Abstract
Cu, Zn superoxide dismutase (SOD1) has been detected within spinal cord mitochondria of mutant SOD1 transgenic mice, a model of familial ALS. The copper chaperone for SOD1 (CCS) provides SOD1 with copper, facilitates the conversion of immature apo-SOD1 to a mature holoform, and influences in yeast the cytosolic/mitochondrial partitioning of SOD1. To determine how CCS affects G93A-SOD1-induced disease, we generated transgenic mice overexpressing CCS and crossed them to G93A-SOD1 or wild-type SOD1 transgenic mice. Both CCS transgenic mice and CCS/wild-type-SOD1 dual transgenic mice are neurologically normal. In contrast, CCS/G93A-SOD1 dual transgenic mice develop accelerated neurological deficits, with a mean survival of 36 days, compared with 242 days for G93A-SOD1 mice. Immuno-EM and subcellular fractionation studies on the spinal cord show that G93A-SOD1 is enriched within mitochondria in the presence of CCS overexpression. Our results indicate that CCS overexpression in G93A-SOD1 mice produces severe mitochondrial pathology and accelerates disease course.
Keywords: amyotrophic lateral sclerosis, motor neuron, neurodegeneration, transgenic, aggregation
There is increasing evidence that SOD1 localization and aggregation within mitochondria may be important for mutant SOD1-induced cellular toxicity (1). Marked mitochondrial pathology characterized by vacuolization was a principal finding in G37R and high-expressing G93A-SOD1 transgenic mouse lines (2, 3). Certain aspects of mitochondrial function, including electron transport chain activity and ATP synthesis rate, are impaired in G93A-SOD1 mice (4, 5). Importantly, both wild-type (WT) and mutant SOD1 proteins have been detected within mitochondria, raising the possibility that mitochondria may have an integral role in SOD1 processing and disease pathogenesis (6–12). The factors that regulate SOD1 entrance, processing, and aggregation within mitochondria are not fully understood, although copper chaperone for SOD1 (CCS) has been hypothesized to influence SOD1's presence within mitochondria.
Human CCS was originally recognized on the basis of its homology to yeast LYS7, a copper chaperone protein that facilitates copper incorporation into SOD1 (13). CCS facilitates the conversion of an immature apo-SOD1 structure to a mature and active holo-SOD1 form by promoting SOD1 disulfide bond formation and insertion of copper (14). The ability of CCS to influence the equilibrium state of apo-and holo-SOD1 levels has been verified in vivo by using CCS knockout (KO) mice (15, 16).
CCS may also affect the subcellular localization of SOD1 within mitochondria. CCS, initially recognized as a cytosolic protein, has been detected within the mitochondrial intermembrane space, with perhaps 5% of total CCS normally found within mitochondria (9). Altering mitochondrial CCS significantly impacts SOD1 levels in mitochondria without changing total SOD1 amount in yeast, where the maturation of SOD1 is totally dependent on CCS (13). However, because SOD1 may be activated via CCS-independent pathways in mammalian cells, SOD1 localization within mitochondria can still occur in the absence of CCS (7, 13, 17, 18).
What role might CCS play in mutant SOD1-related familial ALS? Although SOD1 mutants maintain the ability to interact with CCS, CCS is not required for mutant SOD1-induced disease as demonstrated by studies using CCS KO mice (19, 20). Because mammalian SOD1 can be activated via CCS-independent pathways, the CCS KO study does not exclude an effect of CCS on SOD1 maturation or SOD1-induced disease. To determine whether CCS influences mutant SOD1-induced disease, we generated lines of transgenic mice overexpressing WT human CCS and crossed them with G93A-SOD1 or WT-SOD1 transgenic mice. Our results indicate that CCS overexpression influences the cytosolic/mitochondrial distribution of G93A-SOD1 in vivo and greatly impacts the neurological disease.
Generation and Characterization of CCS Transgenic Mice.
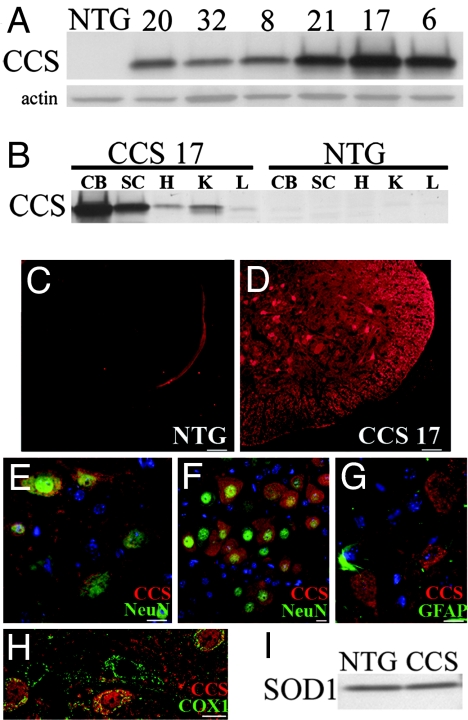
Human WT CCS cDNA was cloned into a mouse prion protein promoter construct shown to express transgenes at levels within the CNS sufficiently high to obtain neurological phenotypes (21). Western blot analysis on spinal cords from 6-week-old mice showed that transgenic lines express CCS at significantly higher levels than nontransgenic mice (Fig. 1A). Line 17 demonstrated the highest level of CCS expression in the spinal cord, followed by lines 21 and 6, whereas lines 8, 20, and 32 showed more modest levels of CCS expression. The tissue distribution of transgene expression was comparable in the multiple CCS lines and is shown for CCS line 17 (Fig. 1B). The highest levels of CCS expression are found within the CNS, particularly in the cerebellum, brainstem, and spinal cord, as well as in the heart and kidney, with less in the liver. Within the spinal cord, immunohistochemical studies confirmed significantly higher levels of CCS protein in transgenics than in nontransgenics (Fig. 1 C and D). CCS was primarily localized within neuronal cell bodies, including ventral horn motor neurons, but there was also CCS expression in the neuropil and white matter tracts (Fig. 1 D and E). Robust CCS expression was observed throughout the brainstem, including hypoglossal motor neurons in the medulla (Fig. 1F). There appeared to be little, if any, transgene expression in astrocytes (Fig. 1G). CCS overexpression was prominent in dorsal root ganglia neurons (data not shown). CCS immunostaining within neurons appeared cytosolic, with punctuate areas of more intense CCS immunoreactivity. On confocal microscopy, many of these punctuate areas of CCS expression colocalized with cytochrome c oxidase subunit 1 (COX1), a mitochondrial marker (Fig. 1H). Endogenous SOD1 levels in the spinal cord were similar in CCS transgenic and nontransgenic littermates, indicating that CCS overexpression does not alter total SOD1 levels in vivo (Fig. 1I). We elected to use CCS lines 17 and 21 for further analysis because they express the highest levels of transgene.
Fig. 1.
Characterization of CCS transgenic mice. (A) Western blot of spinal cord extracts from CCS transgenic lines (20, 32, 8, 21, 17, 6) showing CCS expression levels compared with a nontransgenic (NTG) mouse. Actin was used as loading control (20 μg of protein per lane). (B) Tissue distribution of CCS protein in 24-day-old line 17 CCS transgenic mouse versus NTG mouse on Western blot. CB, cerebellum; SC, spinal cord; H, heart; K, kidney; L, liver (20 μg of protein per lane). (C–H) Immunohistochemical studies of CCS expression in spinal cords of 26-day-old line 17 CCS mouse and NTG littermate. Spinal cord sections from NTG (C) and CCS (D) mice stained for CCS with identical exposure times. Merged images of CCS (red) and NeuN (green) staining in spinal cord ventral horn (E) and hypoglossal nucleus (F) from CCS mouse. Merged image of CCS (red) and GFAP (green) staining in spinal cord ventral horn (G) from CCS mouse. Merged confocal image of CCS (red) and COX1 (green) staining in spinal cord ventral horn (H) from CCS mouse. (I) SOD1 levels in spinal cord extracts from 30-day-old NTG and CCS mice (10 μg protein per lane). (Scale bars: C and D, 50 μm; E, G, and H, 20 μm; F, 10 μm.)
CCS transgenic mice develop normally and demonstrate no overt neurological phenotype. Their survival rate is comparable to that of nontransgenic mice, and we have followed the oldest animals past 2 years at present. On motor testing, line 17 CCS mice and age-matched nontransgenic littermates demonstrate comparable stride lengths (5.7 ± .3 cm for nontransgenic vs. 6.2 ± .3 cm for CCS at 6 months and 6.5 ± .5 cm for nontransgenic vs. 6.2 ± .3 cm for CCS at 12 months; n = 5 for each group) and grip strengths (971 ± 90 dg for nontransgenic vs. 941 ± 59 dg for CCS at 6 months and 959 ± 68 dg for nontransgenic vs. 1,003 ± 50 dg for CCS at 12 months; n = 5 in each group). Histologic examination of line 17 CCS adult mice reveals no evidence of neuronal loss or gliosis (data not shown). These results indicate that the overexpression of WT human CCS in mice does not produce an abnormal motor or neurological phenotype.
CCS Overexpression Accelerates Disease in G93A-SOD1 Transgenic Mice.
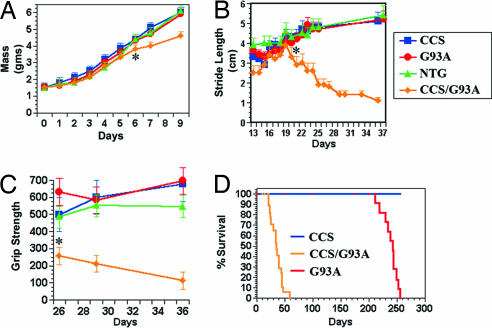
To ascertain the effect of CCS overexpression on disease, we crossed our highest expressing CCS transgenic line 17 with transgenic mice that express G93A-SOD1 under the control of the human SOD1 promoter. This G93A-SOD1 mouse line develops progressive motor deficits beginning at 180 days and succumbs by ≈240 days (22). At birth, CCS/G93A-SOD1 dual transgenic mice are indistinguishable from their nontransgenic, CCS, or G93A-SOD1 littermates. CCS/G93A-SOD1 dual mice grow normally for the first 5 days, but then their growth rate decreases, and by day 6, they weigh significantly less than the other genotypes (Fig. 2A). We could not detect any obvious neurological phenotype in CCS/G93A-SOD1 dual mice during this first week of life, but as early as day 8, CCS/G93A-SOD1 dual mice begin to develop neurological signs of tremor, ataxia, and spasticity with extensor hind-limb posturing. The mean age of onset of symptoms was 11 days. We did not detect any neurological deficits in age-matched nontransgenic, CCS, or G93A-SOD1 mice.
Fig. 2.
CCS overexpression accelerates G93A-SOD1-induced disease. (A) Mass (gms) comparison of line 17 CCS/G93A-SOD1 dual mice with controls. By day 6, CCS/G93A-SOD1 dual mice weigh significantly less (∗, P = 0.016), whereas there is no difference in mass among NTG, CCS, and G93A-SOD1 (P = 0.377) by ANOVA (n = 7–12 for each group; values ± SEM). (B) Stride length measurements in line 17 CCS/G93A-SOD1 dual mice and controls. By day 22, CCS/G93A-SOD1 dual mice have a significantly shorter stride length (∗, P = 0.025), whereas there is no difference in stride length among controls (P = 0.685) by ANOVA (n = 7–10 for each group; values ± SEM). (C) Forepaw grip strength measurements in decigrams of line 17 CCS/G93A-SOD1 dual mice and controls. At day 26, CCS/G93A-SOD1 dual mice have significantly weaker grips (∗, P = 0.003), whereas there is no difference in grip strength among controls (P = 0.165) by ANOVA (n = 4–7 for each group; values ± SEM). (D) Kaplan–Meier survival curves from CCS (n = 15), G93A-SOD1 (n = 14), and CCS/G93A-SOD1 (n = 17) dual mice of line 17. CCS/G93A-SOD1 dual mice have significantly shortened survival (P = 0.001).
We performed tests of motor function to quantify differences between CCS/G93A-SOD1 dual mice and their littermates. Stride length analysis showed that at days 13–15, CCS/G93A-SOD1 dual mice have smaller strides than the other genotypes (Fig. 2B). By day 20, CCS/G93A-SOD1 dual mice attain a maximal stride length, but subsequently their stride length declines as they manifest limb paresis. In contrast, nontransgenic, CCS, and G93A-SOD1 mice continue to increase their stride length as they mature. On grip strength measurements, CCS/G93A-SOD1 dual mice show marked weakness and progressive decline in forelimb strength (Fig. 2C). Such declines in grip strength are not evident in age-matched controls. CCS/G93A-SOD1 dual mice can balance on beam for only 6 ± 2 sec at 18 days and <1 sec at 23 days, whereas age-matched nontransgenic, CCS, or G93A-SOD1 mice were able to maintain balance for the maximum time observed of 30 sec.
Due to their progressive neurological deficits, CCS/G93A-SOD1 dual mice have a mean survival of 36 days, compared with 242 days for G93A-SOD1 mice and >700 days for CCS mice (Fig. 2D). Crossing a lower CCS-expressing line with G93A-SOD1 mice also yielded dual mice that developed neurological deficits including tremor, ataxia, spasticity, and weakness. These line 21 CCS/G93A-SOD1 dual mice had a slightly milder phenotype, with a mean survival of 56 days that is likely related to their overall lower expression level of CCS transgene. Results from these crossing experiments indicate that overexpression of WT human CCS greatly accelerates the neurological phenotype observed in G93A-SOD1 mice.
To ensure that the accelerated phenotype in CCS/G93A-SOD1 dual mice was not just related to the simultaneous overexpression of two related proteins (CCS and SOD1), we also crossed transgenic line 17 CCS with the N29 line of transgenic mice expressing WT human SOD1. This N29 line expresses WT SOD1 protein at levels comparable to the G93A-SOD1 line, but does not develop neurological disease. CCS/WT-SOD1 dual mice develop normally and have not manifested any abnormal neurological phenotype or change in survival at least through 600 days (the oldest such mice currently). Grip strength was comparable even at 1 year of age for CCS/WT-SOD1 dual mice (955 ± 9 dg; n = 5) and age-matched controls (961 ± 16 dg; n = 4). The normal neurological function of CCS/WT-SOD1 dual mice indicates that the accelerated disease course observed in CCS/G93A-SOD1 dual mice is a consequence of CCS overexpression in the context of mutant SOD1 and not due to nonspecific occurrences arising from having both high CCS and SOD1 levels.
CCS Overexpression Promotes Mitochondrial Pathology in G93A-SOD1 Mice.
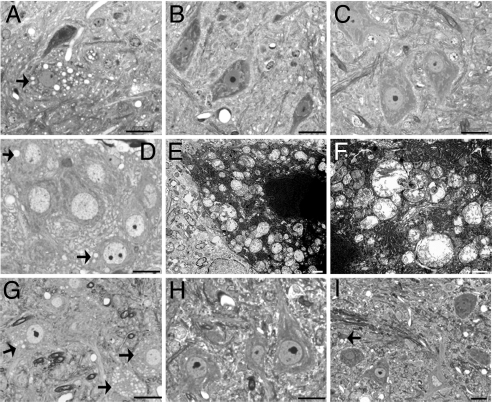
We performed a pathologic examination of line 17 CCS/G93A-SOD1 dual mice to determine the microscopic correlates of the accelerated disease course. By 22 days of age, CCS/G93A-SOD1 dual mice already manifest neurological abnormalities with motor deficits. In the lumbar spinal cord, motor neurons contained many multiple dilated vacuoles (Fig. 3A). In contrast, motor neurons from age-matched CCS or G93A-SOD1 mice show no vacuolar pathology (Fig. 3 B and C). Such vacuoles are also found in many neuronal populations extending throughout the brainstem of these 22-day-old mice, including the hypoglossal motor nucleus within the medulla and in pontine neurons [supporting information (SI) Fig. 7 A and B]. In contrast to the spinal cord, dorsal root ganglia neurons show no vacuolar pathology in CCS/G93A-SOD1 dual mice despite expressing high levels of CCS transgene (SI Fig. 7C). Vacuolar pathology is already evident in spinal motor neurons of CCS/G93A-SOD1 dual mice by postnatal day 7 (Fig. 3D). Electron microscopic analysis of the spinal cord from 22-day-old CCS/G93A-SOD1 dual mice confirmed that these vacuolar structures within motor neurons were swollen in mitochondria (Fig. 3 E and F). We find similar pathology in the spinal cord of the line 21 CCS/G93A-SOD1 dual mice, indicating that vacuoles are also characteristic of other CCS line crosses with G93A-SOD1 mice (Fig. 3G). However, consistent with their normal neurological phenotype, 60-day-old CCS/WT-SOD1 dual mice have no mitochondrial vacuoles, demonstrating that overexpression of CCS in the context of normal SOD1 does not cause pathological changes (Fig. 3H). In contrast to weak CCS/G93A-SOD1 dual mice, 7-month-old weak G93A-SOD1 mice show far fewer vacuolated mitochondria within spinal cord motor neurons (Fig. 3I).
Fig. 3.
CCS overexpression enhances G93A-SOD1-induced mitochondrial pathology. (A–D) Toluidine blue-stained plastic sections of lumbar spinal cord ventral horn from 22-day-old line 17 CCS/G93A-SOD1 dual mouse (A), CCS (B), G93A-SOD1 (C) littermates, and 7-day-old line 17 CCS/G93A-SOD1 dual mouse (D). (E and F) EM of motor neurons in lumbar cord of 22-day-old line 17 CCS/G93A-SOD1 dual mouse. (G–I) Sections of lumbar spinal cord from 22-day-old line 21 CCS/G93A-SOD1 dual mouse (G), 60-day-old line 17 CCS/WT-SOD1 dual mouse (H), and 7-month-old G93A-SOD1 mouse (I). Arrows indicate neurons with vacuoles. (Scale bars: A–D and G–I, 20 μm; E and F, 1 μm.)
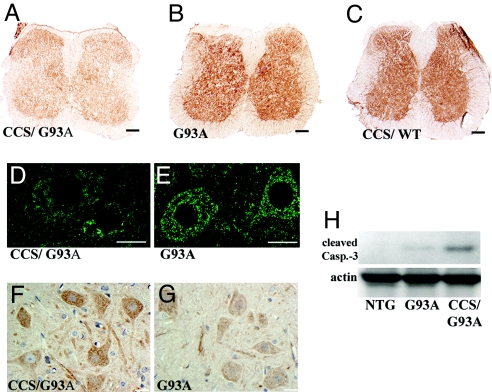
Because of the marked mitochondrial cytopathology, we examined markers of mitochondrial function. By in situ analysis, the COX activity of spinal cord sections from CCS/G93A-SOD1 dual mice was significantly decreased as compared with G93A-SOD1 mice or CCS/WT-SOD1 dual mice (Fig. 4 A–C). Furthermore, the overall pattern of COX1 immunostaining within CCS/G93A-SOD1 neurons was less intense, showing a patchy distribution with clustering, rather than the discrete speckled pattern observed in age-matched G93A-SOD1 mice (Fig. 4 D and E). These results indicate that CCS/G93A-SOD1 dual mice show early deficits in COX function that correlate with the abnormal mitochondrial pathology. The mitochondrial changes also prompted us to assess whether CCS/G93A-SOD1 dual mice showed early changes in expression for proteins related to apoptosis. Both immunohistochemistry and Western blot analysis show increased levels of cleaved caspase-3 expression in the spinal cords of 3-week-old CCS/G93A-SOD1 dual mice compared with controls (Fig. 4 F–H).
Fig. 4.
Line 17 CCS/G93A-SOD1 dual mice show deficits in COX activity and signs of apoptosis. (A–C) In situ COX activity in spinal cord sections of 22-day-old CCS/G93A-SOD1 dual (A), 22-day-old G93A-SOD1 (B), and 33-day-old CCS/WT-SOD1 dual (C) mice. (Scale bars: 200 μm.) (D and E) COX1 immunostaining in 22-day-old mouse spinal cords. (Scale bars: 20 μm.) (F and G) Cleaved caspase-3 immunoreactivity in the ventral horn of 21-day-old mice. (Scale bars: 20 μm.) (H) Western blot shows 17-kD cleaved caspase-3 fragment in spinal cord from 20-day-old mice (50 μg of protein per lane). Actin as loading control.
The anterior horn cell pathology observed in the spinal cord was matched by denervation changes in muscle. Histochemical analysis demonstrated angular fibers and grouped atrophy in a pattern consistent with active and chronic denervation in weak CCS/G93A-SOD1 dual mice, but not in muscle from age-matched controls (SI Fig. 8). There was no mitochondrial vacuolation, ragged red fibers, or changes in in situ COX activity in the muscle of CCS/G93A-SOD1 dual mice that would have suggested the presence of mitochondrial pathology in skeletal muscle. In addition, CCS/G93A-SOD1 dual mice showed no pathology in the kidney or heart where CCS expression is high (data not shown).
Mitochondrial Localization of G93A-SOD1 Is Enhanced with CCS Overexpression.
To study whether CCS overexpression altered the subcellular distribution of SOD1, we isolated mitochondrial and cytosolic fractions from the spinal cords of line 17 CCS/G93A-SOD1 dual mice as well as controls and probed these fractions on Western blots (Fig. 5A). Both CCS/G93A-SOD1 and CCS/WT-SOD1 dual mice had increased levels of CCS in both mitochondrial and cytosolic fractions isolated from the spinal cord. Because CCS/WT-SOD1 dual mice are normal, this finding indicates that an elevated level of CCS in mitochondria does not in itself produce mitochondrial pathology.
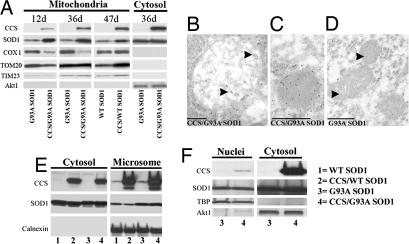
Fig. 5.
CCS overexpression alters the mitochondrial localization of G93A-SOD1 in vivo. (A) Mitochondrial fractions of 12- and 36-day-old CCS/G93A-SOD1 dual mice and G93A-SOD1 littermates or 47-day-old CCS/WT-SOD1 dual mice and WT-SOD1 littermates. Western blots probed for CCS, SOD1, COX1, TOM20, TIM23, and Akt1. Postmitochondrial cytosolic fractions from 36-day-old CCS/G93A-SOD1 dual mice and G93A-SOD1 littermates probed for CCS, SOD1, COX1, and Akt1 (10 μg of protein per lane). (B–D) Mitochondrial labeling of SOD1 in immuno-EM of lumbar spinal cord motor neurons from 28-day-old CCS/G93A-SOD1 dual and G93A-SOD1 mice. Arrowheads highlight gold particles. (Scale bars: 500 nm.) (E) Microsomal and postmicrosomal cytosolic fractions isolated from spinal cords of 21-day-old WT-SOD1 (lane 1), CCS/WT-SOD1 dual (lane 2), G93A-SOD1 (lane 3), and CCS/G93A-SOD1 dual (lane 4) mice probed for CCS, SOD1, and calnexin (endoplasmic reticulum marker) on Western blots (20 μg of protein per lane). (F) Nuclear and postnuclear cytosolic fractions from spinal cords of 24-day-old G93A-SOD1 and CCS/G93A-SOD1 dual mice probed for CCS; SOD1; TATA box-binding protein (TBP), a nuclear marker; and Akt1 on Western blots (10 μg of protein per lane).
Mutant SOD1 is enriched within the mitochondria of CCS/G93A-SOD1 dual mice compared with G93A-SOD1 mice (Fig. 5A). This increased mitochondrial load of SOD1 is present at early as well as later symptomatic stages of disease. However, there is no difference in SOD1 expression levels within the cytosolic fractions of G93A-SOD1 or CCS/G93A-SOD1 dual mice. CCS overexpression appears to slightly increase the mitochondrial localization of SOD1 in CCS/WT-SOD1 dual mice. Immuno-EM with an antibody against SOD1 confirmed that CCS overexpression increases the mitochondrial localization of G93A-SOD1 in motor neurons (Fig. 5 B–D). Within motor neurons, mitochondria (n = 42) from 28-day-old CCS/G93A-SOD1 dual mice demonstrate increased numbers of gold particles compared with mitochondria (n = 56) from age-matched G93A-SOD1 mice (22 ± 3 vs. 5 ± 1 per mitochondria; P = 0.001; Student's t test; mean ± SEM). In CCS/G93A-SOD1 dual mice, there are increased numbers of gold particles within mitochondria at differing stages of vacuolization (Fig. 5 B and C). In contrast, mitochondria in age-matched G93A-SOD1 motor neurons appear homogeneous and contain fewer gold particles (Fig. 5D). These results indicate that CCS overexpression alters the cytosolic/mitochondrial distribution of G93A-SOD1.
We assessed whether the expression of other mitochondrial proteins is altered in CCS/G93A-SOD1 dual mice (Fig. 5A). Levels of COX1, a mitochondrial DNA-encoded protein of the respiratory chain, were decreased in the mitochondria of CCS/G93A-SOD1 dual mice compared with G93A-SOD1 mice, whereas COX1 expression remained unchanged in CCS/WT-SOD1 dual mice. These results likely explain the diminished COX activity observed in CCS/G93A-SOD1 dual mice (Fig. 4). In contrast, levels of two nuclear-encoded mitochondrial proteins, TIM23 and TOM20, did not show significant changes within the mitochondrial fractions of CCS/G93A-SOD1 dual mice, suggesting that there is a selective reduction in mitochondrial protein levels.
To determine whether CCS overexpression altered levels of SOD1 in subcellular compartments other than mitochondria, we performed Western blots on microsomal and nuclear fractions isolated from the spinal cord. CCS levels are comparable in CCS/WT-SOD1 and CCS/G93A-SOD1 dual mice in microsomal fractions containing endoplasmic reticulum and also in postmicrosomal cytosolic fractions (Fig. 5E, lanes 2 and 4). SOD1 is also abundant in these fractions isolated from both WT-SOD1 and G93A-SOD1 transgenics (Fig. 5E, lanes 1 and 3). However, CCS overexpression does not lead to increased G93A-SOD1 levels within microsomal or cytosolic fractions in CCS/G93A-SOD1 dual mice compared with G93A-SOD1 mice (Fig. 5E, lanes 3 and 4). Similarly CCS overexpression does not alter levels of WT-SOD1 in CCS/WT-SOD1 dual mice compared with WT-SOD1 mice in these fractions (Fig. 5E, lanes 1 and 2). In nuclear-enriched fractions, we observed no difference in the levels of G93A-SOD1 expression between CCS/G93A-SOD1 and G93A-SOD1 mice (Fig. 5F).
Aggregation in Line 17 CCS/G93A-SOD1 Mice.
Because ubiquitin- and SOD1-positive aggregates are prominent within the spinal cords of weak G93A-SOD1 mice, we wished to determine whether such aggregates were also present in end-stage CCS/G93A-SOD1 dual mice. Total CCS levels and the banding pattern were comparable in both CCS dual transgenic lines (CCS/WT-SOD1 and CCS/G93A-SOD1), whereas CCS levels were predictably lower in G93A-SOD1 mice (Fig. 6A). Levels of SOD1 monomer are comparable in G93A-SOD1 mice and CCS/G93A-SOD1 dual mice (Fig. 6B). In addition, levels of SOD1 monomer are also comparable between WT-SOD1 and CCS/WT-SOD1 dual mice (Fig. 6C). Thus, CCS overexpression does not increase total SOD1 levels, thereby excluding any marked elevation in total SOD1 expression as the basis for the accelerated phenotype. Spinal cords from CCS/G93A-SOD1 dual mice do show a prominent SOD1-positive band at 50 kDa, which is much less apparent in age-matched G93A-SOD1 mice or CCS/WT-SOD1 dual mice (Fig. 6 B and D, arrowheads). Paralyzed CCS/G93A-SOD1 dual mice do not exhibit SOD1-positive high-molecular-weight protein complexes (HMWPCs) in the spinal cord, which are characteristic of weak and older G93A-SOD1 mice (Fig. 6D). Moreover, on immunostaining, the ubiquitin-positive cellular inclusions observed in spinal cords from end-stage G93A-SOD1 mice are not seen in end-stage CCS/G93A-SOD1 dual mice (Fig. 6 E and F). Although SOD1-positive inclusions are prominent in the spinal cord of weak G93A-SOD1 mice, such inclusions are not observed in the spinal cords of weak CCS/G93A-SOD1 dual mice (SI Fig. 9).
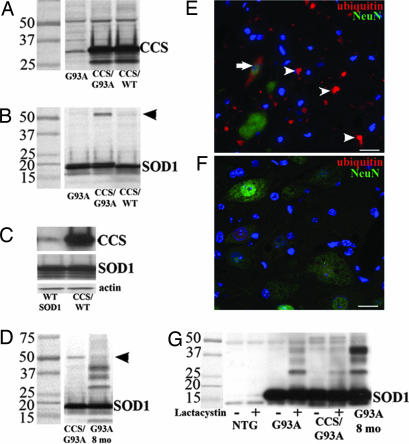
Fig. 6.
Protein aggregation in CCS/G93A-SOD1 dual mice. (A and B) Western blots of spinal cord extracts from 24-day-old G93A-SOD1, CCS/G93A-SOD1 dual, and CCS/WT-SOD1 dual mice probed for CCS (A) and SOD1 (B). (C) Western blot showing CCS and SOD1 expression in spinal cord extracts from 21-day-old WT-SOD1 transgenic and CCS/ WT-SOD1 dual mice. (D) Western blots of spinal cord extracts from 24-day-old CCS/G93A-SOD1 dual and 8-month-old G93A-SOD1 mice probed for SOD1. (E and F) Immunohistochemistry of lumbar spinal cord sections from weak 8-month-old G93A-SOD1 (E) and weak 34-day-old CCS/G93A-SOD1 dual mice (F). Merged images stained for ubiquitin (red), NeuN (green), and DAPI (blue). Arrow shows skein-like inclusions; arrowheads show Lewy-like inclusions. Identical exposure times. (Scale bars: 20 μm.) (G) Western blot of extracts from spinal cord cultures derived from 8-day-old nontransgenic, G93A-SOD1, or CCS/G93A-SOD1 dual mice with (+) or without (−) lactacystin treatment compared with 8-month-old G93A-SOD1 mouse. Western blots probed for SOD1 (20 μg of protein per lane).
To address the possibility that CCS overexpression prevented the formation or accumulation of SOD1 complexes, we used an organotypic spinal cord slice model of SOD1 aggregation (23). In this paradigm, spinal cords from 8-day-old G93A-SOD1 mice rapidly generated SOD1 HMWPCs and cellular inclusions after proteasome inhibition (Fig. 6G). Treatment of spinal cord slices from both G93A-SOD1 and CCS/G93A-SOD1 dual mice showed a similar pattern of SOD1 complex accumulation after proteasome inhibition with lactacystin. This result shows that SOD1 HMWPCs can form within the spinal cord of CCS/G93A-SOD1 dual mice even in the presence of CCS overexpression.
Discussion
Our results indicate that CCS overexpression significantly impacts disease and enhances mitochondrial pathology in G93A-SOD1 mice. We also find that G93A-SOD1 is enriched within mitochondria in the presence of CCS overexpression. How might CCS affect the mitochondrial localization of SOD1? CCS facilitates the conversion of an immature apo-SOD1 structure to a mature holo-SOD1 form via the accelerated oxidation of SOD1 cysteine residues and the formation of disulfide bridges (6, 13, 14). In yeast, immature apo-SOD1 can readily transit across the mitochondrial membrane, whereas mature holo-SOD1 is unable to do so (13). Once within the intermembrane space, apo-SOD1 may heterodimerize with CCS, an interaction that will foster apo-SOD1 conversion to holo-SOD1 and make SOD1 incapable of exiting the mitochondria. However, the conversion of apo- to holo-SOD1 can also occur via CCS-independent pathways that may be found in both mitochondria and cytosol (17, 18, 24). The presence of such pathways may explain why the lack of CCS does not impact mutant SOD1-induced disease.
There are several possible mechanisms for the enhanced mitochondrial cytopathy and accelerated disease course that arise from having increased mutant SOD1 within mitochondria in vivo. First, it is possible that excess nonspecific protein load might somehow disrupt mitochondrial function in dual transgenic mice. However, the fact that CCS/WT-SOD1 dual mice show no pathology argues strongly that the presence of mutant SOD1 is required for mitochondrial dysfunction. It is also possible that high levels of CCS change the reduction potential of G93A-SOD1 and its ability to form correct disulfide bonds, which may foster deleterious protein–protein interactions within mitochondria (6, 12).
Both vacuolar- and inclusion-type pathologies are identified in standard lines of G93A-SOD1 mice. However, their relationship and relevance to disease remain to be determined. Overall, an analysis of differing lines of G93A-SOD1 mice indicates that increasing levels of G93A-SOD1 protein lead to more severe disease and more extensive mitochondrial pathology. The high-expressing G93A-SOD1 line (mean survival = 130 days) develops a pathology characterized predominantly by mitochondrial vacuolation and swelling (2). This line also shows significant cytosolic SOD1-ubiquitin-inclusion pathology with accumulation of SOD1-positive HMWPCs (25, 26). The pathology of a lower expressing G93A-SOD1 line used in this study (mean survival = 240 days) is characterized primarily by cytosolic inclusion-type pathology, although significant mitochondrial pathology is still present (23, 27). Very low-expressing G93A-SOD1 lines, G20 and G5 with survivals of 340 and 400 days, respectively, show almost exclusive ubiquitin-inclusion-type pathology with minimal mitochondrial changes (28). However, because the worsening motor phenotype in G93A-SOD1 mouse lines directly correlates with an increase in total amount of mutant SOD1 (both mitochondrial and cytosolic), it is unclear whether enhanced mitochondrial pathology is responsible for the disease phenotype. Interestingly, WT-SOD1 overexpression in mutant SOD1 transgenic mice increases SOD1 aggregation within mitochondria and accelerates disease (12).
Overexpression of CCS converted a moderate, mostly inclusion-type pathology in our G93A-SOD1 line to a fulminant disease with mitochondrial vacuolar pathology and changed the subcellular distribution of G93A-SOD1. With this accelerated mitochondrial pathology in the CCS/G93A-SOD1 dual mice, we did not observe, even at end stage, any of the typical ubiquitin inclusions, SOD1 inclusions, or SOD1-positive HMWPCs that have been described in G93A or other mutant SOD1 mouse lines (26). However, our results do not exclude the possibility that G93A-SOD1 may also cause nonmitochondrial-based pathology that contributes to cellular dysfunction (29, 30).
Materials and Methods
DNA Constructs and Generation of CCS Transgenic Mice.
WT human CCS cDNA (825 bp) was a gift from Jonathan Gitlin (Washington University, St. Louis, MO). For the generation of transgenic mice, WT CCS cDNA was cloned into MoPrP Xho vector (12 kb) at the unique XhoI site between exons 2 and 3 (JHU-2; Johns Hopkins Special Collection distributed by American Type Culture Collection).
Generation of CCS and SOD1 Dual Transgenic Lines.
Line 17 or line 21 CCS transgenic mice were crossed with transgenic mice expressing the low copy number human G93A-SOD1 mutation (B6SJL-TgNSOD1-G93A; 1Gurdl JR2300) originally obtained from The Jackson Laboratory (Bar Harbor, ME). Line 17 CCS mice were also crossed with a transgenic mouse line expressing WT human SOD1 (line N29, B6SJL- WT-SOD1; The Jackson Laboratory). All animal protocols were approved by our university's Institutional Animal Care and Research Advisory Committee in compliance with National Institutes of Health guidelines.
Survival Analysis and Motor Testing.
Survival analysis, stride lengths (in centimeters), and grip strengths (in decigrams) were done as previously described (22). Mice were weighed daily starting at day of birth. Balance walks were done by recording the time each mouse could stay on a 0.5-inch-wide beam.
Western Blot Analysis and Immunohistochemistry.
Western blots and immunohistochemistry were done as previously described (31). The primary antibodies are listed in SI Text.
EM and Immuno-EM.
Anesthetized mice were perfused with 2% glutaraldehyde, postfixed in 1% osmium tetroxide, and embedded in Eponate 12 (Ted Pella, Redding, CA). One-micrometer sections were stained with toluidine blue. Ultrathin sections (60–90 nm) were stained with uranyl acetate and lead citrate. For immuno-EM, lumbar spinal cords from two CCS/G93A-SOD1 dual mice and two age-matched G93A-SOD1 controls were processed following the method for SOD1 immuno-EM by using a rabbit anti-SOD1, 1/250 (Biodesign, Saco, ME) (30). Motor neurons were identified by their large size (>20 μm) and distinct nuclear/nucleolar morphologies in the ventral horn of the lumbar cord.
Organelle Isolation and Measurement of Enzymatic Activities.
Mouse spinal cord mitochondria was purified through a discontinuous Ficoll gradient as described previously (8). For microsomal isolation, partially purified postnuclear and postmitochondrial fractions of the spinal cord were spun at 100,000 × g for 1 h. Microsome-containing pellets were washed with Tris-EDTA and resuspended in TEN buffer containing 0.5% Nonidet, 0.05% deoxycholic acid, and 0.25% SDS. For nuclear isolation, crude nuclear fractions were obtained from fresh spinal cord tissue by using the Focus SubCell kit (G-Biosciences, St. Louis, MO). Nuclear fractions were then further purified by using a sucrose gradient of PURE Prep Nuclei isolation kit (Sigma–Aldrich, St. Louis, MO).
Supplementary Material
Acknowledgments
This work was supported by the Horace C. Cabe Foundation (J.L.E.), the Jean and Judith Pape Adams Charitable Foundation (J.L.E.), the ALS Research Fund at University of Texas Southwestern (J.L.E.), the Winspear Family Special Center for Research on the Neuropathology of Alzheimer's disease (P.J.B.), the Robert Packard ALS Center (G.M.), and the National Institute of Neurological Disorders and Stroke (G.M.).
Abbreviations
- CCS
copper chaperone for SOD1
- COX
cytochrome c oxidase
- HMWPC
high-molecular-weight protein complex
- KO
knockout
- SOD1
Cu, Zn superoxide dismutase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610923104/DC1.
References
- 1.Manfredi G, Xu Z. Mitochondrion. 2005;5:77–87. doi: 10.1016/j.mito.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Dal Canto MC, Gurney ME. Am J Pathol. 1994;145:1271–1279. [PMC free article] [PubMed] [Google Scholar]
- 3.Wong PC, Pardo CA, Borchelt DR, Lee MK, Copeland NG, Jenkins NA, Sisodia SS, Cleveland DW, Price DL. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 4.Jung C, Higgins CM, Xu Z. J Neurochem. 2002;83:535–545. doi: 10.1046/j.1471-4159.2002.01112.x. [DOI] [PubMed] [Google Scholar]
- 5.Kirkinezos IG, Bacman SR, Hernandez D, Oca-Cossio J, Arias LJ, Perez-Pinzon MA, Bradley WG, Moraes CT. J Neurosci. 2005;25:164–172. doi: 10.1523/JNEUROSCI.3829-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa Y, O'Halloran TV. Antioxid Redox Signal. 2006;8:847–867. doi: 10.1089/ars.2006.8.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Lillo C, Jonsson PA, Vande Velde C, Ward CM, Miller TM, Subramaniam JR, Rothstein JD, Marklund S, Andersen PM, et al. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Vijayvergiya C, Beal MF, Buck J, Manfredi G. J Neurosci. 2005;25:2463–2470. doi: 10.1523/JNEUROSCI.4385-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. J Biol Chem. 2001;276:38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 10.Okado-Matsumoto A, Fridovich I. Proc Natl Acad Sci USA. 2002;99:9010–9014. doi: 10.1073/pnas.132260399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins CM, Jung C, Ding H, Xu Z. J Neurosci. 2002;22:1–6. doi: 10.1523/JNEUROSCI.22-06-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng HX, Shi Y, Furukawa Y, Zhai H, Fu R, Liu E, Gorrie GH, Khan MS, Hung WY, Bigio EH, et al. Proc Natl Acad Sci USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Field LS, Furukawa Y, O'Halloran TV, Culotta VC. J Biol Chem. 2003;278:28052–28059. doi: 10.1074/jbc.M304296200. [DOI] [PubMed] [Google Scholar]
- 14.Furukawa Y, Torres AS, O'Halloran TV. EMBO J. 2004;23:2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong PC, Waggoner D, Subramaniam JR, Tessarollo L, Bartnikas TB, Culotta VC, Price DL, Rothstein J, Gitlin JD. Proc Natl Acad Sci USA. 2000;97:2886–2891. doi: 10.1073/pnas.040461197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartnikas TB, Gitlin JD. J Biol Chem. 2003;278:33602–33608. doi: 10.1074/jbc.M305435200. [DOI] [PubMed] [Google Scholar]
- 17.Carroll MC, Girouard JB, Ulloa JL, Subramaniam JR, Wong PC, Valentine JS, Culotta VC. Proc Natl Acad Sci USA. 2004;101:5964–5969. doi: 10.1073/pnas.0308298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carroll MC, Outten CE, Proescher JB, Rosenfeld L, Watson WH, Whitson LJ, Hart PJ, Jensen LT, Culotta VC. J Biol Chem. 2006;281:28648–28656. doi: 10.1074/jbc.M600138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramaniam JR, Lyons WE, Liu J, Bartnikas TB, Rothstein J, Price DL, Cleveland DW, Gitlin JD, Wong PC. Nat Neurosci. 2002;5:301–307. doi: 10.1038/nn823. [DOI] [PubMed] [Google Scholar]
- 20.Corson LB, Strain JJ, Culotta VC, Cleveland DW. Proc Natl Acad Sci USA. 1998;95:6361–6366. doi: 10.1073/pnas.95.11.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Xu G, Slunt HH, Gonzales V, Coonfield M, Fromholt D, Copeland NG, Jenkins NA, Borchelt DR. Neurobiol Dis. 2005;20:943–952. doi: 10.1016/j.nbd.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Puttaparthi K, Gitomer WL, Krishnan U, Son M, Rajendran B, Elliott JL. J Neurosci. 2002;22:8790–8796. doi: 10.1523/JNEUROSCI.22-20-08790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puttaparthi K, Wojcik C, Rajendran B, DeMartino GN, Elliott JL. J Neurochem. 2003;87:851–860. doi: 10.1046/j.1471-4159.2003.02028.x. [DOI] [PubMed] [Google Scholar]
- 24.Hurd TR, Filipovska A, Costa NJ, Dahm CC, Murphy MP. Biochem Soc Trans. 2005;33:1390–1393. doi: 10.1042/BST0331390. [DOI] [PubMed] [Google Scholar]
- 25.Johnston JA, Dalton MJ, Gurney ME, Kopito RR. Proc Natl Acad Sci USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe M, Dykes-Hoberg M, Culotta VC, Price DL, Wong PC, Rothstein JD. Neurobiol Dis. 2001;8:933–941. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki S, Warita H, Murakami T, Abe K, Iwata M. Acta Neuropathol (Berl) 2004;107:461–474. doi: 10.1007/s00401-004-0837-z. [DOI] [PubMed] [Google Scholar]
- 28.Dal Canto MC, Gurney ME. Acta Neuropathol (Berl) 1997;93:537–550. doi: 10.1007/s004010050650. [DOI] [PubMed] [Google Scholar]
- 29.Urushitani M, Sik A, Sakurai T, Nukina N, Takahashi R, Julien JP. Nat Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. [DOI] [PubMed] [Google Scholar]
- 30.Kikuchi H, Almer G, Yamashita S, Guegan C, Nagai M, Xu Z, Sosunov AA, McKhann GM, II, Przedborski S. Proc Natl Acad Sci USA. 2006;103:6025–6030. doi: 10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Son M, Cloyd CD, Rothstein JD, Rajendran B, Elliott JL. J Biol Chem. 2003;278:14331–14336. doi: 10.1074/jbc.M211698200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.