors may facilitate the development of novel therapies and molecular screening tests.
One of the most important findings of this work is the identification of a large set of 79 genes that were expressed at significantly higher levels in pancreas cancers than the normal pancreas. These genes have not previously been reported in association with pancreatic cancer, and include genes such as POH1, XRCC4, and ADAM9. POH1 is a recently described subunit of the human 26 S proteasome, a multiprotein complex that degrades proteins targeted for destruction by the ubiquitin pathway. POH1 can induce AP-1-dependent drug resistance in fission yeast, and also confers P-glycoprotein-independent resistance to taxol (paclitaxel), doxorubicin, 7-hydroxystaurosporine, and ultraviolet light when transiently overexpressed in mammalian cells. 40 XRCC4 is a nonhomologous end-joining protein used in DNA double-strand break repair, a role required both for normal development and for suppression of tumors. 41 XRCC4 protein expression has not been shown to differ between a limited study of normal and tumor tissues. 42 However, the increased expression of POH1 and XRCC4 in pancreatic cancers, as our data suggest, may contribute to the chemo- and radioresistance often observed for this tumor type. Other genes now recognized as expressed in pancreatic cancer, such as ADAM9, may aid in overcoming this resistance. ADAM9 is one of a family membrane bound metalloproteinases that function in the proteolytic processing of membrane-bound precursors and in modulating cell-cell and cell-matrix interactions, a prominent component of infiltrating pancreatic cancers. 32,43 Thus, the exploitation of this function may offer novel possibilities for prodrug delivery and activation at the tumor surface. 34,44
Numerous genes were also identified in the pancreas cancer-specific clusters that have been previously implicated in therapeutic strategies for pancreatic cancer. These included genes characteristically induced by interferon or retinoic acid stimulation. Both interferons and retinoids have been shown to play a role in the cell growth and differentiation of pancreatic cancer cells. 45,46 Clinical trials are currently ongoing to better evaluate the effects of interferon and retinoic acid therapy on patients with advanced pancreatic carcinoma. 47 The differential expression of these genes among pancreatic cancers suggests that their expression may prove useful in predicting which patients might benefit from interferon or retinoic acid treatment.
The identification of these 79 differentially expressed genes in pancreatic cancer may have diagnostic or therapeutic applications. For example, several of these genes were found to be cell surface or secreted proteins. If so, these proteins may serve as diagnostic markers for primary pancreatic cancers, and may also represent potential targets for the development of a cell-mediated vaccine. 18,24,25 Claudin 4, prostate stem cell antigen, and mesothelin perhaps best exemplify this potential for pancreatic cancer. Toxin-conjugated antibodies targeted to each of these membrane-bound proteins shows efficacy in reducing tumor burden in preliminary studies reported thus far. 18,24,48,49
In attempting to identify overexpressed genes in pancreatic cancers compared to normal pancreas, one has to grapple with the fact that most of the differences in gene expression between tumors and normal pancreas appear to reflect the presence of acinar cells and islets within the normal tissues, and their relative paucity in infiltrating carcinomas because of displacement, atrophy, or destruction by the invasive neoplasm. To circumvent this problem we determined the expression pattern of each gene identified by SAM analysis in the two SAGE libraries of nonneoplastic pancreatic ductal epithelial cells available at the SAGE NCBI website. The utility of this approach can be seen from the confirmation of expression patterns of genes identified by SAM analysis by RT-PCR and immunohistochemistry. Overall, the expression patterns of 16 of the overexpressed genes identified by SAM were determined by RT-PCR and/or immunohistochemical labeling and overexpression could be confirmed for most, although not all, genes analyzed.
Although many of these genes are clearly implicated in processes important in the biology of carcinogenesis, including cell-cell and cell-matrix interactions, transcriptional regulation, or cell-cycle control, further investigation will be required to establish what role the altered expression of many of these overexpressed genes play in the biology of pancreatic cancer.
We chose to further investigate the mechanism underlying the altered regulation of one of these genes, 14-3-3ς, in pancreatic adenocarcinoma. Several lines of evidence suggest that 14-3-3ς can act as a tumor suppressor; 36,50,51 loss of expression of 14-3-3ς has been reported in breast carcinomas, squamous cell carcinomas of the head and neck, 52 primary bladder cancers, 53 lung cancers, 39 and hepatocellular carcinomas. 54 In contrast to breast cancer, 36 we found that hypomethylation of the 14-3-3 sigma promoter appears to be a common phenomenon in pancreatic adenocarcinomas. The significance of the elevated 14-3-3ς expression in pancreatic cancer is unclear, but may relate to the anti-apoptotic role described for this gene. 55
The critical challenges in pancreatic cancer are detecting the disease early enough to allow for curative resection and developing new approaches to treat the disease. The characterization of the gene expression patterns in pancreatic cancer tumor tissues and cell lines by cDNA microarray analysis provides validation of a number of genes with promise for development into novel therapeutic or diagnostic targets, and also provides clues to additional genes and cellular pathways that may play a role in the biology of this deadly tumor.
Footnotes
Address reprint requests to Michael Goggins, M.D., The Johns Hopkins Medical Institutions, Department of Pathology, Ross Building Rm. 632, 720 Rutland Ave., Baltimore, MD 21205-2196. E-mail: mgoggins@jhmi.edu.
Supported by the National Institutes of Health (Specialized Programs of Research Excellence in Gastrointestinal Cancer grant CA62924 to M. G.), the Michael Rolfe Fund for Pancreatic Cancer Research (to M. G.), the Lustgarten Foundation for Pancreatic Cancer Research (to A. W. L.), the Howard Hughes Medical Institute (to P. O. B.), and the National Cancer Institute (grant CA85129 to P. O. B.).
C. A. I.-D. and A. M. contributed equally to this work.
References
- 1.Hruban RH, Iacobuzio-Donahue C, Wilentz RE, Goggins M, Kern SE: Molecular pathology of pancreatic cancer. Cancer J 2001, 7:251-258 [PubMed] [Google Scholar]
- 2.Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH, Kern SE: Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res 1996, 56:5360-5364 [PubMed] [Google Scholar]
- 3.Su GH, Hilgers W, Shekher M, Tang D, Yeo CJ, Hruban RH, Kern SE: Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor-suppressor gene. Cancer Res 1998, 58:2339-2342 [PubMed] [Google Scholar]
- 4.Ebert M, Yokoyama M, Friess H, Kobrin MS, Buchler MW, Korc M: Induction of platelet-derived growth factor A and B chains and over-expression of their receptors in human pancreatic cancer. Int J Cancer 1995, 62:529-535 [DOI] [PubMed] [Google Scholar]
- 5.Friess H, Yamanaka Y, Buchler M, Berger HG, Kobrin MS, Baldwin RL, Korc M: Enhanced expression of the type II transforming growth factor beta receptor in human pancreatic cancer cells without alteration of type III receptor expression. Cancer Res 1993, 53:2704-2707 [PubMed] [Google Scholar]
- 6.Gress TM, Wallrapp C, Frohme M, Muller-Pillasch F, Lacher U, Friess H, Buchler M, Adler G, Hoheisel JD: Identification of genes with specific expression in pancreatic cancer by cDNA representational difference analysis. Genes Chromosom Cancer 1997, 19:97-103 [DOI] [PubMed] [Google Scholar]
- 7.Wallrapp C, Muller-Pillasch F, Micha A, Wenger C, Geng M, Solinas-Toldo S, Lichter P, Frohme M, Hoheisel JD, Adler G, Gress TM: Novel technology for detection of genomic and transcriptional alterations in pancreatic cancer. Ann Oncol 1999, 10(Suppl 4):64-68 [PubMed] [Google Scholar]
- 8.Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD: Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res 2002, 62:2890-2896 [PubMed] [Google Scholar]
- 9.Gardner-Thorpe J, Ito H, Ashley SW, Whang EE: Differential display of expressed genes in pancreatic cancer cells. Biochem Biophys Res Commun 2002, 293:391-395 [DOI] [PubMed] [Google Scholar]
- 10.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, Baron A, Scarpa A, Lemoine NR: Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene 2002, 21:4587-4594 [DOI] [PubMed] [Google Scholar]
- 11.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH: Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol 2002, 160:1239-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gress TM, Muller-Pillasch F, Geng M, Zimmerhackl F, Zehetner G, Friess H, Buchler M, Adler G, Lehrach H: A pancreatic cancer-specific expression profile. Oncogene 1996, 13:1819-1830 [PubMed] [Google Scholar]
- 13.Geng MM, Ellenrieder V, Wallrapp C, Muller-Pillasch F, Sommer G, Adler G, Gress TM: Use of representational difference analysis to study the effect of TGFB on the expression profile of a pancreatic cancer cell line. Genes Chromosom Cancer 1999, 26:70-79 [DOI] [PubMed] [Google Scholar]
- 14.Ryu B, Jones J, Hollingsworth MA, Hruban RH, Kern SE: Invasion-specific genes in malignancy: serial analysis of gene expression comparisons of primary and passaged cancers. Cancer Res 2001, 61:1833-1838 [PubMed] [Google Scholar]
- 15.Ryu B, Jones J, Blades NJ, Parmigiani G, Hollingsworth MA, Hruban RH, Kern SE: Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res 2002, 62:819-826 [PubMed] [Google Scholar]
- 16.Argani P, Iacobuzio-Donahue C, Ryu B, Goggins M, Rosty C, Wilentz RE, Murugesan S, Kaushal M, Leach SD, Jaffee E, Yeo CJ, Cameron JL, Kern SE, Hruban RH: Mesothelin is expressed in the vast majority of adenocarcinomas of the pancreas: identification of a new cancer marker by serial analysis of gene expression (SAGE). Clin Cancer Res 2001, 7:3862-3868 [PubMed] [Google Scholar]
- 17.Argani P, Rosty C, Reiter RE, Wilentz RE, Murugesan SD, Leach SD, Ryu B, Skinner HG, Goggins M, Jaffee EM, Yeo CJ, Cameron JL, Kern SE, Hruban RH: Discovery of new markers of cancer through serial analysis of gene expression (SAGE): prostate stem cell antigen (PSCA) is overexpressed in pancreatic adenocarcinoma. Cancer Res 2001, 61:4320-4324 [PubMed] [Google Scholar]
- 18.Michl P, Buchholz M, Rolke M, Kunsch S, Lohr M, McClane B, Tsukita S, Leder G, Adler G, Gress TM: Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology 2001, 121:678-684 [DOI] [PubMed] [Google Scholar]
- 19.Weber CK, Sommer G, Michl P, Fensterer H, Weimer M, Gansauge F, Leder G, Adler G, Gress TM: Biglycan is overexpressed in pancreatic cancer and induces G1-arrest in pancreatic cancer cell lines. Gastroenterology 2001, 121:657-667 [DOI] [PubMed] [Google Scholar]
- 20.Rosty C, Ueki T, Argani P, Jansen M, Yeo CJ, Cameron JL, Hruban RH, Goggins M: Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am J Pathol 2002, 160:45-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallrapp C, Hahnel S, Muller-Pillasch F, Burghardt B, Iwamura T, Ruthenburger M, Lerch MM, Adler G, Gress TM: A novel transmembrane serine protease (TMPRSS3) overexpressed in pancreatic cancer. Cancer Res 2000, 60:2602-2606 [PubMed] [Google Scholar]
- 22.Nishimura T, Horino K, Nishiura H, Shibuya Y, Hiraoka T, Tanase S, Yamamoto T: Apoptotic cells of an epithelial cell line, AsPC-1, release monocyte chemotactic S19 ribosomal protein dimer. J Biochem (Tokyo) 2001, 129:445-454 [DOI] [PubMed] [Google Scholar]
- 23.Maitra A, Iacobuzio-Donahue C, Rahman A, Sohn TA, Argani P, Meyer R, Yeo CJ, Cameron JL, Goggins M, Kern SE, Ashfaq R, Hruban RH, Wilentz RE: Immunohistochemical validation of a novel epithelial and a novel stromal marker of pancreatic ductal adenocarcinoma identified by global expression microarrays: sea urchin fascin homolog and heat shock protein 47. Am J Clin Pathol 2002, 118:52-59 [DOI] [PubMed] [Google Scholar]
- 24.Hassan R, Viner JL, Wang QC, Margulies I, Kreitman RJ, Pastan I: Anti-tumor activity of K1-LysPE38QQR, an immunotoxin targeting mesothelin, a cell-surface antigen overexpressed in ovarian cancer and malignant mesothelioma. J Immunother 2000, 23:473-479 [DOI] [PubMed] [Google Scholar]
- 25.Jaffee EM, Hruban RH, Biedrzycki B, Laheru D, Schepers K, Sauter PR, Goemann M, Coleman J, Grochow L, Donehower RC, Lillemoe KD, O’Reilly S, Abrams RA, Pardoll DM, Cameron JL, Yeo CJ: Novel allogeneic granulocyte-macrophage colony-stimulating factor-secreting tumor vaccine for pancreatic cancer: a phase I trial of safety and immune activation. J Clin Oncol 2001, 19:145-156 [DOI] [PubMed] [Google Scholar]
- 26.Jaffee EM, Schutte M, Gossett J, Morsberger LA, Adler AJ, Thomas M, Greten TF, Hruban RH, Yeo CJ, Griffin CA: Development and characterization of a cytokine-secreting pancreatic adenocarcinoma vaccine from primary tumors for use in clinical trials. Cancer J Sci Am 1998, 4:194-203 [PubMed] [Google Scholar]
- 27.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, Lashkari D, Shalon D, Botstein D, Brown PO: The transcriptional program in the response of human fibroblasts to serum. Science 1999, 283:83-87 [DOI] [PubMed] [Google Scholar]
- 28.Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998, 95:14863-14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tusher VG, Tibshirani R, Chu G: Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001, 98:5116-5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996, 93:9821-9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schussler MH, Skoudy A, Ramaekers F, Real FX: Intermediate filaments as differentiation markers of normal pancreas and pancreas cancer. Am J Surg Pathol 1992, 140:559-568 [PMC free article] [PubMed] [Google Scholar]
- 32.Iacobuzio-Donahue CA, Ryu B, Hruban RH, Kern SE: Exploring the host desmoplastic response to pancreatic carcinoma: gene expression of stromal and neoplastic cells at the site of primary invasion. Am J Pathol 2002, 160:91-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crnogorac-Jurcevic T, Efthimiou E, Capelli P, Blaveri E, Baron A, Terris B, Jones M, Tyson K, Bassi C, Scarpa A, Lemoine NR: Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene 2001, 20:7437-7446 [DOI] [PubMed] [Google Scholar]
- 34.Ellenrieder V, Hendler SF, Boeck W, Seufferlein T, Menke A, Ruhland C, Adler G, Gress TM: Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res 2001, 61:4222-4228 [PubMed] [Google Scholar]
- 35.Sinha P, Hutter G, Kottgen E, Dietel M, Schadendorf D, Lage H: Increased expression of epidermal fatty acid binding protein, cofilin, and 14-3-3-sigma (stratifin) detected by two-dimensional gel electrophoresis, mass spectrometry and microsequencing of drug-resistant human adenocarcinoma of the pancreas. Electrophoresis 1999, 20:2952-2960 [DOI] [PubMed] [Google Scholar]
- 36.Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H, Marks JR, Lambers AR, Futreal PA, Stampfer MR, Sukumar S: High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci USA 2000, 97:6049-6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW: Gene expression profiles in normal and cancer cells. Science 1997, 276:1268-1272 [DOI] [PubMed] [Google Scholar]
- 38.Terris B, Blaveri E, Crnogorac-Jurcevic T, Jones M, Missiaglia E, Ruszniewski P, Sauvenet A, Lemoine N: Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am J Pathol 2002, 160:1745-1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osada H, Tatematsu Y, Yatabe Y, Nakagawa T, Konishi H, Harano T, Tezel E, Takada M, Takahashi T: Frequent and histological type-specific inactivation of 14-3-3sigma in human lung cancers. Oncogene 2002, 21:2418-2424 [DOI] [PubMed] [Google Scholar]
- 40.Spataro V, Toda T, Craig R, Seeger M, Dubiel W, Harris AL, Norbury C: Resistance to diverse drugs and ultraviolet light conferred by overexpression of a novel human 26 S proteasome subunit. J Biol Chem 1997, 272:30470-30475 [DOI] [PubMed] [Google Scholar]
- 41.Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW: Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 2000, 404:897-900 [DOI] [PubMed] [Google Scholar]
- 42.Sakata K, Matsumoto Y, Tauchi H, Satoh M, Oouchi A, Nagakura H, Koito K, Hosoi Y, Suzuki N, Komatsu K, Hareyama M: Expression of genes involved in repair of DNA double-strand breaks in normal and tumor tissues. Int J Radiat Oncol Biol Phys 2001, 49:161-167 [DOI] [PubMed] [Google Scholar]
- 43.Gress TM, Muller-Pillasch F, Lerch MM, Friess H, Buchler M, Adler G: Expression and in-situ localization of genes coding for extracellular matrix proteins and extracellular matrix degrading proteases in pancreatic cancer. Int J Cancer 1995, 62:407-413 [DOI] [PubMed] [Google Scholar]
- 44.Evans JD, Stark A, Johnson CD, Daniel F, Carmichael J, Buckels J, Imrie CW, Brown P, Neoptolemos JP: A phase II trial of marimastat in advanced pancreatic cancer. Br J Cancer 2001, 85:1865-1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Detjen KM, Farwig K, Welzel M, Wiedenmann B, Rosewicz S: Interferon gamma inhibits growth of human pancreatic carcinoma cells via caspase-1 dependent induction of apoptosis. Gut 2001, 49:251-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Recchia F, Sica G, Casucci D, Rea S, Gulino A, Frati L: Advanced carcinoma of the pancreas: phase II study of combined chemotherapy, beta-interferon, and retinoids. Am J Clin Oncol 1998, 21:275-278 [DOI] [PubMed] [Google Scholar]
- 47.Macdonald JS, Jacobson JL, Modiano M, Moore DF, Gandara DR, Schroder LE, Chapman RA: A phase II trial of etoposide, leucovorin, 5-FU, and interferon alpha 2b (ELFI) + G-CSF for patients with pancreatic adenocarcinoma: a Southwest Oncology Group study (SWOG 9413). Invest New Drugs 2000, 18:269-273 [DOI] [PubMed] [Google Scholar]
- 48.Saffran DC, Raitano AB, Hubert RS, Witte ON, Reiter RE, Jakobovits A: Anti-PSCA mAbs inhibit tumor growth and metastasis formation and prolong the survival of mice bearing human prostate cancer xenografts. Proc Natl Acad Sci USA 2001, 98:2658-2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross S, Spencer SD, Strickland L, Lasky LA, Koeppen H: Prostate stem cell antigen (PSCA) is a potential therapy target for pancreatic cancer. US Can Acad Pathol 2002, 15Abstract 592
- 50.Laronga C, Yang HY, Neal C, Lee MH: Association of the cyclin-dependent kinases and 14-3-3 sigma negatively regulates cell cycle progression. J Biol Chem 2000, 275:23106-23112 [DOI] [PubMed] [Google Scholar]
- 51.Prasad GL, Valverius EM, McDuffie E, Cooper HL: Complementary DNA cloning of a novel epithelial cell marker protein, HME1, that may be down-regulated in neoplastic mammary cells Cell Growth Differ 1992, 3:507-513 [PubMed] [Google Scholar]
- 52.Vellucci VF, Germino FJ, Reiss M: Cloning of putative growth regulatory genes from primary human keratinocytes by subtractive hybridization. Gene 1995, 166:213-220 [DOI] [PubMed] [Google Scholar]
- 53.Ostergaard M, Rasmussen HH, Nielsen HV, Vorum H, Orntoft TF, Wolf H, Celis JE: Proteome profiling of bladder squamous cell carcinomas: identification of markers that define their degree of differentiation. Cancer Res 1997, 57:4111-4117 [PubMed] [Google Scholar]
- 54.Iwata N, Yamamoto H, Sasaki S, Itoh F, Suzuki H, Kikuchi T, Kaneto H, Iku S, Ozeki I, Karino Y, Satoh T, Toyota J, Satoh M, Endo T, Imai K: Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene 2000, 19:5298-5302 [DOI] [PubMed] [Google Scholar]
- 55.Samuel T, Weber HO, Rauch P, Verdoodt B, Eppel JT, McShea A, Hermeking H, Funk JO: The G2/M regulator 14-3-3sigma prevents apoptosis through sequestration of Bax. J Biol Chem 2001, 276:45201-45206 [DOI] [PubMed] [Google Scholar]