Abstract
The proteasome regulates histone lysine methylation and gene transcription, but how it does so is poorly understood. To better understand this process, we used the epistatic miniarray profile (E-MAP) approach to identify factors that genetically interact with proteasomal subunits. In addition to members of the Set1 complex that mediate histone H3 lysine 4 methylation (H3K4me), we found that deleting members of the CCR4/NOT mRNA processing complex exhibit synthetic phenotypes when combined with proteasome mutants. Further biochemical analyses revealed physical associations between CCR4/NOT and the proteasome in vivo. Consistent with the genetic and biochemical interactions linking CCR4/NOT with proteasome and Set1-mediated methylation, we find that loss of Not4 decreases global and gene-specific H3K4 trimethylation (H3K4me3) and decreases 19S proteasome recruitment to the PMA1 gene. Similar to proteasome regulation of histone methylation, loss of CCR4/NOT members does not affect ubiquitinated H2B. Mapping of Not4 identified the RING finger domain as essential for H3K4me3, suggesting a role for ubiquitin in this process. Consistent with this idea, loss of the Not4-interacting protein Ubc4, a known ubiquitin-conjugating enzyme, decreases H3K4me3. These studies implicate CCR4/NOT in the regulation of H3K4me3 through a ubiquitin-dependent pathway that likely involves the proteasome.
Keywords: 19S proteasome, COMPASS, transcription
Histone methylation plays a significant role in chromatin organization, gene transcription, and epigenetic regulation (1). A large body of work now shows that histone lysine methylation functions largely through the recruitment of effector proteins that contain a variety of methyllysine binding domains (2, 3). Because histone lysine residues can receive up to three methyl groups, lysine methylation has the potential to create differential biological outputs that depend on the methyl state of the residue (i.e., mono-, di-, or trimethylation). These attributes make lysine methylation an important contributor to the “histone code,” which has been postulated to govern epigenetic regulation (4).
Of the known sites of histone methylation, one of the best characterized is histone H3 lysine 4 methylation (H3K4me) (5–7). Chromatin immunoprecipitation (ChIP) coupled with whole-genome microarray (ChIP-chip) analysis has revealed that the mono-, di-, and trimethylated H3K4 residues segregate differentially along genes (8, 9). In particular, H3K4 trimethylation (H3K4me3) is localized specifically to the promoter and 5′ ends of genes (10). The enzyme complex responsible for H3K4me, COMPASS, contains the Set1 methyltransferase and a number of other protein subunits that contribute to methylation (5, 6, 11). In particular, the RNA recognition motif (RRM) of Set1 and the Spp1 subunit of COMPASS both contribute specifically to the establishment of H3K4me3, whereas other COMPASS subunits control the occurrence of the individual H3K4me states (12–14). The ability of COMPASS to methylate H3K4 ultimately is controlled by the prior establishment of mono-ubiquitinated histone H2B (ubH2B) in a “trans-tail” regulatory pathway that is poorly understood but also is known to regulate histone H3 lysine 79 (H3K79) methylation mediated by Dot1 (15–17). The ubiquitin-conjugating E2 enzyme, Rad6, and its E3 ubiquitin ligase partner, Bre1, are recruited to chromatin in a mechanism that depends on the PAF transcription elongation complex (18, 19). Once the Rad6/Bre1 complex is localized to chromatin, it mono-ubiquitinates H2B and promotes COMPASS-mediated H3K4me2 and H3K4me3 (15, 16, 20).
The proteasome, in particular the 19S regulatory particle, has been implicated in transcriptional initiation and elongation (21, 22). Recent data also has established the 19S particle as a regulator of nucleosomal histone modifications. In particular, inhibition of 19S function revealed that this complex controls H3K4 and H3K79 methylation at a step after the establishment of H2B ubiquitination (23, 24). How the proteasome regulates chromatin modifications and gene transcription is poorly understood. In this article, we present data that define a previously uncharacterized genetic and biochemical link between the proteasome and the evolutionarily conserved CCR4/NOT complex that may connect these two complexes to the selective regulation of H3K4me3.
Results
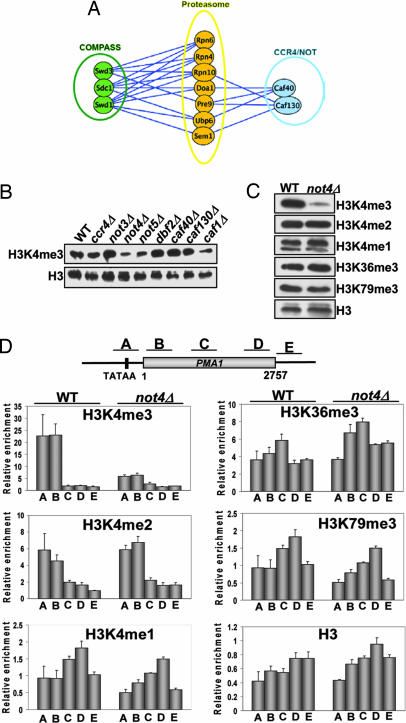
Recent studies have shown that components of the 19S regulatory particle contribute to transcriptional regulation, at least in part, by altering nucleosomal histone modifications. Specifically, H3K4 and H3K79 methylation were shown to depend on 19S function at a step after the establishment of histone H2B ubiquitination (23). To further define this regulatory pathway, we used synthetic genetic array (SGA) technology in high-density epistatic miniarray profile (E-MAP) format (see Materials and Methods for a detailed description) to identify factors that genetically interacted with genes coding for proteasome subunits. As shown in Fig. 1A and consistent with previous results, we found strong growth defects when mutations of proteasomal subunits were combined with deletions of subunits of the H3K4 methyltransferase complex (25). Unexpectedly, components of the CCR4/NOT complex also were identified as having genetic interactions with proteasome mutants (Fig. 1A). This complex is known to regulate multiple cellular processes, including mRNA deadenylation and decapping, protein ubiquitination, and transcription (26, 27). Given its link to transcriptional control, we tested whether components of the CCR4/NOT complex also affected H3K4me. We screened yeast deletion mutants of all of the core factors, excluding not1Δ and not2Δ (which were either inviable or extremely sick) and tested these strains for effects on H3K4me3. We found that not4Δ and not5Δ showed significant reductions in the global levels of H3K4me3, whereas other members had either minor effects (ccr4Δ and caf1Δ) or no detectable changes (caf40Δ and caf130Δ; Fig. 1B). Because Not5, but not Not4, controls overall stability of the CCR4/NOT complex and the protein levels of its members (28), we focused our studies on the mechanism by which Not4 regulates H3K4me3.
Fig. 1.
The CCR4/NOT complex genetically interacts with the proteasome and regulates H3K4me3. (A) The proteasome has genetic interactions with COMPASS and CCR4/NOT complex members. E-MAP and synthetic genetic array technology were used to cross Natr strains harboring individual deletions of proteasome subunits with an array of deletion strains to create sets of Natr Kanr haploid double mutants. Growth rates were assessed as described in ref. 41. Lines connect genes with negative (synthetic sick/lethal) genetic interactions. The lengths of lines and proximity of boxes in this diagram are unrelated to the strengths of the indicated genetic interactions. (B) Loss of CCR4/NOT complex members decreases H3K4me3. Individual CCR4/NOT complex deletion mutants in mid-log phase were screened for effects on H3K4me3 as described in SI Methods. (C) Global H3K4me3 is selectively regulated by CCR4/NOT complex. Wild-type (WT) strain (9XMyc-Set1) and not4Δ (YNL031) strains were grown to mid-log phase, and whole-cell extracts (WCEs) were prepared. Samples were prepared and analyzed by immunoblot analysis. Antibodies used are specified. (D) Nucleosomal H3K4me3, but not other histone methylation marks, are decreased in not4Δ. ChIP was performed with antibodies and strains described in C along the PMA1 gene. A schematic of the PMA1 locus showing the relative locations of primer pairs used in the analyses is shown at the top. Data are normalized to histone H3 levels and are the average and SEM of three independent experiments.
Because H3K4 can be mono-, di-, or trimethylated, we next asked whether loss of Not4 had similar effects on all three H3K4me states. Interestingly, we found not4Δ reduced only H3K4me3, leaving the other H3K4me states, and also histone H3 lysine 36 (H3K36) and H3K79 methylation, unaffected (Fig. 1C). To determine whether gene-specific H3K4me3 was decreased, we used ChIP to examine nucleosomal histones in vivo. In agreement with our results in Fig. 1C, we found dramatic reductions in the levels of H3K4me3, but not other H3K4me forms or H3K36 or H3K79 methylation, on the PMA1 and FMP27 genes (Fig. 1D and data not shown). Importantly, loss of H3K4me3 was not caused by disruption of the COMPASS complex or decreased levels of Set1 mRNA because we found that the stoichiometry of COMPASS is unaffected in not4Δ cells, and Set1 mRNA levels remain normal [supporting information (SI) Fig. 5 A and B]. These data show that the CCR4/NOT complex selectively regulates H3K4me3, a histone modification intimately linked to transcriptional activation, without affecting the integrity of COMPASS.
Our genetic and biochemical analyses suggested a link among COMPASS, the CCR4/NOT complex, and the proteasome (see Fig. 1A). To pursue whether CCR4/NOT was involved in proteasome control of histone methylation, we confirmed that inactivation of the 19S regulatory particle decreased H3K4me (23, 24). Using mutant alleles of two different 19S components, CIM3 (RPT6) and CIM5 (RPT1), we analyzed global H3K4me at either the permissive (24°C) or the nonpermissive (37°C) temperature. Consistent with other reports, we observed a significant decrease in H3K4me3 in the cim3-1 or cim5-1 strains relative to wild type at the nonpermissive temperature (SI Fig. 6A) (23, 24). These results further substantiate a critical role for the 19S proteasome in regulating H3K4me3.
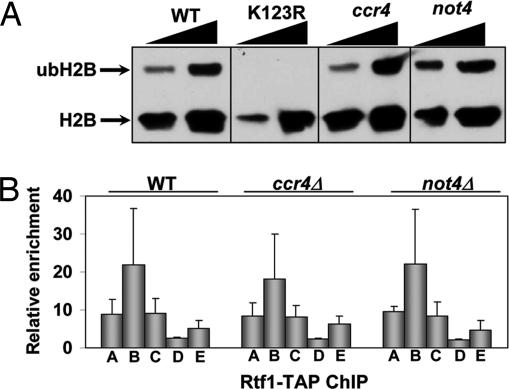
We next tested whether components of the CCR4/NOT complex also regulated H2B mono-ubiquitination because this modification is a prerequisite for the establishment of H3K4me but is not controlled by proteasome. Deletion of either CCR4 or NOT4 had no effect on the ubiquitination levels of histone H2B (Fig. 2A). Instead, we consistently detected a slight, but reproducible, increase in the level of ubH2B in the not4Δ strain (Fig. 2A and Z.-W. Sun, personal communication). Because the PAF complex is required for H2B ubiquitylation and has been linked biochemically to the CCR4/NOT complex (29), we examined its localization on genes by using ChIP in wild-type, ccr4Δ, or not4Δ cells. Consistent with results showing that the CCR4/NOT complex had no effect on H2B ubiquitination (Fig. 2A), we saw no difference in either the relative amount or the distribution of the Rtf1 component of the PAF complex on the PMA1 gene (Fig. 2B). These results indicate that recruitment of PAF to transcribed genes does not depend on the CCR4/NOT complex. Furthermore, these results show that the CCR4/NOT complex regulates H3K4me3 at a step after establishment of H2B ubiquitylation, analogous to that seen for the proteasome.
Fig. 2.
CCR4/NOT regulates H3K4me3 independently of the histone trans-tail pathway. (A) CCR4/NOT complex does not regulate histone H2B mono-ubiquitination. Wild-type (YZS276), K123R (YZS277), ccr4Δ (YNL015), and not4Δ (YNL019) cells were grown to mid-log phase, and H2B ubiquitination was analyzed as described in ref. 25. The lower arrow denotes histone Flag-H2B, and the upper arrow indicates mono-ubiquitinated histone Flag-H2B (ubH2B). (B) The PAF elongation complex is not disrupted on genes in CCR4/NOT complex mutants. Wild-type (Rtf1-TAP), ccr4Δ (YNL032), and not4Δ (YNL033) cells were grown to mid-log phase, and cells then were processed for ChIP analysis. Primers used are for the PMA1 gene and are described in Fig. 1C. Data are the average and SEM of two independent experiments.
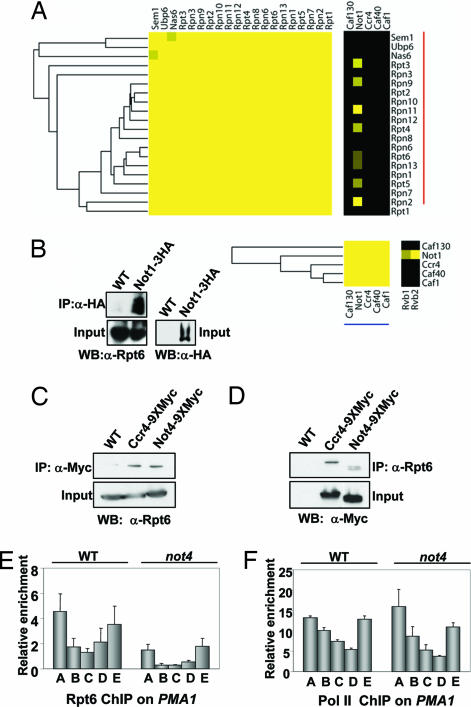
Recently, two large-scale protein–protein interaction maps were generated in Saccharomyces cerevisiae by using a similar affinity-tagging and purification strategy (30, 31). We recently combined these two data sets and, with an algorithm, provided confidence scores for individual interactions (32). Using several independent metrics, we have shown that this recently derived protein–protein interaction data set is of higher quality than those reported previously. In an attempt to generate an accurate portrait of the physical interactome that can be navigated easily, we subjected these data to hierarchical clustering. This clustering analysis accurately recapitulates stable, stoichiometric protein complexes along the diagonal of the clustergram, whereas off-diagonal interactions potentially represent either shared subunits of stable complexes or weaker, possibly transient associations between protein complexes (32) (SI Fig. 7 A and B). We observed one such connection between Not1 and several subunits of the 19S proteasome (Fig. 3A). To confirm this observation, we immunoprecipitated either a 3XHA-tagged version of Not1 or a 9XMyc-tagged version of Ccr4 or Not4 and immunoblotted the precipitates for the presence of the 19S subunit Rpt6 (Fig. 3 B and C). Reciprocal coimmunoprecipitations using an α-Rpt6 antibody pulled down both the Ccr4 and Not4 subunits (Fig. 3D). These experiments confirmed the association of the 19S particle with multiple members of the CCR4/NOT complex. Consistent with a recent report, this proteomic analysis also identified interactions between Not1 and two DNA helicases, Rvb1 and Rvb2, which are known to exist in multiple chromatin remodeling and histone deposition complexes (30, 33). We tested the potential involvement of Rvb1 and Rvb2 in regulating H3K4me but found that strains harboring DamP alleles (decreased alleles by mRNA perturbation) (34) of these genes did not affect any H3K4me state (data not shown).
Fig. 3.
The CCR4/NOT complex interacts with the proteasome and regulates its recruitment to genes. (A) Summary of off-diagonal connections between the 19S proteasome and the NotI component of the CCR4/NOT complex detected from hierarchical clustering of multiple protein interaction data sets (see SI Fig. 7 A and B). Not1 also shares off-diagonal connections with two helicases, Rvb1 and Rvb2, involved in various aspects of chromatin remodeling. The intensity of the yellow corresponds to the confidence of the protein–protein interaction, and black signifies no detected interaction (32). (B) IP of Not1 coprecipitates the 19S proteasome subunit, Rpt6. One milligram of WCE from wild-type (W303) and NotI-3XHA-tagged strains (YNL047) were used in IP experiments that included 3 μl of the appropriate antibody. IP samples and 2% of starting material (Input) were resolved by either SDS/8% PAGE (for Not1-3XHA detection) or SDS/10% PAGE (for Rpt6 detection), and samples were processed for immunoblot analysis by using either anti-Rpt6 or anti-HA antibodies. (C) IP of Ccr4 or Not4 coprecipitates the 19S proteasome. WCEs (1 mg) from wild-type (BY4741), Ccr4–9XMyc (YNL038), and Not4–9XMyc (YNL039) were immunoprecipitated with anti-Myc antibody, resolved by SDS/10% PAGE, transferred to PVDF, and immunoblotted with antibodies to Rpt6. For the input samples, 5% (50 mg) of WCEs from each sample was examined. (D) IP of the 19S coimmunoprecipitates the CCR4/NOT complex. Experiment was performed as described in C except that anti-Rpt6 antibodies were used for the IPs and anti-Myc antibody was used to detect either Ccr4 or Not4. (E and F) 19S proteasome recruitment to the PMA1 gene is disrupted in the not4Δ strain. Anti-Rpt6 (E) or anti-pol II (F) antibodies were used in ChIP analysis. Primers are as described in Fig. 1D. Samples are the average and SEM of three independent experiments.
To address whether CCR4/NOT affects proteasome function, we used antisera against the 19S subunit Rpt6 in ChIP to examine the localization pattern of the 19S particle on PMA1. In wild-type cells, the 19S showed enrichment at both the 5′ and 3′ ends of the PMA1 gene (Fig. 3E). Compared with the wild-type strain, deletion of NOT4 had reduced, but not abolished, recruitment of the 19S across the entire length of the gene (Fig. 3E). The 19S proteasome regulates RNA polymerase II (Pol II) transcription elongation and termination (22, 35). Because CCR4/NOT also interacts with Pol II, we tested whether the loss of Not4 affected the relative amounts and distribution of Pol II on the PMA1 gene as a way to explain the decreased Rpt6 chromatin association. No differences in Pol II levels were detected between the wild-type and not4Δ strains on PMA1 (Fig. 3F). Consistent with a lack of effect on Pol II recruitment and or distribution, neither not4Δ nor ccr4Δ affected PMA1 mRNA levels (SI Fig. 5B). This effect on 19S recruitment was not because of lower expression of Rpt6 or other 19S members in not4Δ cells as the protein levels for these factors are equivalent to wild-type cells (SI Fig. 6B). Although these results reveal 19S recruitment to be defective on PMA1, we note that another gene examined (ADH1) did not show a significant reduction in 19S association (data not shown). These data suggest that Not4 may affect recruitment of the 19S particle to a subset of Pol II-transcribed loci.
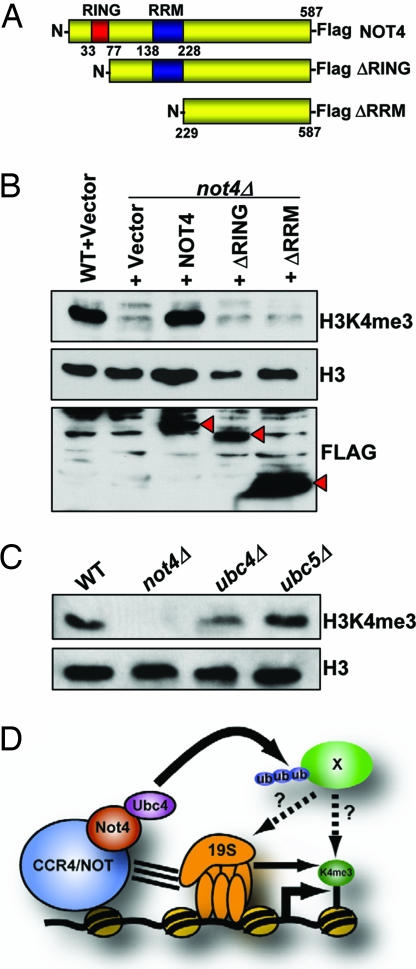
To further define the mechanism of CCR4/NOT proteasome regulation of H3K4me3, we sought to determine which domain of Not4 was responsible for regulating this modification. Not4 contains both a RING and a RRM domain, and although the RRM domain has significant homology to domains in other proteins known to bind RNA, its function in Not4 biology is unknown (36). RING domains are known to mediate protein–protein interactions, and some can act as E3 ubiquitin ligases (37). A previous study has shown that Not4 can mediate ubiquitin transfer to substrates in an in vitro ubiquitin conjugation reaction, but the only known substrates in vivo are members of the nascent polypeptide-associated complex (NAC), none of which affect H3K4me3 (M. Collart, personal communication) (38, 39). We made a series of N-terminal truncation mutants lacking either the RING domain or both the RING and RRM domains and transformed these constructs, or a full-length NOT4 construct, individually into a not4Δ strain (Fig. 4A). As shown in Fig. 4B, exogenous expression of full-length NOT4 fully restored the H3K4me3 defect seen in not4Δ cells. Interestingly, loss of the RING domain, or the RING and RRM domains, failed to rescue H3K4me3. The inability to rescue H3K4me3 by the deletion mutants, however, was not attributable to inadequate protein expression because each mutant was expressed to comparable levels as the full-length construct (Fig. 4B). These data reveal that the RING domain of Not4 is essential for establishing wild-type levels of H3K4me3.
Fig. 4.
A Not4 ubiquitin-dependent pathway regulates H3K4me3. (A) Schematic of full-length and N-terminal truncation constructs of Not4 used in this study. (B) The RING domain of Not4 regulates H3K4me3. Wild-type (9XMyc-Set1) cells were transformed with an empty ADH1 expression vector. not4Δ (YNL031) cells were transformed with either empty vector or the Not4 expression constructs described in A. Cells were grown to mid-log phase in Sc-Ura media and harvested, and WCEs were prepared, normalized to total H3, and analyzed by SDS/15% PAGE. Red arrowheads indicate the occurrence of the Not4 Flag-tagged proteins. (C) Loss of the E2 ubiquitin-conjugating enzyme, Ubc4, decreases H3K4me3. Wild-type (9X-Myc-Set1), not4Δ (YNL031), ubc4Δ (YNL040), and ubc5Δ (YNL041) strains were grown to mid-log phase, processed into WCEs, and analyzed as described in B. (D) Hypothetical model depicting how CCR4/NOT controls H3K4me3. In this model, CCR4/NOT regulates H3K4me3 by recruiting Ubc4 to ubiquitylate an as-yet-unidentified substrate (X) that directly affects 19S proteasome function/recruitment to genes (see Fig. 3E). Although our studies suggest a role for the proteasome in H3K4me3 regulation by Not4, we emphasize that this regulation will likely include other mechanisms in addition to the proteasome (see Discussion).
The requirement of the RING domain in Not4 for regulation of H3K4me3 suggested that Not4-mediated ubiquitin transfer to one or more substrates is critical in this process. To examine this idea further, we again analyzed the recently generated E-MAP data to identify links between Not4 or proteasome members and any known ubiquitin-conjugating E2 enzymes. Because E-MAP analysis is quantitative, one can detect both negative (synthetic sick/lethal) interactions and positive ones (where the double mutant grows better than is expected from growth of the two single mutants) (see SI Methods). We previously showed that these latter interactions can identify cases in which genes are functioning in the same pathway in vivo (40, 47). Interestingly, our E-MAP analysis identified Ubc4, an E2 ubiquitin ligase, as having either positive or negative genetic interactions with proteasomal subunits (RPN10, RPN6, and UBP6) or chaperones (DOA1 and UMP1) (data not shown), suggesting a strong functional link between Ubc4 and the proteasome. These data are consistent with two previously published studies showing that Ubc4 and Not4 copurify and can interact in the yeast two-hybrid system (31, 38). We next tested whether Ubc4 and a related E2 enzyme, Ubc5, regulate H3K4me3. Comparing single deletions of these two enzymes relative to the wild-type strain showed that only ubc4Δ had reduced levels of H3K4me3 (Fig. 4C). We attempted to create a ubc4Δubc5Δ deletion in this background but found that it was synthetically lethal because the double mutant was not able to lose a UBC5 expression plasmid in a plasmid-shuffle assay (data not shown). These results suggest that the ubiquitin-conjugating activity of Not4, perhaps in partnership with Ubc4, regulates H3K4me3 (Fig. 4D).
Discussion
How the 19S proteasome regulates histone methylation currently is not understood. To further define how this complex functions, we used genetic data from an E-MAP that focused on chromosome function and a recently generated physical interaction data set to identify a physical and genetic connection among the CCR4/NOT complex, COMPASS, and the proteasome (41). We found CCR4/NOT specifically regulates H3K4me3 in a fashion that does not alter the integrity of the COMPASS complex because this complex remains intact upon tandem affinity purification (TAP) (see Fig. 1C and SI Fig. 5). By testing individual deletion mutants of CCR4/NOT, we show that the E3 ubiquitin ligase Not4 was the subunit of the complex critical for establishing H3K4me3. Although we do not rule out a role for other CCR4/NOT subunits in the regulation of H3K4 methylation (i.e., Ccr4 and Caf1), our studies identify Not4 as a key regulator of this modification in the CCR4/NOT complex. Similar to what previously has been described for the proteasome, we demonstrated that the CCR4/NOT complex does not affect ubH2B (23, 24). These data suggest that CCR4/NOT and the proteasome are connected to the regulation of H3K4me (see Fig. 4D). Because the RING domain of Not4 and the E2 ubiquitin-conjugating enzyme Ubc4 are required for H3K4me3, they imply a role for Not4 in conjugating ubiquitin to an undefined substrate that regulates this modification, perhaps by altering proteasome localization or function (Fig. 4D).
Although CCR4/NOT is known to interact both genetically and physically with other multiprotein complexes, such as Mediator, SAGA, and Pol II, this complex never has been functionally connected to the proteasome or to chromatin regulation (26, 36). In support of a role for CCR4/NOT in regulating 19S proteasome, we found that loss of Not4 reduced overall levels of 19S proteasome on the PMA1 gene. However, reduced proteasome recruitment cannot be the sole explanation for the decreased H3K4me3 because another test gene examined (ADH1) showed no appreciable reduction in 19S recruitment. Furthermore, H3K79 methylation is unaffected in the not4Δ cells, suggesting that proteasome localization to chromatin cannot be disrupted globally. We speculate that ubiquitin transfer by Not4 partially may regulate 19S chaperone function and/or it may ubiquitylate a factor that is required for full 19S chromatin association and subsequent control of the H3K4me3 activity of COMPASS (see Fig. 4D). Although the precise mechanism of Not4 regulation of H3K4me3 is not known, understanding how the CCR4/NOT complex regulates this mark will depend on finding additional Not4 substrates.
In summary, we have discovered a functional interaction between the CCR4/NOT complex and the proteasome that appears critical for the selective establishment of histone H3K4me3. The data presented directly link CCR4/NOT to both the proteasome and to chromatin regulation. These studies open the way for investigating other possible functions of the CCR4/NOT complex (and potentially other complexes involved in mRNA function) in chromatin biology.
Materials and Methods
Yeast Strains and Cloning.
Yeast strains and their genotypes are listed in SI Table 1. Strains unique to this study that are ccr4Δ or not4Δ were made by amplifying the KanMX cassette from the respective deletion strain (obtained from Open Biosystems, Huntsville, AL) and then by using this integration cassette in a high-efficiency transformation as described in ref. 42. Epitope tagging of factors was performed by using plasmids and techniques as described in ref. 43. De novo deletions of UBC4 and UBC5 were generated by using primers containing gene-specific sequences, along with sequences specific for amplifying a KanMX2 cassette.
The full-length NOT4 ORF and truncation derivatives were cloned by using the restriction sites XbaI and EcoRI as C-terminal mono-Flag fusions into plasmid pN827, which contains an ADH1 promoter driving expression of the inserted sequence (44).
WCE Preparation, Coimmunoprecipitation, and Immunoblot Analysis.
For analysis of H3K4me3 and H3 protein levels (Figs. 1 B and C and 4C), yeast WCEs were prepared as described in ref. 25. After normalizing samples to total H3 content, samples were fractionated by SDS/15% PAGE, transferred to PVDF, and analyzed by immunoblotting. Anti-mono-, di-, and trimethyl H3K4 antibodies, along with the anti-H3 antibodies, were obtained from Upstate Biotechnology (Charlottesville, VA). Anti-trimethyl H3K36 and anti-trimethyl H3K79 were from Abcam (Cambridge, MA).
WCEs were prepared for coimmunoprecipitation analysis by growing cells to mid-log phase and then lysing the cells in IP buffer (10 mM Tris, pH 8.0/150 mM NaCl/0.1% Nonidet P-40/10% glycerol) containing protease and phosphatase inhibitors and 1 mM DTT. IPs were performed in IP buffer with a total of 1 mg of WCEs. To these extracts, 3 μl of respective antibody was added and incubated overnight with rotation. To pellet immune complexes, 10 μl of a 50% slurry of Protein A Sepharose (Amersham, Uppsala, Sweden) was added, and samples were rotated for 1 h at 4°C. Samples were washed three times with 500 μl of IP buffer, resuspended in 10 μl of 2× SDS sample buffer, boiled, fractionated by SDS/8% or 10% PAGE, and immunoblotted with the appropriate antibody. Analysis of ubiquitinated H2B was performed as described in ref. 25.
ChIP Analysis.
ChIP assays were performed and quantitated as previously described by using 3 μl of the anti-protein A antibody (Sigma–Aldrich, St. Louis, MO), anti-Rpb1 (sc-25758; Santa Cruz Biotechnology, Santa Cruz, CA), or the anti-Rpt6 (i.e., Sug1) antibody (a gift from Thomas Kodadek, University of Texas Southwestern Medical Center, Dallas, TX) and 1 mg of WCE (25). Multiplex PCR was performed by using primers specific to target genes (i.e., PMA1, FMP27, and ADH1) and also to a region of chromosome V devoid of ORFs (internal control). The histone modification-specific ChIPs were normalized to histone H3 levels.
E-MAP and Protein–Protein Interaction Analysis.
Synthetic genetic array technology was used to generate a high-density, quantitative E-MAP that focused on various aspects of chromosome function (45–47). The systematic creation of double deletion strains in a 768-colony arrayed format was carried out on a set of 743 essential and nonessential genes involved in processes such as transcriptional regulation, DNA repair, DNA replication, and chromosome segregation (45, 47). Images of the plates containing the colonies corresponding to the double mutants were analyzed with recently developed software designed for E-MAP experiments (41). Briefly, quantitative values are generated corresponding to the relative growth of the double mutant strains. E-MAP analysis therefore can identify not only negative interactions (synthetic sick/lethal pairs) but also positive ones in which the double mutant grows no worse or better (suppression) than the sickest single mutant. A protein–protein interaction map for S. cerevisiae was derived (32) from the raw data of two recent large-scale proteomic analyses (30, 31).
Supplementary Material
Acknowledgments
We thank Vincent Geli (Centre National de la Recherche Scientifique, Marseille, France), Mark Hochstrasser (Yale University, New Haven, CT), Stefan Jentsch (Max Planck Institute, Martinsried, Germany), and Thomas Kodadek (University of Texas Southwestern Medical Center, Dallas, TX) for generous gifts of antibody and yeast strains; Martine Collart, Mary Ann Osley, Ali Shilatifard, Zu-Wen Sun, and Marc Timmers for sharing unpublished data; and members of the Briggs, Krogan, and Strahl laboratories for helpful discussions. This study was supported by National Institutes of Health Grants GM68088 (to B.D.S.) and GM74183 (to S.D.B.) and National Institutes of Health Postdoctoral Fellowship Award GM71106-01A1 (to R.N.L.). J.S.W. is an Investigator of the Howard Hughes Medical Institute, B.D.S. is a Pew Scholar in the Biomedical Sciences, and N.J.K. is a Sandler Family Fellow.
Abbreviations
- E-MAP
epistatic miniarray profile
- H3K4me
histone H3 lysine 4 methylation
- H3K4me3
histone H3 lysine 4 trimethylation
- H3K36
histone H3 lysine 36
- H3K79
histone H3 lysine 79
- RRM
RNA recognition motif
- Pol II
RNA polymerase II
- WCE
whole-cell extract.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607996104/DC1.
References
- 1.Lachner M, O'Sullivan RJ, Jenuwein T. J Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 2.de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. Bioessays. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Strahl BD, Allis CD. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 5.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strahl BD, Ohba R, Cook RG, Allis CD. Proc Natl Acad Sci USA. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 10.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 11.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. Proc Natl Acad Sci USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Schlichter A, Cairns BR. EMBO J. 2005;24:1222–1231. doi: 10.1038/sj.emboj.7600607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fingerman IM, Wu CL, Wilson BD, Briggs SD. J Biol Chem. 2005;280:28761–28765. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun ZW, Allis CD. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 16.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 17.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 18.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 20.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- 22.Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston SA. Mol Cell. 2001;7:981–991. doi: 10.1016/s1097-2765(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 23.Ezhkova E, Tansey WP. Mol Cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 24.Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. Cell. 2005;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 25.Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. Curr Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 26.Collart MA, Timmers HT. Prog Nucleic Acid Res Mol Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 27.Denis CL, Chen J. Prog Nucleic Acid Res Mol Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 28.Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. Mol Cell Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 31.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 32.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Mol Cell Proteomics. 2007 Jan 2; doi: 10.1074/mcp.M600381-MCP200. www.mcponline.org/cgi/reprint/M600381-MCP200v1. [DOI] [PubMed]
- 33.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 34.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 35.Gillette TG, Gonzalez F, Delahodde A, Johnston SA, Kodadek T. Proc Natl Acad Sci USA. 2004;101:5904–5909. doi: 10.1073/pnas.0305411101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collart MA. Gene. 2003;313:1–16. doi: 10.1016/s0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- 37.Pickart CM. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 38.Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, Boelens R, Timmers HT. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panasenko O, Landrieux E, Feuermann M, Finka A, Paquet N, Collart MA. J Biol Chem. 2006;281:31389–31398. doi: 10.1074/jbc.M604986200. [DOI] [PubMed] [Google Scholar]
- 40.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 41.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 44.Mumberg D, Muller R, Funk M. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 45.Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Methods. 2006;40:344–352. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 46.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 47.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Nature. 2007 doi: 10.1038/nature05649. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.