Abstract
A sequence-specific ribozyme (M1GS RNA) derived from the catalytic RNA subunit of RNase P from Escherichia coli was used to target the overlapping exon 3 region of the mRNAs encoding the major transcription regulatory proteins IE1 and IE2 of human cytomegalovirus. A reduction of more than 80% in the expression levels of IE1 and IE2 and a reduction of about 150-fold in viral growth were observed in human cells that stably expressed the ribozyme. In contrast, a reduction of less than 10% in the IE1/IE2 expression and viral growth was observed in cells that either did not express the ribozyme or produced a “disabled” ribozyme that carried mutations that abolished its catalytic activity. Examination of the expression of several other viral early and late genes in the cells that expressed the M1GS ribozyme further revealed an overall reduction of at least 80% in their expression. These results are consistent with the notion that the antiviral effects in these cells are due to the fact that the ribozyme specifically inhibits the expression of IE1 and IE2 and, consequently, abolishes the expression of viral early and late genes as well as viral growth. Our study is the first, to our knowledge, to use M1GS ribozyme for inhibiting human cytomegalovirus replication and demonstrates the utility of this ribozyme for antiviral applications.
Human cytomegalovirus (HCMV) is a ubiquitous herpesvirus that causes mild or subclinical diseases in immunocompetent adults but may lead to severe morbidity or mortality in neonates and immunocompromised individuals (1). Infection by this virus accounts for one of the most common opportunistic diseases in patients with AIDS, CMV retinitis. The emergence of drug-resistant strains of HCMV has posed a need for the development of new drugs and novel treatment strategies (2, 3).
Antisense nucleic acid molecules, including conventional antisense oligonucleotides and antisense ribozymes, are promising gene-targeting agents for specific inhibition of gene expression (4–8). Antisense molecules have been used as anti-HCMV agents to inhibit the expression of HCMV-essential genes and abolish viral replication (4, 9–12). External guide sequences (EGSs; refs. 13 and 14) are antisense oligoribonucleotides that have been used in conjunction with either ribonuclease P (RNase P) or the catalytic RNA subunit of RNase P from Escherichia coli (M1 RNA; ref. 15) to diminish the expression of several genes both in E. coli (16, 17) and in mammalian cells (14, 18–21). The EGS-based technology takes advantage of RNase P or M1 RNA to cleave a targeted mRNA when the EGS hybridizes to the target RNA (refs. 13 and 14; Fig. 1A). The EGSs, when expressed separately from the enzyme, have been shown recently to be effective in inhibiting the gene expression of herpes simplex virus 1 (HSV-1) and influenza virus and, in addition, in abolishing the replication of influenza virus (20, 21). To increase the targeting efficiency, the EGS can be covalently linked to M1 RNA (i.e., its 3′ end) to generate a sequence-specific ribozyme, M1GS RNA (refs. 19, 22; Fig. 1A). We have shown that M1GS RNA efficiently cleaves mRNA substrates that base pair with the EGS in vitro and effectively inhibits the expression of HSV-1 thymidine kinase (TK) in mammalian cells (19, 23). Thus, M1GS RNA represents a distinct class of ribozymes that can be used for gene-targeting applications.
Figure 1.
(A) Schematic representation of a small model substrate (EGS:mRNA) for M1 RNA and a M1GS RNA construct to which a target mRNA has hybridized. (B) Schematic representation of the substrates used in the study. The targeted sequences that bind to the guide sequences of the ribozymes are highlighted. The sites of cleavage by M1 and M1GS RNAs are marked with filled arrows.
Further studies are necessary to develop M1GS ribozyme as a gene-targeting agent and to increase its targeting efficacy. Recently, we have studied in detail how the ribozyme interacts with an mRNA substrate (23–25). In the present study, we provide direct evidence that M1GS ribozyme can function as an antiviral agent and effectively inhibit HCMV replication and gene expression. The target for the ribozyme is the overlapping region of the mRNAs encoding the HCMV immediately early (IE) proteins IE1 and IE2, which are the viral major transcriptional activators responsible for activation of viral gene expression (1). We show that M1GS ribozyme cleaved the target mRNA sequence efficiently in vitro. Moreover, a reduction of 80% in viral IE1/IE2 expression and a reduction of 150-fold in viral growth were observed in cells that expressed the ribozyme. Our study shows the utility of M1GS RNAs as a class of gene-targeting ribozymes for antiviral applications.
Materials and Methods
Viruses, Cells, and Antibodies.
HCMV (strain AD169) was propagated in human foreskin fibroblasts and astrocytoma U373MG cells in DMEM supplemented with 10% (vol/vol) FBS as described (26). The monoclonal antibodies c1201, c1202, and c1203, which react with HCMV proteins gB, UL44, and IE1/IE2, respectively, were purchased from Goodwin Institute for Cancer Research (Plantation, FL). The monoclonal antibodies against gH and human actin were purchased from BioDesign (Kennebunk, ME) and Sigma, respectively.
Ribozyme and Substrate Constructs.
Plasmids pFL117 and pC102 contain the DNA sequence coding for M1 RNA and mutant C102 driven by the T7 RNA polymerase promoter (19, 27). Mutant ribozyme C102 contained several point mutations (e.g., A347C348 → C347U348 and C353C354C355G356 → G353G354A355U356) at the catalytic domain (P4 helix) of the M1 RNA sequence (27). The DNA sequences that encode ribozymes M1-IE and C-IE were constructed by PCR with pFL117 and pC102 as the templates, respectively. The 5′ PCR primer was OliT7 (5′-TAATACGACTCACTATAG-3′), and the 3′ primer was M1IE12 (5′-TGGTGCAACGAGAACCCTGTGGAATTG-3′). The DNA template for transcription in vitro of RNA substrate ie37 was constructed by annealing the T7 promoter-containing oligonucleotide OliT7 with oligonucleotide sIE1 (5′-CGGGATCCTTTCTCGGGGTTCTCGTTGCGATTCCCGGTCCTATAGTGAGTCGTATTA-3′). The DNA template for substrate ie450 was constructed by PCR with an IE2 cDNA clone as the template (28, 29) and 5′ primer ie4505 (5′-TAATACGACTCACTATAGGGGCCCTTCCTCCAAGGTGG-3′) and 3′ primer ie4503 (5′-GATGTCTTCCTGTTTGATGG-3′).
In Vitro Cleavage and Binding of the IE1/IE2 mRNA Substrates by the Ribozymes.
M1GS RNAs and RNA substrates ie37 and ie450 were synthesized in vitro by T7 RNA polymerase and purified further on 8% urea/polyacrylamide gels, as described (19, 24). The M1GS RNAs (10 nM) were incubated with the 32P-labeled mRNA substrate (10 nM) at 37°C in a volume of 10 μl for 25 min in buffer A (50 mM Tris, pH 7.5/100 mM NH4Cl/100 mM MgCl2; refs. 19 and 23). Cleavage products were separated in denaturing gels and quantitated with a STORM840 PhosphorImager (Molecular Dynamics).
The procedures to measure the equilibrium dissociation constants (Kd) of the M1GS-ie37 complexes were modified from Pyle et al. (30). In brief, various concentrations of ribozyme (0.05–50 nM) were preincubated in buffer B [50 mM Tris, pH 7.5/100 mM NH4Cl/100 mM CaCl2/3% (wt/vol) glycerol/0.1% xylene cyanol/0.1% bromophenol blue] for 10 min before mixing with an equal volume of 0.1–0.5 nM RNA substrate preheated under identical conditions. The samples were incubated for 15 min to allow binding, then loaded on a 5% polyacrylamide gel, and run at 10 W with the running buffer (100 mM Tris-Hepes, pH 7.5/10 mM MgCl2; ref. 30). The amount of the bound complex was quantitated with a STORM840 PhosphorImager. The value of Kd was then extrapolated from a graph plotting percentage of product bound versus ribozyme concentration. The values obtained were averages of three experiments.
Construction of the Ribozyme-Expressing Cell Lines.
The DNA sequences coding for the ribozymes were subcloned into retroviral vector LXSN and placed under the control of the U6 RNA promoter (19, 31). The retroviral vector DNAs that contained the ribozyme sequence were transfected into amphotropic PA317 cells by using a mammalian transfection kit purchased from GIBCO/BRL. At 48 h after transfection, culture supernatants that contained retroviruses were collected and used to infect human U373MG cells. At 48–72 h after infection, neomycin (GIBCO/BRL) was added to the culture medium at a final concentration of 600 μg/ml. Cells were selected subsequently in the presence of neomycin for 2 weeks, and neomycin-resistant cells were cloned.
Viral Infection and Preparation of RNA and Protein Extracts.
Cells (n = 1 × 106) were either mock-infected or infected with HCMV at a multiplicity of infection (moi) of 0.05–1 in an inoculum of 1.5 ml of DMEM supplemented with 1% FCS. The inoculum was replaced with DMEM supplemented with 10% (vol/vol) FBS after a 2-h incubation with cells. The infected cells were incubated for a certain period (as stated in the Results section) before harvesting for viral mRNA or protein isolation. To measure the levels of viral immediate-early (IE) transcripts, some of the cells were also treated with 100 μg/ml cycloheximide before and during infection. Total cellular RNA and protein samples were prepared from the cells as described (19).
Northern and Western Analyses to Detect the Expression of M1GS RNAs, Viral mRNAs, and Proteins.
The RNA fractions were separated in 0.8–2.5% agarose gels that contained formaldehyde, transferred to a nitrocellulose membrane, hybridized with the 32P-radiolabeled DNA probes that contained the HCMV or human β-actin DNA sequences, and analyzed with a STORM840 PhosphorImager. The radiolabeled DNA probes used to detect M1GS RNAs, human β-actin mRNA, HCMV IE 5-kilobase (kb) RNA transcript, IE1 mRNA, IE2 mRNA, and US2 mRNA were synthesized from plasmids pFL117, pβ-actin RNA, pCig27, pIE1, pIE2, and pCig38, respectively, by using a random primed labeling kit (Roche Molecular Biochemicals; refs. 19, 28, and 29).
The polypeptides from cell lysates were separated on either SDS/7.5% polyacrylamide gels or SDS/9% polyacrylamide gels cross-linked with N,N′′-methylenebisacylamide, transferred electrically to nitrocellulose membranes, and reacted to the antibodies against HCMV proteins and human actin. The proteins on the membranes were stained subsequently by using a Western chemiluminescent substrate kit (Amersham Pharmacia) and quantitated with a STORM840 PhosphorImager. Quantitation was performed in the linear range of RNA and protein detection.
Assays of the Level of the Inhibition of Viral Replication.
Cells (n = 5 × 105) were infected with HCMV at an moi of 0.5–2. The cells and medium were harvested at 1-day intervals throughout the 7 days after infection, and viral stocks were prepared by adding an equal volume of 10% (vol/vol) skim milk, followed by sonication. The titers of the viral stocks were determined by infecting 1 × 105 human foreskin fibroblasts and counting the number of plaques 10–14 days after infection. The values obtained were averages from triplicate experiments.
Results
Construction of M1GS Ribozymes and in Vitro Studies of Their Targeting Activity.
Previous studies have shown that IE1 and IE2, expressed predominantly as IE (α) gene products during infection, function as the major transcriptional activators and play a key role in regulating the expression of viral early (β) and late (γ) genes (1). It has been shown that antisense oligonucleotides targeting IE2 mRNA abolish viral replication (9, 10). Meanwhile, a HCMV mutant with a deletion in the exon 4 of IE1 failed to express viral early and late genes (e.g., UL44) and to replicate in human cells at a low moi (32). IE1 and IE2 share 85 amino-terminal amino acids because of alternative splicing and polyadenylylation of transcripts initiating at a strong promoter enhancer (1, 33). Therefore, targeting the overlapping region of the mRNAs (i.e., exon 3) coding for IE1 and IE2 should simultaneously shut down the expression of both proteins and may yield a more effective inhibition of viral replication.
Because most mRNA species inside cells are usually associated with proteins and are present in a highly organized and folded conformation, it is critical to choose a targeted region that is accessible to binding of M1GS ribozymes to achieve efficient targeting. In vivo mapping with dimethyl sulfate has been used extensively to determine the accessibility of mRNA and structure of RNAs in cells (19, 34). The sequences in the overlapping regions of the IE1 and IE2 mRNAs that were accessible to dimethyl sulfate modification were mapped by this method. A position 39 nucleotides upstream from the 3′ end of exon 3 was chosen as the cleavage site for M1GS ribozyme. This site seemed to be one of the sequences that are most accessible to dimethyl sulfate modification (data not shown). This region is also highly conserved among different HCMV strains (e.g., AD169 and Towne; ref. 1). Moreover, its flanking sequence has features that need to be present to interact with a M1GS ribozyme to achieve optimal cleavage. These features include having a guanosine and a pyrimidine as the nucleotide 3′ and 5′ adjacent to the site of cleavage, respectively (35).
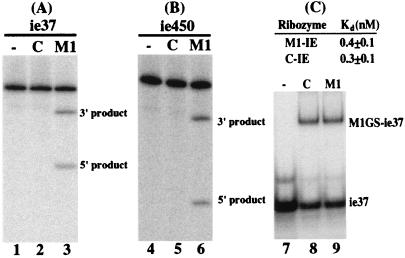
Two M1GS ribozymes, M1-IE and C-IE RNAs, were constructed by covalently linking the 3′ termini of M1 and C102 RNAs to a guide sequence of 13 nucleotides that is complementary to the targeted mRNA sequence. C102 RNA, a mutant derived from M1 RNA, contains several point mutations at the P4 catalytic domain and is catalytically inactive (23, 27). RNA substrates ie37 and ie450 contain the targeted HCMV mRNA sequence of 37 and 450 nucleotides, respectively (Fig. 1B). Efficient cleavage of these two substrates by M1-IE was observed and yielded products of 12 and 25 nucleotides and 150 and 300 nucleotides, respectively (Figs. 1B and 2, lanes 3 and 6). In contrast, cleavage of the same substrates by C-IE was barely detected (Fig. 2, lanes 2 and 5).
Figure 2.
Cleavage of ie37 (A) and ie450 (B) and binding of ie37 by M1GS ribozymes (C). (A and B) No ribozymes were added to the reaction mixture in lanes 1 and 4; 10 nM of M1-IE (lanes 3 and 6) and C-IE (lanes 2 and 5) were incubated with 32P-labeled IE RNA substrate (10 nM) at 37°C in a volume of 10 μl for 25 min in buffer A. Cleavage products were separated on 15% (A) and 4% (B) polyacrylamide gels that contained 8 M urea. (C) Substrate ie37 at a concentration of 0.1 nM was incubated either alone (lane 7) or in the presence of 0.2 nM ribozyme C-IE (lane 8) and M1-IE (lane 9) in buffer B for 15 min to allow binding and then loaded on a 5% polyacrylamide gel.
To determine further whether the differential cleavage efficiencies observed with M1-IE and C-IE RNAs are possibly due to their different binding affinities to the mRNA sequence, the affinities of the ribozymes to substrate ie37 were determined by a gel-shift binding assay. Similar amounts of the complexes formed by ribozymes and substrate ie37 were observed when the same amounts of M1GS RNAs were used (Fig. 2C, lanes 8 and 9). Indeed, the binding affinity of M1-IE RNA to substrate ie37, measured as the value of dissociation constant (Kd), was similar to that to C-IE RNA (Fig. 2C). However, a very little amount of cleavage product was observed even in the presence of high concentrations of C-IE RNA and a prolonged incubation period (Fig. 2, lanes 2 and 5 and data not shown). These observations suggested that the mutations in C-IE RNA do not significantly affect the binding affinity of the ribozyme to the mRNA sequence but abolish its catalytic activity. Thus, C-IE RNA may be used as a control for the antisense effect in our experiments in cultured cells.
Expression of M1GS RNA in Human Cell Culture.
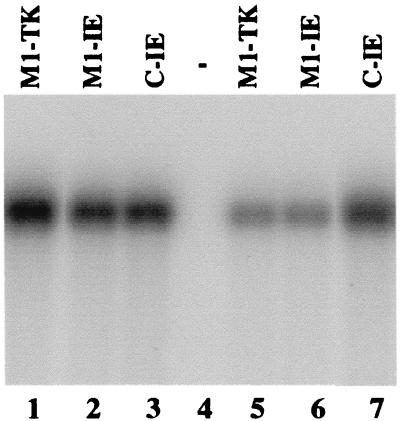
The DNA sequences coding for M1-IE and C-IE ribozymes were subcloned into retroviral vector LXSN and placed under the control of the small nuclear U6 RNA promoter, which has been shown to express M1GS RNA and other RNAs steadily (14, 19, 31, 36). Human U373MG cell lines that contained these retroviral DNA sequences and expressed the ribozymes were constructed and cloned (see Materials and Methods). An additional cell line was also constructed that expressed a ribozyme, M1-TK, that targeted the HSV-1 TK mRNA (19). No cleavage of substrate ie37 and ie450 by M1-TK was observed in vitro (data not shown). This cell line was used to determine whether M1GS RNA with an incorrect guide sequence can target the IE1/IE2 mRNA in tissue culture. The level of M1GS RNA expression in each individual cell clone was determined by Northern analysis with a DNA probe that is complementary to M1 RNA. Fig. 3 shows the result from two sets of cloned cell lines that expressed M1-TK (lanes 1 and 5), M1-IE (lanes 2 and 6), or C-IE (lanes 3 and 7). The different levels of M1GS RNA among each individual clone (e.g., Fig. 3, compare lane 1 and 5) are presumably due to the incorporation of the LXSN-M1GS sequence into different locations of the host chromosome, and its expression is influenced by the flanking sequence at the insertion site. Only the cell lines that expressed similar levels of these ribozymes were used for further studies in tissue culture.
Figure 3.
The expression of M1GS ribozymes in human cells. Northern analyses were carried out with RNA isolated from parental U373MG cells (−, lane 4) and two sets of cloned cell lines that expressed M1-TK (lanes 1 and 5), M1-IE (lanes 2 and 6), and C-IE (lanes 3 and 7) ribozymes. Equal amounts of each RNA sample (30 μg) were separated on 2.5% agarose gels that contained formaldehyde, transferred to a nitrocellulose membrane, and hybridized to a 32P-radiolabeled probe that contained the DNA sequence coding for M1 RNA. The hybridized products corresponding to the full-length retroviral transcripts (≈6 kb), transcribed from the long terminal repeat promoter, are at the top of the gel and are not shown.
Inhibition of HCMV IE1/IE2 Expression in M1GS-Expressing Cells.
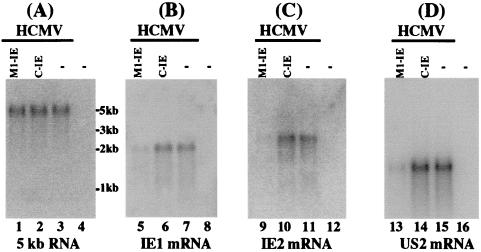
Cells were infected with HCMV at an moi of 0.05–1. Total RNAs were isolated from cells that were pretreated with 100 μg/ml of cycloheximide and then were either mock-infected or infected with HCMV. Under this condition, only viral IE mRNAs were synthesized (1). The levels of IE1/IE2 mRNAs in the infected cells were determined by Northern analyses. The levels of the 5-kb-long viral IE transcript (5-kb RNA), the expression of which is not regulated by IE1/IE2 under the assay conditions (29), were used as internal controls for the quantitation of expression of IE1 and IE2 mRNAs. Fig. 4 shows the results (which are summarized in Table 1) of the Northern analysis experiments with the IE1 (Fig. 4B), IE2 (Fig. 4C), and 5-kb RNA (Fig. 4A) probes. A reduction of about 85% (average of three experiments) in the levels of IE1 and IE2 mRNAs was observed in cells that expressed M1-IE, whereas cells that expressed C-IE only had a reduction of less than 10%. The low level of inhibition found in cells that expressed C-IE RNA was probably due to an antisense effect, because C-IE RNA contained the same guide sequence as M1-IE but did not have catalytic activity.
Figure 4.
Levels of HCMV mRNAs as determined by Northern analysis. Cells (n = 1 × 106) were either mock-infected (lanes 4, 8, 12, and 16) or infected with HCMV (moi = 1) (lanes 1–3, 5–7, 9–11, and 13–15) and were harvested at either 8 (A–C) or 24 h (D) after infection. Viral infection was carried out in either the absence (D) or the presence (A–C) of 100 μg/ml cycloheximide. Northern analyses were carried out with RNA isolated from parental U373MG cells (−, lanes 3, 4, 7, 8, 11, 12, 15, and 16) and cell lines that expressed M1-IE (lanes 1, 5, 9, and 13) and C-IE (lanes 2, 6, 10, and 14). Equal amounts of each RNA sample (30 μg) were separated on agarose gels that contained formaldehyde, transferred to a nitrocellulose membrane, and hybridized to a 32P-radiolabeled probe that contained the cDNA sequence of the HCMV 5-kb transcript (lanes 1–4), IE1 mRNA (lanes 5–8), IE2 mRNA (lanes 9–12), and US2 mRNA (lanes 13–16). The hybridized products corresponding to the 5-kb RNA, IE1, IE2, and US2 mRNAs were about 5, 1.9, 2.2, and 1.5 kb, respectively (1, 28, 29, 33).
Table 1.
Levels of inhibition of the mRNA and protein expression of different viral genes in cells that expressed M1-TK, C-IE, or M1-IE, as compared to the levels of inhibition of cells that did not express a ribozyme (U373-MG)
| HCMV gene | Viral gene class | Level of inhibition, %
|
|||
|---|---|---|---|---|---|
| U373-MG | M1-TK | C-IE | M1-IE | ||
| IE1 mRNA | α | 0 | 1 | 8 | 85 ± 8 |
| IE2 mRNA | α | 0 | 2 | 7 | 85 ± 8 |
| US2 mRNA | β | 0 | 1 | 5 | 80 ± 6 |
| IE1/IE2 protein | α | 0 | 2 | 4 | 88 ± 10 |
| UL44 protein | β, γ | 0 | 1 | 3 | 80 ± 5 |
| Glycoprotein B | β, γ | 0 | 0 | 3 | 80 ± 9 |
| Glycoprotein H | γ | 0 | 2 | 1 | 80 ± 10 |
Values shown are the means from triplicate experiments. The values of standard deviation that are less than 5% are not shown.
The levels of IE1 and IE2 proteins in M1GS-expressing cells are expected to reduce because of the decreased level of IE1/IE2 mRNAs. Proteins were isolated from cells at 24 h after infection, separated in SDS/polyacrylamide gels, and transferred to two identical membranes. One membrane was stained with an anti-IE1/IE2 antibody (anti-IE1/IE2; Fig. 5B), and the other was stained with a monoclonal antibody against human actin (anti-actin; Fig. 5A). The latter serves as an internal control for the quantitation of IE1/IE2 protein expression. The results of three independent experiments are summarized in Table 1: a reduction of 88% in the level of IE1 and IE2 proteins was observed in cells that expressed M1-IE, whereas a reduction of less than 10% was found in cells that expressed C-IE or M1-TK.
Figure 5.
Levels of human actin and HCMV proteins as determined by Western blot analysis. Protein samples were isolated from cells that expressed specified ribozymes and either mock-infected (lanes 4, 8, 12, 16, and 20) or infected with HCMV (moi = 0.5–1; lanes 1–3, 5–7, 9–11, 13–15, and 17–19) for 36 h (A and B) or 48 h (C–E), separated in either SDS/9% polyacrylamide gels (A) or SDS/7.5% polyacrylamide gels (B–E), and then transferred to membranes. One membrane was allowed to react with a monoclonal antibody (anti-actin) against human actin (A), whereas the others were stained with monoclonal antibodies anti-IE1/IE2, anti-UL44, anti-gB, and anti-gH against HCMV IE1/IE2, UL44, gB, and gH proteins, respectively (B–E).
Inhibition of Viral Gene Expression and Growth by the Ribozyme-Mediated Reduction of IE1/IE2 Expression.
Inhibition of IE1 and IE2 expression is expected to result in a reduction of the expression of both viral early (β) and late (γ) genes (1). To determine whether this reduced expression is the case, cells were infected with HCMV at an moi of 0.05–1 for 48 h. The level of the US2 mRNA (an early mRNA; Fig. 4D) as well as the protein levels of UL44 (an early protein), gB (an early and late protein), and gH (a late protein) (Fig. 5 C–E) were determined. The level of the HCMV 5-kb transcript and the protein level of human actin were used as the internal controls. A reduction of 80% in the expression levels of these genes was observed in cells that expressed M1-IE RNA, but no significant reduction was detected in cells that expressed C-IE or M1-TK (Figs. 4 and 5; Table 1). These results suggested an overall inhibition of viral early and late gene expression in the M1-IE-expressing cells.
To determine whether viral growth was also inhibited in the ribozyme-expressing cells, cells were infected with HCMV at an moi of 0.5–2. Virus stocks were prepared from the infected cultures at 1-day intervals for the 7 days after infection, and the plaque-forming unit count was determined by measurement of the viral titer in human fibroblasts. After 5 days of infection, a reduction of about 150-fold in viral yield was observed in cells that expressed M1-IE ribozyme, whereas no significant reduction was found in those that expressed C-IE or M1-TK (Fig. 6).
Figure 6.
Growth analysis of HCMV in parental U373MG cells and cell lines that expressed M1-IE and C-IE. Cells (n = 1 × 105) were infected with HCMV at a moi of 1. Virus stocks were prepared from the infected cells at 1-day intervals for the 7 days after infection, and the plaque-forming unit (PFU) count was determined by measurement of the viral titer on human fibroblasts. These values are the means from triplicate experiments. SDs are indicated by the error bars.
Discussion
The M1GS-based technology represents an attractive approach for gene inactivation, because it has most of the properties of the conventional antisense targeting method and, in addition, catalytic and irreversible cleavage of the target RNA. The hammerhead and hairpin ribozymes (6–8) as well as the EGSs (14, 21) have been shown to be promising gene-targeting agents for inhibition of viral gene expression and replication. By tethering the EGS to M1 RNA, one of most efficient catalytic RNAs that are found in nature and hydrolyze RNA, M1GS RNA represents a distinct class of gene-targeting ribozymes and may be more effective than EGSs in diminishing the expression of a target mRNA. Our study is, to our knowledge, the first to use M1GS ribozyme for inhibiting viral replication. Several criteria must be satisfied if successful targeting with ribozymes is to be achieved. Among these are high efficiency of cleavage, sequence specificity of the ribozymes, and efficient delivery of the reagents. We have constructed a M1GS RNA that targets the overlapping region of HCMV IE1 and IE2 mRNAs and shown that the ribozyme cleaves the target mRNA sequence efficiently in vitro. Moreover, a reduction of about 85% in the expression level of IE1 and IE2 and a 150-fold reduction in viral growth were observed in cells that expressed the ribozyme. In contrast, a reduction of less than 10% in the levels of IE1/IE2 expression and viral growth was observed in cells that expressed C-IE or M1-TK. Similar differences in the level of inhibition of IE1/IE2 expression and viral growth between cells that expressed M1-IE and those that expressed C-IE or M1-TK were also observed when different sets of cell lines that expressed these ribozymes were used (data not shown). Because M1-TK targets an unrelated mRNA, these results suggested that the observed inhibitory effect in the M1-IE-expressing cells is not due to the nonspecific effect of the ribozyme sequence. Although C-IE is catalytically inactive, this ribozyme contained the identical guide sequence and had a binding affinity to the mRNA sequence similar to that of M1-IE. Thus, our results suggest that the overall observed inhibition of viral gene expression and growth with M1-IE RNA was primarily due to the targeted cleavage by the ribozyme as opposed to the antisense effect or other nonspecific effects of the guide sequence of the ribozyme.
The activity of RNase P ribozymes seems to be specific. First, the ribozymes did not show significant cytotoxicity, because cells expressing ribozymes are indistinguishable from the parental cells in terms of cell growth and viability for up to 2 months (data not shown). Moreover, the antiviral effect of the ribozyme (inhibition of viral growth) seems to be caused by the reduction of the IE1 and IE2 expression, because the expression of all of the viral early and late genes examined, including US2, UL44, gB, and gH, was found to be reduced significantly in cells that expressed M1-IE but not in those that expressed C-IE or M1-TK. The extent of the observed inhibition of the expression of most of these viral early and late genes correlated with that of the inhibition of the IE1 and IE2 expression. However, no reduction in the levels of other viral IE transcripts (e.g., 5-kb RNA and UL36 mRNA) was found in M1GS-expressing cells (Fig. 4A and data not shown). Thus, M1GS ribozyme is highly specific in inhibiting the expression of its target mRNAs. Furthermore, this notion is consistent with the fact that only the growth of HCMV, but not HSV-1, a related herpesvirus, was inhibited in M1-IE-expressing cells (data not shown).
We showed that the M1GS RNAs introduced into human cells were stably expressed. The expression cassette that produces these M1GSs is driven by the promoter for small nuclear U6 RNA. This promoter has been used to express EGSs and ribozymes in mammalian cells, and the transcript from this promoter is quite stable (14, 19, 36). M1GS ribozymes may also increase their activity in cultured cells by interacting with the cellular proteins, possibly including the protein components of human RNase P (15, 19). Several “RNA chaperone” proteins have been shown to stimulate the activity of hammerhead and group I ribozymes by facilitating either folding of the ribozyme or dissociation of the product (37–39). Further studies on potential interactions between M1GS RNAs and cellular proteins will provide insight into how a M1GS ribozyme functions in cultured cells.
HCMV is a member of the human herpesvirus family, which includes seven other different viruses such as HSV and Epstein–Barr virus (1, 40). All of these viruses can engage in lytic replication as well as establish latent infections. HCMV IE1 and IE2 are among the first viral proteins expressed during lytic infection (1). Their homologous proteins have been found in every other herpesvirus and are among those that are major targets for drug development (9–11). To evaluate the anti-HCMV activity of M1GS RNA further, the ribozymes can be delivered into the monocyte/macrophage-lineage cells where HCMV is believed to establish latent infection (1). These experiments will determine whether the ribozymes can abolish the IE1/IE2 expression in these cells and prevent HCMV reactivation from latent infection into lytic replication. These studies as well as the in vitro studies on how to construct highly active ribozymes will facilitate the development of M1GS ribozymes as gene-targeting agents for antiviral applications.
Acknowledgments
We thank Dr. Edward Mocarski for advice and critical reading of the manuscript. P.T. is partially supported by a Block Grant Predoctoral Fellowship from the Graduate Division of the University of California, Berkeley. F.L. is a Pew Scholar in Biomedical Sciences and a recipient of a Basil O'Connor Award (March of Dimes) and a Regent's Junior Faculty Fellowship (University of California). This research has been supported by University of California AIDS Research Program Grant R98-B-146, March of Dimes National Birth Defects Foundation, and National Institutes of Health Grants AI41927 and GM54875.
Abbreviations
- HCMV
human cytomegalovirus
- EGS
external guide sequence
- TK
thymidine kinase
- moi
multiplicity of infection
- kb
kilobase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100101797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100101797
References
- 1.Mocarski E S. In: Virology. Fields B N, Knipe D M, editors. Vol. 2. New York: Raven; 1996. pp. 2447–2492. [Google Scholar]
- 2.Baldanti F, Simoncini L, Talarico C L, Sarasini A, Biron K K, Gerna G. AIDS. 1998;12:816–818. [PubMed] [Google Scholar]
- 3.Palella F J, Jr, Delaney K M, Moorman A C, Loveless M O, Fuhrer J, Satten G A, Aschman D J, Holmberg S D. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 4.Marvick C. J Am Med Assoc. 1998;280:871. (lett.). [PubMed] [Google Scholar]
- 5.Zamecnik P C, Stephenson M L. Proc Natl Acad Sci USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altman S. Proc Natl Acad Sci USA. 1993;90:10898–10900. doi: 10.1073/pnas.90.23.10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi J J. Chem Biol. 1999;6:R33–R37. doi: 10.1016/S1074-5521(99)80001-5. [DOI] [PubMed] [Google Scholar]
- 8.Poeschla E, Wong-Staal F. Curr Opin Oncol. 1994;6:601–606. doi: 10.1097/00001622-199411000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Anderson K P, Fox M C, Brown-Driver V, Martin M J, Azad R F. Antimicrob Agents Chemother. 1996;40:2004–2011. doi: 10.1128/aac.40.9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azad R F, Driver V B, Tanaka K, Crooke R M, Anderson K P. Antimicrob Agents Chemother. 1993;37:1945–1954. doi: 10.1128/aac.37.9.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulamba G B, Hu A, Azad R F, Anderson K P, Coen D M. Antimicrob Agents Chemother. 1998;42:971–973. doi: 10.1128/aac.42.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pari G S, Field A K, Smith J A. Antimicrob Agents Chemother. 1995;39:1157–1161. doi: 10.1128/aac.39.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forster A C, Altman S. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Y, Hwang E, Altman S. Proc Natl Acad Sci USA. 1992;89:8006–8010. doi: 10.1073/pnas.89.17.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altman S, Kirsebom L A. In: The RNA World. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 351–380. [Google Scholar]
- 16.Guerrier-Takada C, Salavati R, Altman S. Proc Natl Acad Sci USA. 1997;94:8468–8472. doi: 10.1073/pnas.94.16.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerrier-Takada C, Li Y, Altman S. Proc Natl Acad Sci USA. 1995;92:11115–11119. doi: 10.1073/pnas.92.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma M, Benimetskaya L, Lebedeva I, Dignam J, Takle G, Stein C A. Nat Biotechnol. 2000;18:58–61. doi: 10.1038/71924. [DOI] [PubMed] [Google Scholar]
- 19.Liu F, Altman S. Genes Dev. 1995;9:471–480. doi: 10.1101/gad.9.4.471. [DOI] [PubMed] [Google Scholar]
- 20.Kawa D, Wang J, Yuan Y, Liu F. RNA. 1998;4:1397–1406. doi: 10.1017/s1355838298980918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plehn-Dujowich D, Altman S. Proc Natl Acad Sci USA. 1998;95:7327–7332. doi: 10.1073/pnas.95.13.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank D, Harris M, Pace N R. Biochemistry. 1994;33:10800–10808. doi: 10.1021/bi00201a030. [DOI] [PubMed] [Google Scholar]
- 23.Kilani A F, Trang P, Jo S, Hsu A, Kim J, Nepomuceno E, Liou K, Liu F. J Biol Chem. 1999;275:10611–10622. doi: 10.1074/jbc.275.14.10611. [DOI] [PubMed] [Google Scholar]
- 24.Kilani A F, Liu F. RNA. 1999;5:1235–1247. doi: 10.1017/s1355838299990672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trang P, Hsu A W, Liu F. Nucleic Acids Res. 1999;27:4590–4597. doi: 10.1093/nar/27.23.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen D H, Jiang H, Lee M, Liu F, Zhou Z H. Virology. 1999;260:10–16. doi: 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- 27.Kim J J, Kilani A F, Zhan X, Altman S, Liu F. RNA. 1997;3:613–623. [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins D E, Martens C L, Mocarski E S. J Gen Virol. 1994;75:2337–2348. doi: 10.1099/0022-1317-75-9-2337. [DOI] [PubMed] [Google Scholar]
- 29.Zhu H, Cong J P, Shenk T. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pyle A M, McSwiggen J A, Cech T R. Proc Natl Acad Sci USA. 1990;87:8187–8191. doi: 10.1073/pnas.87.21.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller A D, Rosman G J. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 32.Greaves R F, Mocarski E S. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stenberg R M, Witte P R, Stinski M F. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ares M, Igel A H. Genes Dev. 1990;4:2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- 35.Liu F, Altman S. Nucleic Acids Res. 1996;24:2690–2696. doi: 10.1093/nar/24.14.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertrand E, Castanotto D, Zhou C, Carbonnelle C, Lee N S, Good P, Chatterjee S, Grange T, Pictet R, Kohn D, et al. RNA. 1997;3:75–88. [PMC free article] [PubMed] [Google Scholar]
- 37.Coetzee T, Herschlag D, Belfort M. Genes Dev. 1995;8:1575–1588. doi: 10.1101/gad.8.13.1575. [DOI] [PubMed] [Google Scholar]
- 38.Bertrand E L, Rossi J J. EMBO J. 1995;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuchihashi Z, Khosla M, Herschlag D. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- 40.Roizman B, Sears A E. In: Virology. Fields B N, Knipe D M, editors. Vol. 2. New York: Raven; 1996. pp. 2231–2296. [Google Scholar]