Abstract
We report here the cloning, expression, and characterization of a dual-substrate, cAMP and cGMP, cyclic nucleotide phosphodiesterase (PDE) from mouse. This PDE contains the consensus sequence for a PDE catalytic domain, but shares <50% sequence identity with the catalytic domains of all other known PDEs and, therefore, represents a new PDE gene family, designated PDE10A. The cDNA for PDE10A is 3,370 nt in length. It includes a full ORF, contains three in-frame stop codons upstream of the first methionine, and is predicted to encode a 779-aa enzyme. At the N terminus PDE10A has two GAF domains homologous to many signaling molecules, including PDE2, PDE5, and PDE6, which likely constitute a low-affinity binding site for cGMP. PDE10A hydrolyzes cAMP with a Km of 0.05 μM and cGMP with a Km of 3 μM. Although PDE10A has a lower Km for cAMP, the Vmax ratio (cGMP/cAMP) is 4.7. RNA distribution studies indicate that PDE10A is expressed at highest levels in testis and brain.
Intracellular levels of the second messengers cAMP and cGMP are regulated by both their rates of synthesis by cyclases and their hydrolysis by phosphodiesterases. By regulating the duration and amplitude of these messenger signals, phosphodiesterases (PDEs) play critical regulatory roles in a wide variety of signal transduction pathways. Previous studies have demonstrated that PDEs regulate such processes as learning and memory, vision, olfaction, platelet aggregation, aldosterone synthesis, insulin secretion, T cell activation, and penile erection (1–7). The diversity of processes regulated by PDEs is reflected by, and a result of, the diversity of PDE isoforms encoded within by the mammalian genome. Past investigations have established the existence of nine different phosphodiesterase gene families, each encoding PDEs with unique substrate specificities, kinetics, allosteric regulators, tissue-expression profiles, and pharmacological sensitivities (1, 8–12). Because of the unique characteristics of each PDE isoform and their importance in many different physiologic processes, PDEs represent likely drug targets for pharmacological intervention. To further our understanding of the diversity of PDEs and their physiologic regulatory roles, it is imperative to identify the full repertoire of PDEs present in the mammalian genome and understand the biochemical properties they each possess. Toward this end, we report here the cloning and characterization of a previously unknown dual-substrate PDE, PDE10A, from mouse.
MATERIALS AND METHODS
Database Searching for Expressed Sequence Tag (EST) PDE Sequences.
The amino acid sequences of PDE2, PDE5, and PDE6 were used as queries to search the database of ESTs (13, 14). The program used was the Basic Local Alignment Search Tool (blast) (15), accessed from the database search and analysis “Search Launcher” (16). This search resulted in many EST sequences with homology to PDEs. Each of these EST sequences then were used as queries in a blastn search of GenBank to determine whether they represented different but known PDEs or whether the EST sequence represented a truly unknown PDE. EST clone ID 760844 was isolated in this manner as a sequence that appeared to represent a novel PDE.
Other Databases or Programs Used.
Homology of PDE10A N terminus to other GAF-containing proteins was detected by Position-Specific Iterated BLAST (psi-blast) searches of the nonredundant GenBank database and by use of the Multiple Alignment Construction and Analysis Workbench (MACAW). The GAF domain boundaries of PDE10A were defined by searching of the Simple Modular Architecture Research Tool (SMART) database. Alignments of GAF domains were constructed by using clustalx and by manual adjustment.
Sources of EST Clones.
Clone 760844 was ordered from Genome Systems (St. Louis).
DNA Sequencing and Sequence Assembly.
Plasmid DNA was prepared by using the SNAP kit (Invitrogen). Primers were designed by using the program amplify (freeware by William Engels, Genetics Department, University of Wisconsin, Madison). Sequencing was done by using Applied Biosystems Prism dye terminator cycle-sequencing kit (Perkin–Elmer), and sequencing reactions were purified by using Centri-sep columns (Princeton Separations, Adelphia, NJ). Sequences were assembled by using the program sequencher 3.0 (Gene Codes, Ann Arbor, MI).
Primers Used.
Primers used were 10strt.2 (catggaaaaattatatggtttgacggatg), 10/14ASsq.2 (ccacggagtagccgacgtctgaac), and 10strt.3 (gcatggaaaaattatatggtttgacg).
DNA Probe Synthesis and Northern Blotting.
DNA probes were generated from PCR clone 10(2) by using Prime-It RmT random primer labeling kit (Stratagene). [α-32P]dCTP at 6,000 Ci/mmol was used, and the reaction product was purified by using Centri-sep columns. Multiple-tissue mRNA and mRNA dot blots were purchased from CLONTECH. Prehybridization and hybridization were done at 65°C by using ExpressHyb (CLONTECH). Northern blots were washed first at room temperature with 2× SSC and then at 50°C with 0.1× SSC and 0.1% SDS. RNA dot blots were washed first at room temperature in 2× SSC and 1% SDS followed by washing at 65°C in 0.1× SSC and 0.5% SDS.
5′ and 3′ Rapid Amplification of cDNA Ends (RACE).
Marathon-adapted cDNA and Advantage polymerase PCR mix was purchased from CLONTECH. Reactions were set up as follows: 0.5 ng of adapted cDNA, 0.2 μM AP1 or AP2 primer, 0.2 μM gene-specific primer, 5 μl of 10× reaction buffer (supplied with Advantage polymerase), 0.2 mM dNTP, 1 μl of Advantage KlenTaq Polymerase mix, in a final volume of 50 μl. Reaction cycles were as follows: 94°C for 1 min; 5 cycles of 94°C for 30 sec, 72°C for 4 min; 5 cycles of 94°C for 30 sec, 70°C for 4 min; 25 cycles of 94°C for 30 sec, 68°C for 4 min.
Generation of Full-Length MMPDE10A.
Three independent PCR clones [10(14), 10(2), and 10(3)] of PDE10A were amplified in independent PCRs from mouse testis marathon-adapted cDNA (CLONTECH) and subcloned into the PCRII-topo vector (Invitrogen). Clone 10(14) was amplified by using primers strt.2 and AP2; clone 10(2) was amplified by using primers strt2 and 10(14)Assq2; and clone 10(3) was amplified by using primers strt.3 and 10(14)Assq2. Clones 10(14) and 10(2) were amplified by using Advantage polymerase mix (CLONTECH), and clone 10(3) was amplified by using PfuTurbo (Stratagene). Each independent clone was sequenced to verify the correct cDNA sequence and to ensure no PCR errors were present. Clone 10(14) contained two PCR artifacts (T to C at 1273 and A to C at 1559) that were not found in clone 10(2) or 10(3). Clone 10(2) was subcloned into pFastBac1 (GIBCO/BRL), and 10(3) was subcloned into pFastBacHt for baculoviral expression.
Expression and Purification of PDE10A and PDE2A1.
One microgram of 10(2) or 10(3) was electroporated into 106 Sf9 cells in a 0.4-cm electroporation cuvette at 71 μF and infinite resistance by using 300 V, 50 W, and 50 mA and plated out into 60-mm dishes. After 4–5 days, the medium was harvested and used to infect a 50-ml culture of 5 × 105 cells per ml for 5 days, after which the medium again was harvested. For kinetics and inhibitor assays, viral titer of the amplified virus stock was determined and used to infect a 50-ml culture of 2 × 106 cells per ml at a multiplicity of infection of 3. After 3 days cells were pelleted and harvested in homogenization buffer containing 40 mM Tris⋅HCl (pH 7.5), 15 mM benzamidine, 15 mM 2-mercaptoethanol, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, and 5 mM EDTA. This was homogenized in a Dounce homogenizer on ice with 25 strokes, and glycerol was added to a final concentration of 20–30%. This preparation was stored at −20°C in aliquots and did not lose appreciable activity over 3 weeks. For nickel nitrilotriacetate (Ni-Nta) (Qiagen) purification, 1.5-liter cultures were infected with 10(3) and harvested as above. These cultures then were pelleted and homogenized as above in 5–10 vol of buffer containing 50 mM Tris⋅HCl (pH 8.5), 5 mM 2-mercaptoethanol, 5 mM imidazole, 1 tablet/7 ml of EDTA-free protease inhibitor (BMB), and 100 mM KCl. 10(3) was purified according to the protocol provided with the pFastBac vectors by using Ni-Nta resin, except for the following modification. The cleared lysate was passed through a 1-ml column twice, washed, and then eluted with elution buffer containing 200 mM imidazole. Unlike the PDE10A homogenate, purified PDE10A was not stable on ice or at −20°C. Thus eluted protein was run through a Centri-sep column equilibrated in 50 mM Tris⋅HCl, pH 7.5/5 mM 2-mercaptoethanol/50% glycerol. This preparation then was stored at −70°C and did not lose activity over at least 1 week. Bovine PDE2A1 was also expressed in Sf9 cells by using the above procedure. Recombinant PDE2 was purified by using a cGMP affinity resin as described previously (17). Purified PDE2 was stored as described for purified PDE10A.
Kinetic and Inhibitor Studies.
All PDE assays were done according to the method of Hansen and Beavo (18) in a buffer containing 40 mM Mops (pH 7.5), 0.8 mM EGTA, 15.0 mM magnesium acetate, 0.2 mg/ml BSA, and [3H]cAMP or [3H]cGMP (50,000 cpm/reaction) in a final volume of 250 μl. All assays were carried out in triplicate, and reaction times and enzyme amounts were kept such that the lowest substrate concentration gave no more than 30% hydrolysis. PDE10A activity is defined as the total activity in PDE10A-infected Sf9 cells minus the background activity of control Sf9 cells. Hydrolytic activity was significantly higher in PDE10A-infected cells compared with control Sf9 cells because no activity above background was present in the control Sf9 cells at the dilutions of PDE10A used for kinetic and inhibitor assays. Thus, the basal hydrolytic activity in SF9 cells was not a significant contributor to the total activity in PDE10A-infected cells. Inhibitor studies were done without added unlabeled cGMP to keep the substrate concentration low (22.6 nM) so that IC50 values would approximate the Ki. Vmax assays on PDE10A purified to near homogeneity were done at substrate concentrations of 40 μM cAMP and 300 μM cGMP. 3-Isobutyl-1-methylxanthine (IBMX), erythro-9-(2-hydroxy-3-nonyl)adenine (EHNA), and dipyridamole were obtained from Sigma. Zaprinast was a gift from May & Baker (Dagenham, U.K.). Rolipram was obtained from BioMol (Plymouth Meeting, PA). SCH 51866 {(+)-cis-5,6a,7,8,9,9a-hexahydro-2-[4-[trifluoromethyl]phenylmethyl]-5-methyl-cylopent[4,5]imidazo[2,1-b]purin-4(3H)one} was a gift from Schering- Plough, and sildenafil was a gift from Pfizer Central Research (Sandwich, U.K.). Enoximone was a gift from the Merrell Dow Research Institute (Cincinnati).
cGMP-Binding Assays.
Binding assays were done by incubating partially purified PDE10A with 9.2 μM cGMP and 200 cpm/pmol of [3H]cGMP in a buffer containing 1 mM EDTA, 25 mM 2-mercaptoethanol, 5 mM NaH2PO4, and 5 mM Na2HPO4. Reactions were incubated for 45 min at room temperature. Less than 1% hydrolysis of cGMP occurs under these conditions. At the end of the incubation, the reaction mixtures were spun through Centri-sep columns preequilibrated with the above reaction buffer, and the radioactivity in the total volume was measured. By using this procedure, binding was measured under conditions of equilibrium similar to the procedure of Hummel and Dreyer (19). Bound cGMP was calculated by measuring the [3H]cGMP that eluted with PDE2 or PDE10A above that which eluted with reactions containing BSA alone.
RESULTS
Cloning and Tissue Distribution of PDE10A.
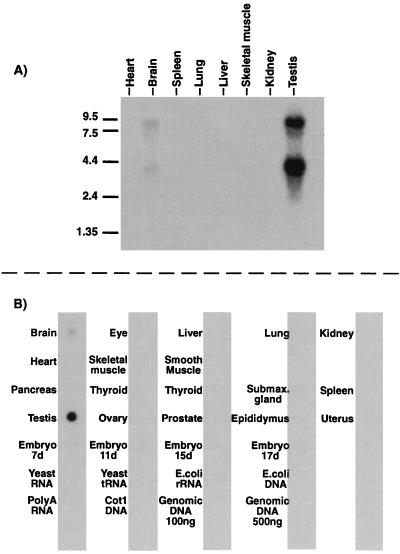
Using bioinformatic approaches for cloning previously unknown PDEs, we have isolated several ESTs that represent new PDE gene families (8, 9). We report here the isolation of an EST (clone ID 760844) that contained sequences with similarity to the noncatalytic cGMP-binding domains of PDE2, PDE5, and PDE6 (20, 21). This EST cDNA clone was used to probe a mouse Northern blot, and the initial data suggested that this gene was expressed at highest levels in mouse testis (data not shown). Oligonucleotide primers were synthesized based on the sequence of this EST clone and were used to perform both 5′ and 3′ RACE from mouse testis to clone the cDNA corresponding to this EST clone. 5′ RACE yielded several clones, two of which (1 and 6) were 592 bp in length, contained the sequence of EST clone 760844, and extended this sequence 548 nt. These clones contained a start methionine with three in-frame, upstream stop codons, indicating the full N-terminal coding sequence was present (Fig. 1). 3′ RACE yielded a single clone [clone 10(14)], which was approximately 3.0 kb in size and extended the sequence of EST clone 760844 to a poly(A) tail, indicating that the full-coding C-terminal end of this cDNA was present. Sequencing of clones 1, 6, and 10(14) demonstrated that the contiguous sequence contained not only domains similar to the noncatalytic cGMP-binding domains of PDEs 2, 5, and 6, but also contained a region of sequence homologous to the catalytic domain of all previously characterized mammalian PDEs. To generate a cDNA representing the full ORF of this new PDE, primer 10strt2 or 10strt3 was used in combination with primer 10(14)Assq2 to directly PCR-amplify the full coding sequence of PDE10A from testis cDNA. This was done in two independent PCRs and yielded clones 10(2) and 10(3). Sequencing of the independent clones 1, 6, 10(2), 10(3), and 10(14) yielded a consensus sequence for the full coding region [two apparent PCR errors were found in clone 10(14) that were not present in clones 10(2) and 10(3)]. The cDNA for MMPDE10A is 3,370 bp in length and is predicted to encode an enzyme 779 aa in length, with a predicted molecular mass of 88,517 Da. Because this catalytic domain shares a sequence identity of less than 50% with other known PDEs, this PDE does not belong within any of the previously described nine PDE gene families and, therefore, is designated MMPDE10A. Northern blot analysis of PDE10A RNA expression in mouse tissues was preformed by using two separate probes. Probe 1 was generated from an XhoI (nucleotide 1856)-to-EcoRI (nucleotide 2288) digest that corresponded to the catalytic domain. This probe hybridized to two bands in both brain and testis at approximately 9 kb and 4 kb (Fig. 2A). A second probe, probe 2, corresponding to the N terminus of PDE10A, was made by digesting with HindIII (from vector) and EcoRV (nucleotide 733 of PDE10A), also hybridized to two bands of the same size in testis and brain on a second, independent Northern blot (data not shown). Probe 1 also was used to assess the tissue distribution of PDE10A in 22 mouse tissues by RNA dot blot (Fig. 2B). In agreement with the Northern blot, a hybridization signal was observed in testis and brain, with testis having the highest level of signal.
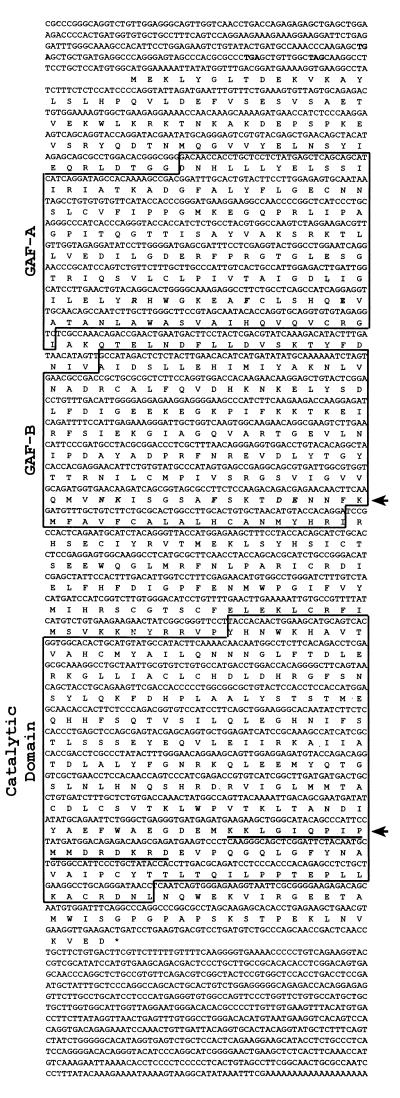
Figure 1.
Full-length sequence of MMPDE10A. cDNA sequence of PDE10A is shown. Bold codons above the predicted start methionine are in-frame stop codons indicating the cDNA contains the entire ORF. Boxed amino acid regions indicate domains identified by sequence similarity to known domains in other proteins. Bold, italic amino acids in GAFA and GAFB domains correspond to residues thought to be important for the support of cGMP binding in PDE5. Top arrow points to the consensus motif [N(K/R)XnFX3DE] found in full in GAFB. Lower arrow points to a predicted bipartite nuclear localization signal found within the C terminus of PDE10A. Asterisk indicates the final stop codon.
Figure 2.
Northern blot and RNA dot blot analyses of PDE10A expression. (A) Northern blot analysis of poly(A) RNA from mouse tissues. Two bands, 9 kb and 4 kb, are detected in both testis and brain that hybridize to PDE10A probes. (B) RNA dot blot analysis of 22 mouse tissues. Consistent with Northern data, mouse testis and brain appear to express PDE10A, with highest levels of expression in testis.
Baculovirus Expression of PDE10A and Characterization of PDE10A Activity.
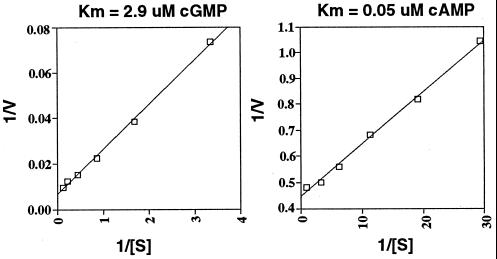
Full-length PDE10A [clone 10(2) or 10(3)] was subcloned into a baculovirus expression vector [pFastBac1 for 10(2) or pFastBac Ht for 10(3)] and transfected into Sf9 cells to produce recombinant virus. All Km and inhibitor studies used the 10(2) clone, which does not have any additional sequences attached to the expressed protein. Vmax and binding assays were done by using clone 10(3), which contains an N-terminal hexahistidine fusion. Sf9 cells infected with PDE10A recombinant virus were assayed for PDE activity compared with that found in control, uninfected Sf9 cell homogenates 3 days post infection. Assays for both cAMP and cGMP PDE activity were performed with a range of substrate concentrations from 0.01 to 9.2 μM. Sf9 cell homogenates infected with recombinant PDE10A virus had significantly higher cAMP and cGMP hydrolytic activity than the control Sf9 cells. Two separate Sf9 cell infections were performed with 10(2) to prepare two independent batches of PDE10A expressing Sf9 cell homogenates, and kinetic curve assays were done by using either cAMP or cGMP as substrate. Background PDE activity, if any, was measured in noninfected Sf9 cells and subtracted from activities measured in PDE10A homogenates. The Km of PDE10A is 0.05 μM for cAMP and 2.9 μM for cGMP (Fig. 3) (average of three separate experiments on two independent enzyme preparations). PDE10A also was purified to near homogeneity by using histidine-tagged clone 10(3). Using these preparations, Vmax assays were preformed by using substrate concentrations of either 40 μM cAMP or 300 μM cGMP. Hydrolysis was linear with time for both cAMP and cGMP (not shown). The highest specific activity measured in these assays was 0.74 μmol/min per mg (cAMP) and 3.5 μmol/min per mg (cGMP). Although this specific activity is low compared with some PDEs, it is similar to other low Km PDEs (11). The Vmax ratio (cGMP/cAMP) is 4.7, indicating a higher specific activity for cGMP.
Figure 3.
PDE10A kinetics. Shown are Lineweaver–Burk plots for both cAMP and cGMP. The displayed Km is the calculated average number.
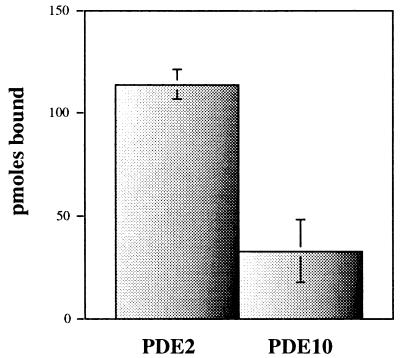
cGMP-binding assays also were done by using purified PDE10A. A modified Hummel and Dreyer procedure (19), using Sephadex G-50 spin columns equilibrated to the same cGMP concentration as that used in the binding assays, were used. Under these conditions, using 9.2 μM cGMP and approximately 200 pmol of protein, near stoichiometric binding of cGMP is detected for purified PDE2; however, much less binding is detected for PDE10A (Fig. 4).
Figure 4.
cGMP-binding assays for PDE2 and PDE10A. Binding is detected for PDE2 at 9.2 μM, whereas much less binding is detected for PDE10A. Results are average picomoles bound, with range shown for PDE2 (n = 2) and SD shown for PDE10 (n = 6).
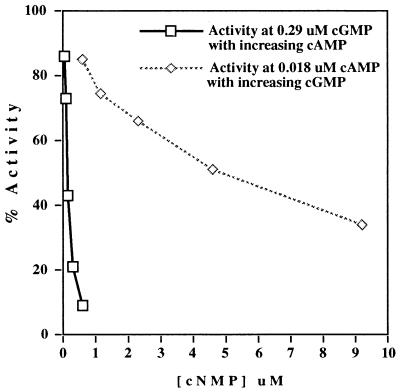
Because PDE2 cAMP hydrolysis is stimulated by cGMP, the effect of cGMP on cAMP hydrolysis and vice versa also was assayed in homogenates expressing clone 10(2). For these studies, [3H]cAMP hydrolysis was measured in the presence of 0.018 μM cAMP and a range of cGMP concentrations from 0.6 to 9.2 μM. The reverse also was done with [3H]cGMP (and 0.29 μM cGMP) and 0.052–0.6 μM cAMP. Neither cGMP nor cAMP stimulated hydrolytic activity of the other (Fig. 5).
Figure 5.
Effect of cGMP on cAMP hydrolysis and effect of cAMP on cGMP hydrolysis. [3H]cAMP hydrolysis was measured in the presence of 0.018 μM cAMP and a range of cGMP concentrations from 0.6 to 9.2 μM. The reverse also was done with [3H]cGMP (and 0.29 μM cGMP) and 0.052–0.6 μM cAMP. cAMP may function as a potent inhibitor of cGMP hydrolysis.
The effects of several PDE inhibitors also were determined for PDE10A (Table 1). In contrast to the other newly described PDEs, PDE8 and PDE9, PDE10A is inhibited by the nonselective PDE inhibitor IBMX. PDE10A was not inhibited well by EHNA, sildenafil (Viagra), or Enoximone within dose ranges considered to be specific for these inhibitors. PDE10A, however, was inhibited by dipyridamole and SCH 51866 within 10-fold of the doses previously considered to be specific for these inhibitors. Zaprinast inhibited PDE10A at a dose 14- and 72-fold higher than that considered specific for PDE5 and PDE6, and rolipram inhibited PDE10A at an approximately 24-fold higher dose than the IC50 for PDE4.
Table 1.
Inhibitor profile for PDE10A
| Inhibitor | Selective for PDE type (IC50) | IC50 for PDE10, μM (n = 3) |
|---|---|---|
| IBMX | Nonselective (2–50 μM) | 2.6 ± 0.6 |
| Zaprinast | PDE5/6 (0.76/0.15 μM) | 10.8 ± 1.4 |
| EHNA | PDE2 (1 μM) | 69 ± 2.8 |
| Enoximone | PDE3 (1 μM) | >100 |
| Dipyridamole | PDE5/6/8 (0.9/0.38/4.5 μM) | 1.1 ± 0.1 |
| SCH 51866 | PDE1 and 5/9 (0.1/1.5 μM) | 1 ± 0.3 |
| Sildenafil | PDE5 (3.9 nM) | >1 |
| Rolipram | PDE4 (2.0 μM) | 47.3 ± 8.2 |
DISCUSSION
The cDNA presented here demonstrates the existence of a previously unknown cyclic nucleotide PDE present in the genome of Mus musculus. This 3,370-bp cDNA represents a full ORF of PDE10A because it contains three in-frame stop codons upstream of the predicted start methionine (Fig. 1). PDE10A is expected to consist of 779 aa, with a predicted molecular mass of 88,517 Da and a conserved catalytic domain at the C terminus homologous to all class I PDEs.
Previous work has identified the existence of 18 PDE genes clustered to 9 gene families within mammalian genomes (1, 8–12). Comparison of all known PDE genes reveals that at least 70% sequence identity within the catalytic domain is conserved between PDEs within the same gene family (not shown). This conservation falls to below 50% sequence identity when comparing PDEs that do not belong to the same gene family. Comparison of the amino acid sequence within the catalytic domain of PDE10A to all other previously identified PDEs reveals less than 50% sequence identity (not shown), thus establishing PDE10A as a member of a 10th gene family.
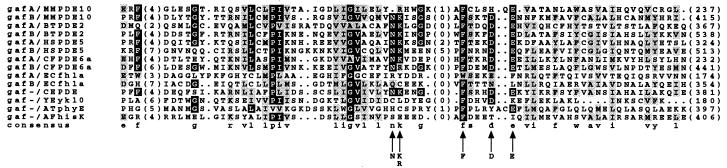
In addition to a homologous catalytic domain, PDE10A also contains two imperfect repeats at the N terminus similar to the N termini of PDE2, PDE5, and PDE6 (Fig. 6). These tandem sites encode a noncatalytic cGMP-binding site in PDE2, PDE5, and PDE6 (20, 21). In the case of PDE2, this site acts as an allosteric activator of the cAMP hydrolytic activity of this enzyme (22). For PDE5, cGMP binding may regulate phosphorylation of the enzyme (23), and, for PDE6, cGMP binding may regulate the interaction between the catalytic subunits and the inhibitory γ-subunit and transducin (24–27). Structure/function studies, by alanine mutagenesis, of this domain in PDE5 have identified a series of residues necessary for the support of cGMP binding (28). This motif [N(K/R)XnFX3DE] is found in each repeat of PDE2, PDE5, and PDE6. Interestingly, PDE10A is unique from these PDEs because it contains only three of five residues of this motif in the first repeat (Fig. 6). That two of these residues are missing in the first repeat of PDE10A may indicate either that this site does not participate in cGMP binding, or that the substituted residues can functionally replace those conserved in other PDEs (in contrast to alanine), or that this site serves some function other than cGMP binding (see below). In PDE5, mutagenesis of the conserved aspartic acid found in the above motif to alanine in each repeat also has demonstrated that each repeat binds cGMP with different affinities, the first site having a high affinity (Kd = 0.5 μM) and the second site having a lower affinity (Kd > 10 μM) (29). In PDE10A this aspartic acid is missing in the first repeat. Thus, if PDE10A is analogous to PDE5 and the first site does not bind cGMP, then it may be predicted that PDE10A will have a low affinity for binding cGMP. In addition to the above PDEs, it also has been realized recently that these conserved repeats are present in many signaling molecules from eubacterial, archaeal, and eukaryotic organisms (30). Reflecting the diverse signaling molecules known to contain this domain, each repeat now has been termed a GAF (for cGMP binding and stimulated phosphodiesterases, Anabaena adenylyl cyclases, and Escherichia coli FhlA) domain (30). Similar to the mammalian PDEs, many of these proteins contain two GAF domains at the N terminus. Several, however, contain only one GAF domain (including two PDEs from Caenorhabditis elegans), and the GAF domain may be the only identifiable domain in a polypeptide (for instance, YEykl0; see Fig. 6) (30). Although this domain does bind cGMP in PDE2, PDE5, and PDE6, it is likely that this is not its function in all cases. For instance, several E. coli proteins contain GAF domains, yet the E. coli genome does not encode for an identifiable guanylyl cyclase (unpublished observation). Instead, it is possible that the GAF domain folds with a similar tertiary topology, which allows it to act as a binding domain for divergent small molecules. In support of this possibility, E. coli FhlA, a transcriptional regulatory protein, binds formate within the N terminus, which contains two (see Fig. 6) GAF domains (31).
Figure 6.
Multiple sequence alignment of the C-terminal PDE10A GAF domains to GAF domains of other proteins. Numbers in parentheses at the end of the sequences indicate the amino acid numbers shown for each protein. Numbers in parentheses within sequence alignment indicate the number of amino acids omitted from the alignment for clarity. Arrows mark the motif described as supporting cGMP binding in PDE5. Shading reflects at least 50% conservation. BTPDE2, Bos taurus PDE2 (accession no. M73512); HSPDE5, Homo sapiens PDE5 (AF043731); CFPDE6A, Canis familiaris PDE6 alpha (Y13282); EcfhlA, E. coli FhlA (X52227); CEPDE, C. elegans gene R0807.6 “similar to phosphodiesterase” (Z12017); YEykl0, Saccharomyces cerevisiae ORF YKL069W (Z28069.1); AtphyE, Arabidopsis thaliana phytochrome E (X76610); AFHisK, Archaeoglobus fulgidus gene AF1483 “putative histidine kinase” (AE001000).
Apparently, the function of these GAF domains in PDE10A is different from those of PDE2 in that cGMP does not stimulate PDE10A activity (Fig. 5). Studies utilizing radioimmunoassays and purified histidine-tagged PDE10A suggest that, unlike PDE6, PDE10A does not purify with bound cGMP (not shown). In vitro binding assays using purified PDE2 and PDE10A also were done by using a procedure similar to that of Hummel and Dryer for measuring binding under equilibrium conditions (19). At relatively high ligand concentrations (9.2 μM), using ≈200 pmol of either PDE2 or PDE10A, binding is detected easily for PDE2, but much less binding is detected for PDE10A. It is probable, therefore, that as predicted by mutagenesis studies of PDE5, PDE10A has a Kd for cGMP well above 9.2 μM. Alternatively, the binding conditions used to date may be missing some activating component, the enzyme may be mostly inactive, or PDE10A may bind another small molecule with a higher affinity. To date, experiments to test these possibilities have been hampered by very low purification yields.
Searching PDE10A sequence with psort (a program for detecting sorting and subcellular localization motifs) (32) reveals the presence of a bipartite nuclear localization signal (NLS) (Fig. 1). Interestingly, this NLS is located within the catalytic domain at the C-terminal end. Very little is known regarding the presence and function (if any) of cyclic nucleotide PDEs within the nucleus (33). Further studies will be necessary to determine whether PDE10A indeed is localized to the nucleus in vivo.
Because PDE10A was cloned without prior insight into its hydrolytic activities, homogenates from Sf9 cells expressing PDE10A were used to measure the activity of this new enzyme. These assays showed that PDE10A hydrolyzes both cAMP and cGMP, with apparent Km values of 0.05 and 3.0 μM, respectively (Fig. 3). The Km for cAMP and cGMP for PDE10A is approximately 10-fold lower than the Km of PDE2 for both cAMP and cGMP (34). PDE10A also differs from PDE2 in that cGMP does not stimulate cAMP hydrolysis (Fig. 5). The kinetic data indicate PDE10A may regulate both cAMP and cGMP pathways in vivo. However, because PDE10A has a very low Km and Vmax for cAMP, it is also possible that in vivo PDE10A functions as a cAMP-inhibited cGMP PDE (see also Fig. 5). In this case cAMP may act to potentiate cGMP. This would be analogous to PDE3, which has a similar kinetic profile but is, instead, a cGMP-inhibited cAMP PDE. Recent studies suggest cGMP indeed does potentiate cAMP in vivo by inhibition of PDE3 (35).
PDE10A is also unique from all previously characterized PDE families pharmacologically (Table 1). IBMX, a nonselective inhibitor of PDE families 1–6, is also a potent inhibitor of PDE10A, with an IC50 of 2.6 μM. Inhibitors selective for PDE2 (EHNA) or PDE5 (sildenafil) did not inhibit PDE10A within dose ranges considered to be specific. Dipyridamole, an inhibitor of PDE5, PDE6, and PDE8, also inhibits PDE10A with an IC50 of 1.1 μM, approximately 10-fold higher than the Km for PDE5 and PDE6. The emerging data from this study and studies of PDE8 (9, 11) indicate that this inhibitor should no longer be considered specific for the PDE5 and PDE6 families. SCH 51866, an inhibitor of PDE1, PDE5, and PDE9, also inhibits PDE10A with an IC50 of 1.0 μM. Zaprinast, an inhibitor of PDE5 and PDE6, and rolipram, an inhibitor of PDE4, both inhibit PDE10A at higher doses; IC50 of 10.8 and 47.3 μM, respectively. Because dipyridamole and SCH 51866 both inhibit PDE10A with the highest potency of any inhibitors tested thus far, these inhibitors may suggest structural features useful for the development of more selective inhibitors of PDE10A.
PDE10A RNA expression was surveyed in 22 mouse tissues by both Northern blot and RNA dot blot analyses. Northern blot analysis indicates that PDE10A expression is highest in testis and brain, with two bands of approximately 9 kb and 4 kb detected in both tissues. Probes from both the catalytic domain and the N terminus detect both bands in these tissues in independent blots (Fig. 2 and data not shown). Why two bands are detected in both testis and brain RNA is unknown, but presumably reflects heterogeneous RNA species corresponding to PDE10A. RNA dot blot analysis confirms the Northern blot data and further indicates that PDE10A mRNA in mouse does not appear to be expressed at high levels in the following tissues: pancreas, intestinal smooth muscle, eye, thymus, skeletal muscle, ovary, uterus, epididymis, spleen, heart, liver, lung, kidney, thyroid, prostate, and embryo (days 7–17).
CONCLUSION
In conclusion, data describing a 10th PDE gene family, PDE10A, are presented here. PDE10A hydrolyzes both cAMP and cGMP. The kinetics suggest in vivo, however, that cAMP may regulate cGMP hydrolysis. PDE10A represents a previously unrecognized means by which cyclic nucleotides are likely to be regulated in both mouse testis and brain. PDE10A also contains two conserved GAF domains at the N terminus that are similar to and different from the GAF domains of PDE2, PDE5, and PDE6. These domains may mediate a low-affinity binding for cGMP.
Acknowledgments
We thank Jacquelyn Soderling for her expertise and advice regarding baculovirus expression. This work was supported by National Institutes of Health Grants DK21723 and GM07750.
ABBREVIATIONS
- PDE
phosphodiesterase
- IBMX
3-isobutyl-1-methylxanthine
- RACE
rapid amplification of cDNA ends
- EST
expressed sequence tag
- SCH 51866
(+)-cis-5,6a,7,8,9,9a-hexahydro-2-[4-[trifluoromethyl]phenylmethyl]-5-methyl-cylopent[4,5]imidazo[2,1-b]purin-4(3H)-one
- EHNA
erythro-9-(2-hydroxy-3-nonyl)adenine
Note Added in Proof:
A human PDE10 has recently been identified by both Pfizer Central Research (J. Lanfear, personal communication) and ICOS Corporation (36).
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF110507).
References
- 1.Beavo J A. Physiol Rev. 1995;75:725–748. doi: 10.1152/physrev.1995.75.4.725. [DOI] [PubMed] [Google Scholar]
- 2.Zhao A Z, Zhao H, Teague J, Fujimoto W, Beavo J A. Proc Natl Acad Sci USA. 1997;94:3223–3228. doi: 10.1073/pnas.94.7.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao A Z, Bornfeldt K E, Beavo J A. J Clin Invest. 1998;102:869–873. doi: 10.1172/JCI3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Yee C, Beavo J A. Science. 1999;283:848–851. doi: 10.1126/science.283.5403.848. [DOI] [PubMed] [Google Scholar]
- 5.Ballard S A, Gingell C J, Tang K, Turner L A, Price M E, Naylor A M. J Urol. 1998;159:2164–2171. doi: 10.1016/S0022-5347(01)63299-3. [DOI] [PubMed] [Google Scholar]
- 6.Boolell M, Allen M J, Ballard S A, Gepi-Attee S, Muirhead G J, Naylor A M, Osterloh I H, Gingell C. Int J Impot Res. 1996;8:47–52. [PubMed] [Google Scholar]
- 7.Chuang A T, Strauss J D, Murphy R A, Steers W D. J Urol. 1998;160:257–261. [PubMed] [Google Scholar]
- 8.Soderling S H, Bayuga S J, Beavo J A. J Biol Chem. 1998;273:15553–15558. doi: 10.1074/jbc.273.25.15553. [DOI] [PubMed] [Google Scholar]
- 9.Soderling S H, Bayuga S J, Beavo J A. Proc Natl Acad Sci USA. 1998;95:8991–8996. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi M, Matsushima K, Ohashi H, Tsunoda H, Murase S, Kawarada Y, Tanaka T. Biochem Biophys Res Commun. 1998;250:751–756. doi: 10.1006/bbrc.1998.9379. [DOI] [PubMed] [Google Scholar]
- 11.Fisher D A, Smith J F, Pillar J S, St. Denis S H, Cheng J B. Biochem Biophys Res Commun. 1998;246:570–577. doi: 10.1006/bbrc.1998.8684. [DOI] [PubMed] [Google Scholar]
- 12.Fisher D A, Smith J F, Pillar J S, St. Denis S H, Cheng J B. J Biol Chem. 1998;273:15559–15564. doi: 10.1074/jbc.273.25.15559. [DOI] [PubMed] [Google Scholar]
- 13.Boguski M S, Lowe T M, Tolstoshev C M. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 14.Hillier L D, Lennon G, Becker M, Bonaldo M F, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- 15.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Smith R F, Wiese B A, Wojzynski M K, Davison D B, Worley K C. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- 17.Martins T J, Mumby M C, Beavo J A. J Biol Chem. 1982;257:1973–1979. [PubMed] [Google Scholar]
- 18.Hansen R S, Beavo J A. Proc Natl Acad Sci USA. 1982;79:2788–2792. doi: 10.1073/pnas.79.9.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hummel J P, Dreyer W J. Biochim Biophys Acta. 1962;63:530–532. doi: 10.1016/0006-3002(62)90124-5. [DOI] [PubMed] [Google Scholar]
- 20.McAllister-Lucas L M, Sonnenburg W K, Kadlecek A, Seger D, Trong H L, Colbran J L, Thomas M K, Walsh K A, Francis S H, Corbin J D, Beavo J A. J Biol Chem. 1993;268:22863–22873. [PubMed] [Google Scholar]
- 21.Charbonneau H, Prusti R K, LeTrong H, Sonnenburg W K, Mullaney P J, Walsh K A, Beavo J A. Proc Natl Acad Sci USA. 1990;87:288–292. doi: 10.1073/pnas.87.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stroop S D, Charbonneau H, Beavo J A. J Biol Chem. 1989;264:13718–13725. [PubMed] [Google Scholar]
- 23.Thomas M K, Francis S H, Corbin J D. J Biol Chem. 1990;265:14971–14978. [PubMed] [Google Scholar]
- 24.Cote R H, Bownds M D, Arshavsky V Y. Proc Natl Acad Sci USA. 1994;91:4845–4849. doi: 10.1073/pnas.91.11.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arshavsky V Y, Dumke C L, Bownds M D. J Biol Chem. 1992;267:24501–24507. [PubMed] [Google Scholar]
- 26.Yamazaki A, Bartucca F, Ting A, Bitensky M W. Proc Natl Acad Sci USA. 1982;79:3702–3706. doi: 10.1073/pnas.79.12.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamazaki A, Bondarenko V A, Dua S, Yamazaki M, Usukura J, Hayashi F. J Biol Chem. 1996;271:32495–32498. doi: 10.1074/jbc.271.51.32495. [DOI] [PubMed] [Google Scholar]
- 28.Turko I V, Haik T L, McAllister-Lucas L M, Burns F, Francis S H, Corbin J D. J Biol Chem. 1996;271:22240–22244. doi: 10.1074/jbc.271.36.22240. [DOI] [PubMed] [Google Scholar]
- 29.McAllister-Lucas L M, Haik T L, Colbran J L, Sonnenburg W K, Seger D, Turko I V, Beavo J A, Francis S H, Corbin J D. J Biol Chem. 1995;270:30671–30679. doi: 10.1074/jbc.270.51.30671. [DOI] [PubMed] [Google Scholar]
- 30.Aravind L, Ponting C P. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 31.Korsa I, Bock A. J Bacteriol. 1997;179:41–45. doi: 10.1128/jb.179.1.41-45.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakai K, Horton P. Trends Biochem Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 33.Lupidi G, Eufemi M, Luciani S, Ferraro A, Riva F. Ital J Biochem (Engl Ed) 1990;39:30–37. [PubMed] [Google Scholar]
- 34.Beavo J, Houslay M D. Cyclic Nucleotide Phosphodiesterases: Structure, Regulation and Drug Action. New York: Wiley; 1990. [Google Scholar]
- 35.Maurice D H, Haslam R J. Mol Pharmacol. 1990;37:671–681. [PubMed] [Google Scholar]
- 36.Loughney, K., Snyder, P. B., Uher, L., Rosman, G. J., Ferguson, K. & Florio, V. A. (1999) Gene, in press. [DOI] [PubMed]