Abstract
Phosphodiesterases (PDEs) hydrolyze the second messengers cAMP and cGMP. It remains unknown how individual PDE families selectively recognize cAMP and cGMP. This work reports structural studies on substrate specificity. The crystal structures of the catalytic domains of the D674A and D564N mutants of PDE10A2 in complex with cAMP and cGMP reveal that two substrates bind to the active site with the same syn configuration but different orientations and interactions. The products AMP and GMP bind PDE10A2 with the anti configuration and interact with both divalent metals, in contrast to no direct contact of the substrates. The structures suggest that the syn configurations of cAMP and cGMP are the genuine substrates for PDE10 and the specificity is achieved through the different interactions and conformations of the substrates. The PDE10A2 structures also show that the conformation of the invariant glutamine is locked by two hydrogen bonds and is unlikely to switch for substrate recognition. Sequence alignment shows a potential pocket, in which variation of amino acids across PDE families defines the size and shape of the pocket and thus determines the substrate specificity.
Keywords: crystal structure, cyclic nucleotides cAMP and cGMP
Cyclic nucleotide phosphodiesterases (PDEs) are enzymes hydrolyzing the second messengers adenosine and guanosine 3′,5′-cyclic monophosphates (cAMP and cGMP). The human genome encodes 21 PDE genes that are categorized into 11 families (1, 2). Selective inhibitors against individual PDE families have been developed as therapeutics for treatment of various human diseases (3–8). The best known examples are the PDE5 inhibitors sildenafil (Viagra), vardenafil (Levitra), and tadalafil (Cialis) that have been used for treatment of male erectile dysfunction (5). Sildenafil (Revatio) has also been approved for treatment of pulmonary hypertension (9).
PDE10 was independently identified by three groups in 1999 and shows a dual activity on hydrolysis of both cAMP and cGMP (10–12). PDE10 is highly expressed in brain striatum (13–16). Reduction of PDE10A mRNA and protein levels in striatum of transgenic mice implies a role of PDE10A in Huntington's disease (17, 18). Knockout mice experiments suggest that PDE10A is involved in regulating striatal output, possibly by reducing the sensitivity of medium spiny neurons to glutamatergic excitation (19). The PDE10 inhibitor papaverine is effective in improving executive function deficits associated with schizophrenia, and therefore inhibition of PDE10 may represent an approach to treatment of psychosis (20, 21).
PDE families contain a variable N-terminal regulatory domain and a conserved C-terminal catalytic domain. Individual PDE families show different substrate preferences. Crystal structures have been reported for the catalytic domains of seven PDE families in the unliganded form or in complex with inhibitors or products: PDE1B, PDE2A, PDE3B, PDE4B/4D, PDE5A, PDE7A, and PDE9A (22–34). However, it remains a puzzle how the conserved catalytic pocket of the PDE families selectively recognizes cAMP and cGMP. On the basis of the crystal structures of PDE4-AMP and PDE5-GMP, a “glutamine switch” mechanism was proposed for the substrate specificity (31). However, this mechanism is challenged by the mutagenesis experiments, in which the Q817A mutation in PDE5A1 did not significantly impact cAMP binding (35).
We have performed a systematic structural study on the substrate specificity of PDE10A2. Reported here are eight crystal structures of the catalytic domains of the wild-type PDE10A2 in the unliganded state or complex with products AMP and GMP, the D674A mutant and its complexes with substrates cAMP and cGMP, and the D564N mutant and its complex with cAMP. These structures reveal the conformation and interactions of substrates cAMP and cGMP in PDE10A2 and suggest a pocket that may determine the substrate specificity.
Results
Kinetic Properties of the Catalytic Domain of PDE10A2.
The catalytic domain of PDE10A2 (449–789) has a Km and kcat of 56 nM and 0.33 s−1 for cAMP and 4.4 μM and 1.2 s−1 for cGMP. The kcat/Km values are 5.9 and 0.27 s −1·μM−1, respectively, for cAMP and cGMP, and the specificity constant (kcat/Km)cAMP/(kcat/Km)cGMP is 22. These numbers suggest a dual substrate specificity of PDE10A2, although the enzyme is more effective toward cAMP. The Vmax values of our PDE10A2 catalytic domain are 507 nmol/min per mg for cAMP and 1,860 nmol/min per mg for cGMP and are comparable with those for the full-length PDE10A1 (10). There are two divalent metal ions at the active site of wild-type PDE10A2. The first ion coordinates with His529, His563, Asp564, and Asp674, and two waters, and has the same interactions as zinc in PDE4 (23). The second metal coordinates with Asp564 and five water molecules.
The D674A mutant lost at least 4 orders of magnitude in the catalytic activity, as shown by an assay in which no activity was observed at 40 μg/ml D674A mutant, compared with only 5 ng/ml wild-type enzyme required for a normal assay. A consequence of the D674A mutation is loss of zinc ion, as confirmed by no electron density for the zinc site in the structures of unliganded D674A, D674A-cAMP, and D674A-cGMP. This observation implies that zinc is not critical for assembly of the PDE subdomains (23) but is essential for the catalysis. The D564N mutant has the catalytic activity <1/1,000 of the wild-type PDE10A2, although both divalent metals are found in the D564N structures.
Architecture of PDE10 Structures.
Although the PDE10A2 catalytic domain with residues 449–789 was used in the crystallization, only 449–770 can be traced unambiguously in the maps. The C-terminal residues 771–789 in all of the crystals and residues 571–586 of molecule B in the structures of PDE10A-GMP, D674A-cAMP, D674A-cGMP, and D564N-cAMP cannot be traced. Three residues from the expression vector, Ser446, His447, and Met448, have clear electron density and are included in the structures.
The catalytic domain of PDE10A2 contains 15 α-helices and no β-strands (Fig. 1). The topological folding of PDE10A2 is the same as those of known structures of seven PDE families. The superposition of the PDE10A2 catalytic domain over the cAMP-specific PDE4D2 and the cGMP-specific PDE5A1 yielded rmsd values of 1.2 Å for the backbone atoms of 291 comparable PDE4D2 residues and 1.5 Å for 270 comparable PDE5A1 residues, indicating the overall similarity. The main difference between PDE10A2 and PDE4D2 occurs at the N terminus, in which H1 in PDE4D2 becomes a coil in PDE10A2 and H2 in PDE4D2 corresponds to a 310 helix. The short helix H4 in PDE4 is missing in PDE10A2, probably resulting from the absence of 2 aa. However, PDE10A2 contains an extra helix at the N terminus, which occupies a location similar to that in PDE5A1 with a positional difference ≈3 Å. The H-loop of PDE10A2 (residues 571–592) contains two short α-helices and is comparable with that of PDE4D2 but not PDE5A1, in which the H-loop has variable conformations (34).
Fig. 1.
Structures of the catalytic domain of PDE10A2. (A) Ribbon diagram. Divalent metals zinc and magnesium are shown in red and purple. Green balls–sticks represent cAMP. (B) Superposition of PDE10 (green ribbons) over PDE4 (gold) and PDE5 (salmon). Cyan ribbons represent comparable regions among three structures.
All eight crystal structures have the same space group with similar cell dimensions (Table 1) and contain two catalytic domains in their asymmetric units, which are related by a rotation axis of ≈100°. Because this angle differs from a twofold axis, the association of two PDE10A2 molecules appears to be crystallographic packing but is not biologically relevant. Superposition of molecules A over B yielded rmsd values of 1.4–1.6 Å for the eight structures, suggesting conformational changes of some loops. Indeed, residues 689–718 of the M-loop shift their Cα atoms as much as 7.5 Å, whereas other portions of the molecule remain comparable. The movements appear to be forced by the crystal lattice packing, as shown by the fact that the transposition of molecule A to B results in a clash with symmetry-related molecules.
Table 1.
Statistics on diffraction data and structure refinement
| Data | 10A native | 10A-AMP | 10A-GMP | D674A | D674A-cAMP | D674A-cGMP | D564N | D564N-cAMP |
|---|---|---|---|---|---|---|---|---|
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| a, Å | 51.1, | 51.4 | 49.8 | 51.4 | 49.3 | 48.7 | 51.4 | 49.4 |
| b, Å | 82.0 | 82.0 | 81.9 | 82.0 | 82.3 | 82.0 | 82.2 | 82.3 |
| c, Å | 155.4 | 155.5 | 156.7 | 155.4 | 153.2 | 154.0 | 155.2 | 155.9 |
| Resolution, Å | 1.56 | 1.56 | 1.90 | 1.45 | 1.45 | 1.52 | 1.56 | 1.90 |
| Reflections | 85,847 | 87,893 | 48,795 | 110,590 | 101,420 | 95,665 | 91,666 | 47,078 |
| Redundant | 12.0 | 10.2 | 6.7 | 10.1 | 5.8 | 9.6 | 7.4 | 10.0 |
| Complete, % | 91.6 (58.5)* | 93.3 (62.1) | 94.8 (67.4) | 94.4 (71.6) | 90.9 (50.0) | 99.9 (99.1) | 97.0 (77.5) | 92.3 (64.2) |
| Average I/σ | 9.7 (2.2) | 8.2 (2.0) | 7.9 (2.0) | 8.9 (2.1) | 11.1 (2.2) | 10.0 (2.5) | 12.7 (3.3) | 7.7 (2.1) |
| Rmerge | 0.067 (0.47) | 0.094 (0.44) | 0.071 (0.33) | 0.077 (0.39) | 0.073 (0.37) | 0.082 (0.45) | 0.064 (0.28) | 0.078 (0.32) |
| Structure refinement | ||||||||
| Rfactor | 0.197 | 0.200 | 0.213 | 0.209 | 0.218 | 0.204 | 0.209 | 0.217 |
| Rfree | 0.223 (10)† | 0.222 (10) | 0.249 (10) | 0.228 (10) | 0.236 (10) | 0.222 (10) | 0.228 (10) | 0.253 (10) |
| Reflections | 81,787 | 83,987 | 47,053 | 101,832 | 96,206 | 92,068 | 89,422 | 45,261 |
| rmsd for | ||||||||
| Bond, Å | 0.0075 | 0.0064 | 0.0054 | 0.0066 | 0.0045 | 0.0063 | 0.0045 | 0.0058 |
| Angle | 1.3° | 1.2° | 1.2° | 1.3° | 1.3° | 1.3° | 1.1° | 1.2° |
| Average Bfactor, Å2 | ||||||||
| Protein | 22.9 (5,266)‡ | 19.5 (5,285) | 27.1 (5,148) | 20.6 (5,306) | 19.6 (5,176) | 19.7 (5,190) | 19.9 (5,292) | 26.9 (5,182) |
| Ligand | 20.1 (23) | 29.2 (24) | 16.0 (22) | 22.2 (69) | 42.6 (22) | |||
| Waters | 29.6 (425)‡ | 26.2 (399) | 30.2 (322) | 27.3 (425) | 24.9 (437) | 25.0 (366) | 27.9 (458) | 29.9 (320) |
| Zn | 21.3 (2)‡ | 22.8 (2) | 25.8 (2) | 26.8 (2) | 34.0 (2) | |||
| Mg | 15.5 (2)‡ | 13.8 (2) | 24.9 (2) | 15.3 (2) | 16.0 (2) | 15.1 (2) | 19.7 (2) | 31.2 (2) |
*The numbers in parentheses are for the highest resolution shell.
†The percentage of reflections is omitted for calculation of Rfree.
‡The no. of atoms in the crystallographic asymmetric unit.
No Significant Conformational Changes on Mutation or Substrate Binding.
Structural superposition of the D564N and D674A mutants over the unliganded PDE10A2 yielded rmsd values of 0.10 and 0.12 Å for the Cα atoms of molecule A, 0.18 and 0.17 Å for molecule B, indicating no conformational changes on the mutations. The binding of cAMP, cGMP, or GMP caused 2.5–4.7% shrinking in the crystallographic a axis of D674A-cAMP, D564N-cAMP, D674A-cGMP, and PDE10A2-GMP (Table 1) and disorder of the H-loop residues 671–685 in molecule B of these structures. However, superposition of the unliganded PDE10A2 structure over PDE10A2-AMP, PDE10A2-GMP, D674A-cAMP, D564N-cAMP, and D674A-cGMP yielded rmsd values of 0.13, 0.24, 0.33, 0.28, and 0.29 for molecule A, and 0.16, 0.28, 0.72, 0.53, and 0.80 for molecule B, implying overall conformational similarity. A careful examination showed that residues Lys705–Ile708 of the M-loop shift as much as 4 times the average rmsd in the five substrate–product complex structures. These changes appear to be the consequence of lattice contacts promoted by the hydrogen bond of cAMP/cGMP with the carbonyl oxygen of Leu706 of a symmetry-related molecule. In addition, the N-terminal residues 462–470 and 480–482 in molecule A of D674A-cGMP showed Cα positional changes 2–4 times the average difference because of the additional binding of cGMP to the N terminus. The only biologically significant change is the reorientation of the side chain of Leu635 of molecule A in the structures of D674A-cAMP and D564N-cAMP to avoid clashing with the ribose of cAMP.
Different Binding of Substrates cAMP and cGMP.
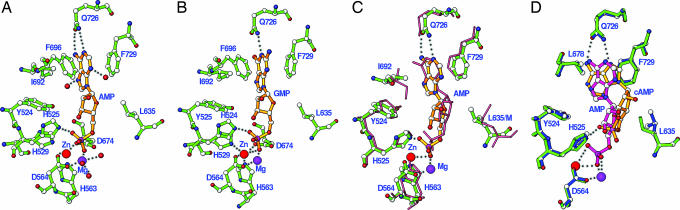
Substrates cAMP and cGMP bind only at the active site of molecule A but not B of PDE10A2 (Fig. 2). The failure of substrate binding to B is probably because of the movement of both helix H14 and the M-loop into the active site [supporting information (SI) Fig. 5]. For example, Ile692 of H14 is located ≈4 Å to Phe729 in molecule B and thus blocks the interactions of the purines of cAMP and cGMP with Phe729 and Gln726. The cAMP binding in the D674A-cAMP and D564N-cAMP structures is the same. The adenine of cAMP is sandwiched by Phe729 on one side and Ile692 and Phe696 on another side. N6 and N7 of adenine of cAMP form hydrogen bonds with OE1 and NE2 of Gln726, respectively. The cyclic phosphate group of cAMP forms one hydrogen bond with His525 and three with water molecules. The ribose O5′ of cAMP hydrogen-bonds with a water molecule. O2′ forms two hydrogen bonds with a water molecule and the carbonyl oxygen of Leu706 from symmetry-related molecule B. In addition, cAMP interacts through van der Waals forces with residues Tyr524, Leu635, Leu675, Val678, Phe696, and Met713 but does not directly contact the divalent metals. Substrate cAMP has a syn configuration and a 3′ endo ribose.
Fig. 2.
Binding of cAMP and cGMP. (A) Interaction of cAMP (golden bonds) with PDE10A2 residues (green bonds) of the D674A mutant. The dotted lines represent hydrogen bonds. Small isolated red balls are water molecules. (B) Interaction of cGMP (gold) with D674A residues. (C) Superposition of cAMP (gold) over cGMP (pink). The residues from D674A-cAMP and D674A-cGMP are shown in blue and green, respectively. (D) Hydrogen bonds of Gln726 with Tyr693 and a water molecule bound to Trp762 and Tyr730.
Three molecules of cGMP bind to the PDE10A2 catalytic domain: one at the active site of molecule A and two in a pocket formed by the N-terminal residues of molecule A and symmetry-related molecules. The cGMP binding at the active site resembles that of cAMP in several aspects (Fig. 2), including the same syn configuration, shrinkage in a axis of the crystal, disorder of residues 571–586 in molecule B, and a hydrogen bond with the carbonyl oxygen of Leu706 from a symmetry-related molecule. However, significant differences between the binding of cAMP and cGMP are observed. The orientation of cGMP is flipped by ≈180° from that of cAMP, whereas their bases and phosphates occupy a similar location (Fig. 2). As result, the hydrogen bond of His525 with phosphate O2 of cAMP is swapped to O5′ of cGMP, and Gln726 forms only one hydrogen bond to N7 of cGMP.
An unexpected observation is that two molecules of cGMP bind to the N terminus of molecule A in D674A-cGMP. At this site, cGMP interacts with Ile450–Ser453 of molecule A, symmetry-related residues of Leu547, Leu654–Leu656, Gln743–Pro747 of molecule A, and symmetry-related Leu466–Lys470 of molecule B. The guanines of two cGMPs stack against one another and form three hydrogen bonds with Arg467 and Gln743 from symmetry-related molecules. The phosphate O2 forms two hydrogen bonds with Ser453 and Leu656. Because this pocket does not exist in the structures in complex with cAMP, AMP, and GMP, it appears to be an artifact of the crystal packing. Nevertheless, the additional cGMP binding may lead to the positional shift of residues 462–471 and 480–483 of molecule B in D674A-cGMP, which is 2- to 4-fold the overall rmsd.
Products AMP and GMP Do Not Simulate Substrate Binding.
The products AMP and GMP bind only to the active site of molecule A but not to molecule B. Both AMP and GMP have the same interactions and the same anti configuration and 3′ endo puckering (Fig. 3). The adenine of AMP and guanine of GMP are sandwiched by hydrophobic residues Phe729 on one side and Ile692 and Phe696 on another side. The adenine of AMP forms two hydrogen bonds with the side chain of Gln726 compared with only one between GMP and Gln726. The interactions of AMP with PDE10A2 are the same as those in the structure of PDE4-AMP, in terms of hydrogen-bonding pattern, nucleotide conformation, and hydrophobic contact (Fig. 3C).
Fig. 3.
Binding of products. (A) Interaction of AMP (gold) with PDE10A2 residues (green). (B) Interaction of GMP with PDE10A2 residues. (C) Superposition of PDE10A2-AMP over PDE4D2-AMP (salmon sticks) (27). (D) Superposition of AMP (pink) over cAMP (gold).
However, the contact pattern of the products AMP/GMP with the invariant glutamine is different from those of the substrates cAMP/cGMP although all of the ligands have the same stacking against Phe729 (Fig. 3D). The products have the anti configuration in contrast to syn of the substrates. Gln726 of PDE10A2 forms two hydrogen bonds with N1 and N6 of AMP and one with N1 of GMP. However, N6 and N7 of cAMP and N7 of cGMP are involved in hydrogen bonds with Gln726. In addition, the phosphate groups of the products coordinate directly with both divalent metals and form hydrogen bonds with metal-bound residue Asp674 (Fig. 3). In comparison, the closest distance from the substrate atoms to the divalent metal ions is ≈4 Å, indicating no direct interaction.
Discussion
The Binding of cAMP and cGMP Simulates the Catalytic Process.
Because the D674A and D564N mutants are basically inactive, it needs to be addressed whether the substrate binding in the crystal structures simulates the enzymatic process. The following observations suggest that the structural information is biologically relevant. First, the crystals of our PDE10A2 catalytic domain possess enzymatic activity, as shown by the fact that substrates cAMP and cGMP were hydrolyzed to AMP and GMP when the unmutated PDE10A2 crystals were soaked in the substrate solutions. Second, the wild-type PDE10A2 and the inactive mutants of D674A and D564N have almost identical conformations for the active-site residues, implying that substrate binding in the mutants simulates the biological process. Finally, the same conformation and interactions of cAMP in the mutant structures of D674A-cAMP and D564N-cAMP suggest that the metal ions do not directly impact the binding and conformation of the substrates. This hypothesis is supported by the fact that the closest distance of the substrate atoms to the metals is ≈4 Å in the D564N-cAMP structure. Therefore, we postulate that the substrate conformation and interaction in these crystal structures resemble those in the catalysis.
It has been reported that cAMP and cGMP exist in equilibrium between syn and anti configurations in solution, with syn:anti ratios of 30:70 and 95:5, respectively, for cAMP and cGMP (36). The (Fo − Fc) and (2Fo − Fc) maps, which were calculated from the structures before cAMP and cGMP were built in, show only the syn configuration of cAMP and cGMP bound to the active site of PDE10A2 (SI Fig. 6). Modeling of anti cAMP and cGMP into the pocket showed that the phosphate oxygen of the substrates clashes with two water molecules coordinated with zinc and magnesium, supporting that the anti conformers do not bind. Thus, the structures suggest that the syn conformer of cAMP and cGMP is the biological substrate for PDE10A2. However, it remains unknown which conformer of cAMP and cGMP is the genuine substrate for other PDE families. Early studies suggested that the preferable substrates are: syn cAMP and cGMP for PDE1 and PDE2, anti cAMP for PDE3 and PDE4, and anti cGMP for PDE3 and PDE5 (37, 38).
Because the substrates and products have different nucleotide configurations: syn for cAMP and cGMP, and anti for AMP and GMP, it would be interesting to know whether the enzyme takes an additional step to perform the syn to anti conversion during catalysis. To address this question, we carried out an activity assay in the presence of AMP and observed no significant inhibition by 5 mM AMP. On the basis of such poor binding of AMP, we hypothesize that the syn products leave the active site right after the catalysis, and the syn to anti conversion is not catalyzed by the enzyme but is an automatic process in solution. The occupancy of the anti AMP and GMP at the active site in the crystals may reflect the forced binding of the products at high concentration because they are predominant in solution (39). This argument is supported by the fact that only the syn configuration of 8-bromo-AMP was observed in the crystal when its syn configuration is predominant in solution (24, 39).
Implications for Substrate Specificity.
The issue of the substrate specificity has not been extensively elucidated. The structures of PDE4D2-AMP, PDE4B2-AMP, and PDE5A1-GMP (24, 28, 31) show that the side chain of the invariant glutamine is fixed in opposite orientations in PDE4 and PDE5 and form two hydrogen bonds with products AMP and GMP. Because chemical formulas of AMP and GMP differ only in the phosphate portion from cAMP and cGMP, the conformation and interactions of nucleosides of the products have been assumed to simulate those of the substrates. This belief has led to a currently popular model, glutamine switch for the substrate specificity (31). In this mechanism, the glutamine is assumed to form two hydrogen bonds with cAMP but one with cGMP in cAMP-specific PDE families, and similarly cGMP-specific PDEs would have one hydrogen bond difference for the two substrates. For PDE families with dual substrate specificity, the side chain of the invariant glutamine would freely rotate to form two hydrogen bonds with cAMP or cGMP.
The glutamine switch mechanism is supported by the structure of dual cAMP/cGMP-specific PDE1B (31), in which the glutamine is unbonded and could be freely rotated. However, it was challenged by two lines of evidence. First, the Q817A mutation of PDE5 weakened Km for cGMP by 60-fold but did not significantly change Km for cAMP (35). Second, the structure of dual-specific PDE2A3 shows that the side chain of Gln859 is fixed by a hydrogen bond with Tyr827 (22). Thus, for the glutamine to switch, this preexisting hydrogen bond has to be broken, and the net gain of energy will be zero.
The glutamine switch is even less likely in view of our PDE10A2 structure, in which the side chain of Gln726 is locked by two hydrogen bonds with Tyr693 and a water bound to Tyr730 and Trp762 (Fig. 2). In addition, two hydrogen bonds between Gln726 and N6 and N7 of cAMP in the PDE10A2 structure are in a totally different pattern from the bidentate bonds of N1 and N6 of the model cAMP predicted by the glutamine switch (31). The single contact between Gln726 and N7 of cGMP (Fig. 2) differs entirely from the two predicted hydrogen bonds between the glutamine and N1 and O6 of cGMP (31). Most importantly, different conformations and interactions between the products and the substrates suggest that the products are not a reasonable model for the substrate binding, at least in PDE10, although it is unknown how similar they are in other PDE families.
The present PDE10A2 structures reveal that cAMP and cGMP bind to the same pocket but have different orientations and interactions, suggesting that the substrate specificity is determined by a different binding of the substrates in the PDE10 family. The structure-based sequence alignment of the nucleoside-binding pocket, that we tentatively term “substrate specificity pocket” or S-pocket, shows dramatic variation of amino acids across PDE families (Table 2 and Fig. 4). Apparently, the amino acid variation not only represents changes of the chemical groups in the PDE families but also defines the shape and size of the binding pocket. To match the chemical nature and shape of the pocket, substrates may bind to individual PDE families in the same or different orientations and configurations with different affinity. In some extreme cases, poor substrate binding, such as Km = 3.9 mM for cGMP in PDE7 (33) and Km > 100 μM for cAMP in PDE9 (2), would explain their preferred cAMP or cGMP specificity. The invariant glutamine in the S-pocket provides hydrogen bonds for binding of substrates but may not be a key element in distinguishing substrates. Instead, multiple elements at the S-pocket must work together to determine the substrate specificity. Each amino acid in the S-pocket may contribute to the specificity differently but should not exclusively dominate the substrate specificity.
Table 2.
Alignment of amino acids at the substrate-binding pocket
| 678 | 685 | 689 | 692 | 693 | 713 | 725 | 726 | 729 | 730 | 762 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PDE10A2 | V | T | A | I | Y | M | G | Q | F | Y | W |
| PDE1B | P | H | T | L | M | L | S | Q | F | I | N |
| PDE2 | Q | T | A | I | Y | M | L | Q | F | M | W |
| PDE3 | P | H | T | I | V | F | L | Q | F | I | W |
| PDE11 | V | S | A | V | T | F | L | Q | W | I | W |
| PDE4 | P | Y | T | I | M | M | S | Q | F | I | Y |
| PDE7 | P | S | S | V | T | L | I | Q | F | M | W |
| PDE8 | P | C | A | I | S | V | S | Q | F | I | W |
| PDE5 | I | Q | A | V | A | M | M | Q | F | I | W |
| PDE6 | I | Q | A | V | A | M | L | Q | F | I | W |
| PDE9 | E | A | V | L | L | F | A | Q | F | I | Y |
Fig. 4.
A potential S-pocket. (A) The PDE10A2 residues (green bonds) are superimposed over the PDE4D2 residues (thinner salmon sticks). (B) Surface presentation of the S-pocket in PDE10.
Methods
Protein Expression and Purification of PDE10A2.
The cDNA of the catalytic domain of human PDE10A2 (GenBank BAA84467) was purchased from American Type Culture Collection (Manassas, VA). The coding region for amino acids 449–789 of PDE10A2 was amplified by PCR and subcloned into the expression vector pET15b. The resultant plasmid pET-PDE10 was transferred into Escherichia coli strain BL21 (CodonPlus) for overexpression. The E. coli cell carrying pET-PDE10 was grown in 2× YT medium at 37°C to absorption A600 = 0.7, and then 0.1 mM isopropyl β-d-thiogalactopyranoside was added for further growth at 20°C overnight. The recombinant PDE10 protein was purified on columns of nickel-nitrilotriacetic acid (Qiagen, Valencia, CA), Q-Sepharose, and Sephacryl S300 (Amersham Biosciences, Piscataway, NJ). A typical purification yielded ≈10 mg of PDE10A2 with a purity >95% from a 2-liter cell culture. The D674A and D564N mutants were produced by a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and verified by DNA sequencing. Overexpression and purification of the mutants used the same protocols for the wild-type protein.
Enzymatic Assay.
Enzymatic activity of the catalytic domains of PDE10A2 and its mutants was assayed by using [3H]cAMP or [3H]cGMP (20,000 cpm per assay) as substrates in a reaction mixture of 20 mM Tris·HCl, pH 7.5/1.0 mM DTT/10 mM MgCl2 at room temperature for 15 min (33). The reaction was terminated by the addition of 0.2 M ZnSO4/Ba(OH)2. Radioactivity of unreacted [3H]cAMP or [3H]cGMP in the supernatant was measured by a liquid scintillation counter. The turnover rate was measured at nine concentrations of substrate and at hydrolysis of 15–50% substrate. Each measurement was repeated three times. The parameters of Km, kcat, and Vmax were calculated following steady-state kinetics.
Crystallization and Structure Determination.
All crystals were grown by hanging drop and have the space group P212121 with similar cell dimensions (Table 1). The unliganded PDE10A2 (449–789) was crystallized against a well buffer of 0.1 M Hepes, pH 7.5/0.2 M MgCl2/50 mM 2-mercaptoethanol (2-ME)/16% PEG 3350. Crystals of the D564N and D674A mutants were grown against a well buffer of 0.1 M Hepes, pH 7.5/0.1 M MgCl2/100 mM 2-ME/13% PEG 3350. The complexes of PDE10A2-AMP, PDE10A2-AMP, D674A-cAMP, D674A-cGMP, and D564N-cAMP were prepared by soaking the unliganded crystals in 20 mM cAMP or 30 mM cGMP in a buffer of 16% PEG 8000/0.1 M Hepes, pH 7.5/0.1 M MgCl2/60 mM 2-ME at 4°C for 1.5–6 h. The crystallization buffer plus 20% ethylene glycol or 15% PEG 400 was used as the cryosolvent. Diffraction data were collected on beamline X29 at Brookhaven National Laboratory and processed by program HKL (Table 1) (40).
The structure of the unliganded PDE10A2 was solved by the molecular replacement program AMoRe (41), using the PDE4D2 catalytic domain as the initial model. The rotation and translation searches yielded two solutions that are obviously distinct from background and have correlation coefficients of 11.2 and 10.4 and R factors of 0.538 and 0.539 for 4543 reflections between 4 and 8 Å resolution. When the two solutions were added together, the correlation coefficient and R factor were improved to 0.21 and 0.51. The electron density map was improved by the density modification package of CCP4. The atomic model was rebuilt by program O (42) and refined by program CNS (Table 1) (43). The refined catalytic domain of the unliganded PDE10A2 was used to solve other structures.
Supplementary Material
Acknowledgments
We thank beamline X29 at National Synchrotron Light Source for collection of diffraction data, Drs. Richard Wolfenden and Charles Carter for proofreading the manuscript, and Xiuyan Qin for assistance on cell culture and protein expression. This work was supported in part by National Institutes of Health Grant GM59791 (to H.K.).
Abbreviations
- PDE
cyclic nucleotide phosphodiesterase
- S-pocket
substrate specificity pocket.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2OUN, 2OUP, 2OUQ, 2OUR, 2OUS, 2OUU, 2OUV, and 2OUY).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700279104/DC1.
References
- 1.Bender AT, Beavo JA. Pharmacol Rev. 2006;58:488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- 2.Mehats C, Andersen CB, Filopanti M, Jin SL, Conti M. Trends Endocrinol Metab. 2002;13:29–35. doi: 10.1016/s1043-2760(01)00523-9. [DOI] [PubMed] [Google Scholar]
- 3.Truss MC, Stief CG, Uckert S, Becker AJ, Wafer J, Schultheiss D, Jonas U. World J Urol. 2001;19:344–350. doi: 10.1007/s003450100221. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Shakur Y, Yoshitake M, Kambayashi JJ. Cardiovasc Drug Rev. 2001;19:369–386. doi: 10.1111/j.1527-3466.2001.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 5.Corbin JD, Francis SH. Int J Clin Pract. 2002;56:453–459. [PubMed] [Google Scholar]
- 6.Lipworth BJ. Lancet. 2005;365:167–175. doi: 10.1016/S0140-6736(05)17708-3. [DOI] [PubMed] [Google Scholar]
- 7.Castro A, Jerez MJ, Gil C, Martinez A. Med Res Rev. 2005;25:229–244. doi: 10.1002/med.20020. [DOI] [PubMed] [Google Scholar]
- 8.Menniti FS, Faraci WS, Schmidt CJ. Nat Rev Drug Discov. 2006;5:660–670. doi: 10.1038/nrd2058. [DOI] [PubMed] [Google Scholar]
- 9.Galie N, Ghofrani HA, Torbicki A, Barst RJ, Rubin LJ, Badesch D, Fleming T, Parpia T, Burgess G, Branzi A, et al. N Engl J Med. 2005;353:2148–2157. doi: 10.1056/NEJMoa050010. [DOI] [PubMed] [Google Scholar]
- 10.Soderling SH, Bayuga SJ, Beavo JA. Proc Natl Acad Sci USA. 1999;96:7071–7076. doi: 10.1073/pnas.96.12.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujishige K, Kotera J, Michibata H, Yuasa K, Takebayashi S, Okumura K, Omori K. J Biol Chem. 1999;274:18438–18445. doi: 10.1074/jbc.274.26.18438. [DOI] [PubMed] [Google Scholar]
- 12.Loughney K, Snyder PB, Uher L, Rosman GJ, Ferguson K, Florio VA. Gene. 1999;234:109–117. doi: 10.1016/s0378-1119(99)00171-7. [DOI] [PubMed] [Google Scholar]
- 13.Seeger TF, Bartlett B, Coskran TM, Culp JS, James LC, Krull DL, Lanfear J, Ryan AM, Schmidt CJ, Strick CA, et al. Brain Res. 2003;985:113–126. doi: 10.1016/s0006-8993(03)02754-9. [DOI] [PubMed] [Google Scholar]
- 14.Kotera J, Sasaki T, Kobayashi T, Fujishige K, Yamashita Y, Omori K. J Biol Chem. 2004;279:4366–4375. doi: 10.1074/jbc.M308471200. [DOI] [PubMed] [Google Scholar]
- 15.Xie Z, Adamowicz WO, Eldred WD, Jakowski AB, Kleiman RJ, Morton DG, Stephenson DT, Strick CA, Williams RD, Menniti FS. Neuroscience. 2006;139:597–607. doi: 10.1016/j.neuroscience.2005.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coskran TM, Morton D, Menniti FS, Adamowicz WO, Kleiman RJ, Ryan AM, Strick CA, Schmidt CJ, Stephenson DT. J Histochem Cytochem. 2006;54:1205–1213. doi: 10.1369/jhc.6A6930.2006. [DOI] [PubMed] [Google Scholar]
- 17.Hebb AL, Robertson HA, Denovan-Wright EM. Neuroscience. 2004;123:967–981. doi: 10.1016/j.neuroscience.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Hu H, McCaw EA, Hebb AL, Gomez GT, Denovan-Wright EM. Eur J Neurosci. 2004;20:3351–3363. doi: 10.1111/j.1460-9568.2004.03796.x. [DOI] [PubMed] [Google Scholar]
- 19.Siuciak JA, McCarthy SA, Chapin DS, Fujiwara RA, James LC, Williams RD, Stock JL, McNeish JD, Strick CA, Menniti FS, Schmidt CJ. Neuropharmacology. 2006;51:374–385. doi: 10.1016/j.neuropharm.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Siuciak JA, Chapin DS, Harms JF, Lebel LA, McCarthy SA, Chambers L, Shrikhande A, Wong S, Menniti FS, Schmidt CJ. Neuropharmacology. 2006;51:386–396. doi: 10.1016/j.neuropharm.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Rodefer JS, Murphy ER, Baxter MG. Eur J Neurosci. 2005;21:1070–1076. doi: 10.1111/j.1460-9568.2005.03937.x. [DOI] [PubMed] [Google Scholar]
- 22.Iffland A, Kohls D, Low S, Luan J, Zhang Y, Kothe M, Cao Q, Kamath AV, Ding YH, Ellenberger T. Biochemistry. 2005;44:8312–8325. doi: 10.1021/bi047313h. [DOI] [PubMed] [Google Scholar]
- 23.Xu RX, Hassell AM, Vanderwall D, Lambert MH, Holmes WD, Luther MA, Rocque WJ, Milburn MV, Zhao Y, Ke H, Nolte RT. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]
- 24.Xu RX, Rocque WJ, Lambert MH, Vanderwall DE, Luther MA, Nolte RT. J Mol Biol. 2004;337:355–365. doi: 10.1016/j.jmb.2004.01.040. [DOI] [PubMed] [Google Scholar]
- 25.Sung BJ, Hwang KY, Jeon YH, Lee JI, Heo YS, Kim JH, Moon J, Yoon JM, Hyun YL, Kim E, et al. Nature. 2003;425:98–102. doi: 10.1038/nature01914. [DOI] [PubMed] [Google Scholar]
- 26.Huai Q, Wang H, Sun Y, Kim HY, Liu Y, Ke H. Structure (London) 2003;11:865–873. doi: 10.1016/s0969-2126(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 27.Huai Q, Colicelli J, Ke H. Biochemistry. 2003;42:13220–13226. doi: 10.1021/bi034653e. [DOI] [PubMed] [Google Scholar]
- 28.Huai Q, Liu Y, Francis SH, Corbin JD, Ke H. J Biol Chem. 2004;279:13095–13101. doi: 10.1074/jbc.M311556200. [DOI] [PubMed] [Google Scholar]
- 29.Huai Q, Wang H, Zhang W, Colman R, Robinson H, Ke H. Proc Natl Acad Sci USA. 2004;101:9624–9629. doi: 10.1073/pnas.0401120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scapin G, Patel SB, Chung C, Varnerin JP, Edmondson SD, Mastracchio A, Parmee ER, Singh SB, Becker JW, Van der Ploeg LH, Tota MR. Biochemistry. 2004;43:6091–6100. doi: 10.1021/bi049868i. [DOI] [PubMed] [Google Scholar]
- 31.Zhang KY, Card GL, Suzuki Y, Artis DR, Fong D, Gillette S, Hsieh D, Neiman J, West BL, Zhang C, et al. Mol Cell. 2004;15:279–286. doi: 10.1016/j.molcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Card GL, England BP, Suzuki Y, Fong D, Powell B, Lee B, Luu C, Tabrizizad M, Gillette S, Ibrahim PN, et al. Structure (London) 2004;12:2233–2247. doi: 10.1016/j.str.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Liu Y, Chen Y, Robinson H, Ke H. J Biol Chem. 2005;280:30949–30955. doi: 10.1074/jbc.M504398200. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Liu Y, Huai Q, Cai J, Zoraghi R, Francis SH, Corbin JD, Robinson H, Xin Z, Lin G, Ke H. J Biol Chem. 2006;281:21469–21479. doi: 10.1074/jbc.M512527200. [DOI] [PubMed] [Google Scholar]
- 35.Zoraghi R, Corbin JD, Francis SH. J Biol Chem. 2006;281:5553–5558. doi: 10.1074/jbc.M510372200. [DOI] [PubMed] [Google Scholar]
- 36.Yathindra N, Sunderalingam M. Biochem Biophys Res Commun. 1974;56:119–126. doi: 10.1016/s0006-291x(74)80323-2. [DOI] [PubMed] [Google Scholar]
- 37.Butt E, Beltman J, Becker DE, Jensen GS, Rybalkin SD, Jastorff B, Beavo JA. Mol Pharmacol. 1995;47:330–339. [PubMed] [Google Scholar]
- 38.Butt E, Beltman J, Becker DE, Jensen GS, Rybalkin SD, Jastorff B, Beavo JA. Mol Pharmacol. 1995;47:340–347. [PubMed] [Google Scholar]
- 39.Lee CH, Evans FE, Sarma RH. J Biol Chem. 1975;250:1290–1296. [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Navaza J, Saludjian P. Methods Enzymol. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 42.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 43.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.