Abstract
Many eukaryotic and viral mRNAs, in which the first transcribed nucleotide is an adenosine, are decorated with a cap-1 structure, 7MeG5′-ppp5′-A2′OMe. The positive-sense RNA genomes of flaviviruses (Dengue, West Nile virus) for example show strict conservation of the adenosine. We set out to produce GpppA- and 7MeGpppA-capped RNA oligonucleotides for non-radioactive mRNA cap methyltransferase assays and, in perspective, for studies of enzyme specificity in relation to substrate length as well as for co-crystallization studies. This study reports the use of a bacteriophage T7 DNA primase fragment to synthesize GpppACn and 7MeGpppACn (1 ≤ n ≤ 9) in a one-step enzymatic reaction, followed by direct on-line cleaning HPLC purification. Optimization studies show that yields could be modulated by DNA template, enzyme and substrate concentration adjustments and longer reaction times. Large-scale synthesis rendered pure (in average 99%) products (1 ≤ n ≤ 7) in quantities of up to 100 nmol starting from 200 nmol cap analog. The capped RNA oligonucleotides were efficient substrates of Dengue virus (nucleoside-2′-O-)-methyltransferase, and human (guanine-N7)-methyltransferase. Methyltransfer reactions were monitored by a non-radioactive, quantitative HPLC assay. Additionally, the produced capped RNAs may serve in biochemical, inhibition and structural studies involving a variety of eukaryotic and viral methyltransferases and guanylyltransferases.
INTRODUCTION
The cap is a unique structure at the 5′-end of viral and cellular eukaryotic mRNAs (1,2). It is critical for both mRNA stability and binding to the ribosome during translation. Cap 0 (7MeG5′-ppp5′-N) formation is a co-transcriptional modification resulting from three enzymatic activities: RNA triphosphatase, guanylyltransferase and S-adenosyl-l-methionine (AdoMet) dependent (guanine-N7)-methyltransferase (N7MTase). Further methylation at the 2′O position of the ribose of the first nucleotide by an AdoMet-dependent (nucleoside-2′-O-)-methyltransferase (2′OMTase) leads to a cap 1 structure (7MeG5′-ppp5′-N2′OMe). Capped RNA molecules are required as substrates for the biochemical characterization of mRNA cap MTases, as well as structural studies on guanylyltransferases and MTases complexed with products or substrates, and they are not commercially available.
The single-stranded positive-sense RNA genome of flaviviruses is decorated with a cap 1 structure. The genus Flavivirus contains at least 70 viruses (3), among them emerging human pathogens such as Dengue, West Nile and Japanese encephalitis virus (4), all of which bear a 7MeG5′-ppp5′-A2′OMe cap structure with a strictly conserved adenosine as first nucleotide. We set out to produce (7Me)GpppA-capped (i.e. GpppA- or 7MeGpppA-) RNA molecules to aid our studies on the capping machinery of flaviviruses (5,6).
In order to get GpppA-capped RNAs, several approaches can be taken that differ widely in their efficiency. They can be synthesized chemically starting from mono- or diphosphate RNA (7,8). A 7MeGpppA cap can also be added to di- or triphosphate RNA using vaccinia virus capping enzyme that contains RNA triphosphatase, guanylyltransferase and N7MTase activities (9,10). However, diphosphate and triphosphate RNAs are not commercially available. (7Me)GpppA can be incorporated upon in vitro transcription by bacteriophage T7, T3 or SP6 DNA-dependent RNA polymerases (DdRp) (e.g. (11)) using their respective promoters and an adenosine in position +1 or +2 (12,13). The efficiency of these reactions is rather low because these promoters require a guanine as starting nucleotide. Very recently, an alternative, the bacteriophage T7 class II ϕ2.5 promoter (14) which requires an adenosine as first transcribed nucleotide, was successfully applied to generate GpppA-capped transcripts (15). (7Me)GpppA was reported to be incorporated upon in vitro transcription by E. coli DdRp using the bacteriophage λ PL promoter (16), a method that has not been exploited since. Finally, very short capped oligonucleotides (7Me)GpppACC were produced efficiently using bacteriophage T7 DNA primase-helicase (17).
Here we report the adaptation of the latter approach to prepare large amounts of capped RNA oligonucleotides 7MeGpppACn and GpppACn with a range of chain lengths (1 ≤ n ≤ 7). Instead of the full-length bacteriophage T7 DNA primase-helicase, we used a recombinant N-terminal fragment of 30 kD bearing the primase activity (18,19). The use of the fragment offers several advantages regarding enzyme production and purification compared to the full-length protein namely a 10 to 20 times higher relative expression yield, higher solubility and the fact that it does not co-purify with DNA (19). The DNA primase fragment was reported to show similar activity to the full-length protein when using high DNA template concentrations (19). Nevertheless, production of short-capped RNA oligonucleotides 7MeGpppAC2 that has been shown for full-length protein (17), has never been demonstrated for the DNA primase fragment. In this work, we synthesized 7MeGpppAC2 and GpppAC2 as well as longer products using the DNA primase fragment. After undertaking optimization studies, we designed a robust and effective method to produce high amounts of pure (7Me)GpppACn (1 ≤ n ≤ 7). Given the lack of commercial availability of capped RNA oligonucleotides we expect this method to be useful for many researchers working in the still emerging field of characterization of viral and mammalian RNA capping machineries. In addition, we demonstrate that the produced GpppA-capped RNAs are substrates of Dengue virus 2′OMTase and human mRNA cap N7MTase. Both methyltransferase reactions were monitored by a non-radioactive, quantitative assay based on HPLC and the reaction products were characterized.
MATERIAL AND METHODS
Protein expression and purification
T7 DNA primase fragment
The T7 DNA primase expression vector pET19b/PrD was kindly provided by Charles C. Richardson (Harvard Medical School, Boston, USA). E. coli BL21(DE3) cells (500 ml) transformed with pET19b/PrDT were grown in Luria-Bertani medium containing ampicillin. At an OD600 of 0.6 isopropyl β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 200 μM, and expression was allowed to proceed for 2 h at 37°C. The cellular pellet was resuspended in 10 ml lysis buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF)) supplemented with 1 mM benzamidine, 10 mM imidazole, 100 μg/ml lysozyme and 1 μg/ml DNAse I. Since the expressed protein bears a His-tag at the N-terminus, immobilized-metal-affinity chromatography (IMAC) was used for the first purification step (chelating sepharose fast flow resine (Amersham Biosciences) loaded with Ni2+). The protein was eluted with lysis buffer containing 250 mM imidazole. Fractions were then directly loaded onto a HiTrap DEAE Sepharose FF 1-ml column (Amersham Biosciences) equilibrated with 50 mM HEPES, pH 7.5, containing 0.1 mM DTT, and eluted with a gradient of NaCl (0–500 mM). The protein elutes at 200 mM NaCl. The sample was diluted to lower the NaCl concentration to 50 mM and loaded onto a HiTrap Blue Sepharose 1-ml column (Amersham Biosciences) equilibrated with 50 mM HEPES, pH 7.5, containing 1 mM DTT and 1 mM EDTA. Elution was carried out with a gradient of NaCl (0–800 mM). Glycerol was added to reach 50% and the protein stored at −20°C.
Dengue virus MTase domain (NS5MTaseDV)
Recombinant NS5MTaseDV was produced as described in (6).
Human MTase (hMTase)
The cDNA coding for hMTase was a kind gift from Aaron J. Shatkin (Center of Advanced Biotechnology and Medicine, Piscataway, USA.). It was amplified by PCR using primers RBSATGLys6HishMTs 5′-CCATGAAACATCACCATCACCATCACGCAAATTCTGCAAAAGCAGAAG-3′ and hMTstopattb2as 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTACTGCTGTTTCTCAAAGGCAAAC-3′. A second PCR was then used to add the attb1 recombination site to the 5′ using primers attB1RBSATGLys6His 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTAAGGAGGTAGAACCATGAAAC ATCACCATCACCATCAC-3′ and hMTstopattb2as given above. The PCR product was then cloned into the expression vector pDest14 (Invitrogen) using the Gateway technology. E. coli Rosetta(DE3) cells were transformed with pDest14/6His-hMTase and grown in Luria-Bertani medium containing ampicillin and chloramphenicol. At an OD600 of 0.6, IPTG and ethanol were added to a final concentration of 100 μM and 2%, respectively, and expression was allowed to proceed for 18 h at 17°C. The cellular pellet was resuspended in 10 ml lysis buffer (50 mM Tris, pH 8.5, 300 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, antiprotease cocktail (Complete®, Roche)) supplemented with 10 mM imidazole, 100 μg/ml lysozyme and 1 μg/ml DNAse I and 0.5% triton X100. After lysis by sonication and clarification, immobilized-metal-affinity chromatography (IMAC) was used for the first purification step (chelating sepharose fast flow resine (Amersham Biosciences) loaded with Ni2+). The hMTase was eluted with lysis buffer pH 7.5 containing 250 mM imidazole. Fractions were then diluted 5 times in 50 mM Bis-Tris pH 6.8; 50 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, and loaded onto a 5-ml heparine column (Amersham Biosciences). The hMTase was then eluted with a gradient of NaCl (0–1 M) using a buffer at pH 7.5. The protein elutes at between 150 and 350 mM NaCl. It was finally dialyzed against 50 mM Tris, pH 7.5, 300 mM NaCl, 50% glycerol, 5 mM β-mercaptoethanol and stored at −20°C.
HPLC set-up
A Waters model 600 gradient HPLC system equipped with two 600 pumps, a 717 plus Autosampler injector, a 996 photodiode array detector and an in-line degasser AF was employed for reverse-phase chromatography. The column assembly consisting of a pre-column (Delta-pak C18 100 Å, 5 µm, 3.9 × 20 mm) and the separation column (Nova-pak C18, 4 μm, 3.9 × 150 mm) was protected by a filter insert. For the on-line cleaning procedure, both columns were installed in parallel on a two 7000 Rheodyne valve system (Interchim). As buffer stock solution, a 1 M solution of triethylammonium bicarbonate (TEAB) was prepared by adding dry-ice to a 1 M triethylamine solution until the pH reached 7.4 and filtered through 0.22 μM GV-type membranes (Millipore). HPLC eluents were freshly prepared. Eluent A was a 0.05 M solution of TEAB (pH 7.4) and eluent B was a 1:1, v/v, mixture of acetonitrile (HPLC grade, SDS) and TEAB (final concentration 0.05 M, pH 7.4). Separations were run at a flow rate of 1 ml/min and started with a 5-min elution (100% eluent A) on the pre-column to remove proteic material. The applied gradients for analytical and preparative separation of capped RNAs as well as for analysis of methyltransferase reactions are explained in the corresponding sections (see below).
Mass spectrometry analysis
MALDI-TOF mass spectra were recorded on a Voyager DE mass spectrometer (Perseptive Biosystems) equipped with an N2 laser (337 nm). MALDI conditions were: accelerating potential, 24 000 V; guide wire, 0.05% of accelerating voltage; grid voltage, 94% of accelerating voltage; delay extraction time, 550 ns. Spectra were obtained in negative mode and were not smoothed. The oligonucleotides (100 pmol) were suspended in 10 μl of water and desalted using drop dialysis through a membrane filter WSWP 0.025 μm, 13 mm (Millipore) floating on a 0.1 M ammonium citrate solution for 30 min. When drop dialysis did not allow to remove totally salt traces, the samples were further treated with a few beads of DOWEX 50 W X8 resin (ammonium form) before spotting them on the MALDI target. Samples of 0.5 μl were mixed with 0.5 μl of the matrix 2,4,6-trihydroxyacetophenone (THAP, 45 mg, ammonium citrate, 4 mg in 500 μl acetonitrile/water, 1:1, v/v) and the mixtures were spotted on the stainless steel MALDI target and left to dry under air before MALDI analysis.
Capped RNA synthesis
Analytical scale
Experiments were carried out in 20–40 µl reaction volume containing T7 DNA primase reaction buffer (40 mM Tris pH 7.5, 10 mM MgCl2, 50 mM potassium glutamate, 1 μM ZnCl2), 10 mM DTT and 50 μg/ml BSA as well as CTP (Amersham Biosciences), cap analog (New England Biolabs), DNA oligonucleotides (Invitrogen, HPLC grade) and enzyme at concentrations given in table and figure legends. Reaction mixtures were set up and incubated at 37°C for time intervals given in table and figure legends. Reactions were stopped by heating the samples at 95°C for 5 min and stored at −20°C. For HPLC analysis 20 μl of sample were mixed with 200 μl of TEAB (0.05 M) and analyzed. The analytical gradient started after 5 min at 100% eluent A with an increase to 10% eluent B after 25 min, to 30% eluent B after 35 min and to 50% eluent B after 40 min. Reaction yields were calculated from HPLC peak areas considering differences in absorbance due to molar absorptivities ε increasing with increasing chain length. The ε260 values (M−1 cm−1) of GpppA and 7MeGpppA were calculated as 26900 (11500G + 15400A) and 25500, respectively. ε260 values of G (11500), 7MeG (10100) and A (15400) were taken from http://www.owcsarzy.net/emethod.htm and (20). ε260 values of products were calculated using the nearest neighbor method (see http://www.owcsarzy.net/emethod.htm and (21)). ε260 of GpppACC was calculated as 31200 (11500G + 21000ApC + 14200CpC − 7200C). Values of methylated and longer products were calculated likewise. Yields of total product are given as percentage of substrate (cap analog) conversion. Additionally, from these values, yields were converted into pmol product, which can then be related to the amount of enzyme (pmol) used to generate the determined amount of product. Yields of single products are given in percentage of substrate (cap analog) conversion or in percentage of total product (explained in figure or table legends).
Preparative scale
Samples were set up in 200 μl reaction volume containing T7 DNA primase reaction buffer, 10 mM DTT and 50 μg/ml BSA, 5 mM CTP, 1 mM cap analog, 10 μM DNA template and 4 μM T7 DNA primase. Reactions were incubated at 37°C during time intervals given in Table 3, stopped by heating at 95°C for 5 min and stored at −20°C. Samples of 50 μl were mixed with 200 µl of TEAB (0.05 M) and analyzed. Preparative gradient started after 5 min at 100% eluent A with an increase to 10% eluent B after 30 min and to 30% after 45 min. After peak separation and collection, the samples were lyophilized three times and resuspended in water. Their purity was verified by HPLC, their concentration determined by measuring OD260 values and their molecular mass measured by mass spectrometry. Yields of preparative reactions are given as isolated yields based on measured amounts of products (absorbance measurements of pure products after lyophilization using molar absorptivities calculated as given above) and the amount of substrate used (Table 3). Samples were stored at −20°C.
Table 3.
Large-scale synthesis of short (7Me)GpppA-capped RNAs starting from 200 nmol of (7Me)GpppA. DNA templates dAC2, dAC3 and dAC4 correspond to CCCCCGGTCT25, CCCCGGGTCT25 and CCCGGGGTCT25, respectively. For experimental conditions and MALDI-TOF characterization, see Materials and Methods. Yields are given as isolated yields (percentage of substrate conversion and nmol or µg product produced from 200 nmol of cap analog) based on absorbance measurements of pure products after lyophilization. Given retention times correspond to the use of the preparative gradient given in Material and Methods
| Cap analog | DNA template/Reaction time | HPLC retention time | Purity | Isolated yields | m/z exp. | m/z calc. | ||
|---|---|---|---|---|---|---|---|---|
| (min) | (%) | (%) | (nmol) | (µg) | negative mode | |||
| GpppAC | dAC4/48 h | 27.9 | 100 | 2 | 4 | 12.9 | 1076.22 | 1076.58 |
| GpppAC2 | dAC2/15 h | 29.6 | 100 | 37 | 74 | 16.6 | 1382.23 | 1381.76 |
| GpppAC3 | dAC3/21 h | 31.2 | 100 | 42 | 84 | 20.2 | 1688.24 | 1686.94 |
| GpppAC4 | dAC4/48 h | 31.1 | 99 | 23 | 46 | 23.9 | 1991.42 | 1992.12 |
| GpppAC5 | dAC4/48 h | 33.3 | 100 | 21 | 42 | 27.6 | 2297.55 | 2297.31 |
| GpppAC6 | dAC4/48 h | 35.0 | 99 | 8 | 15 | 31.2 | 2601.81 | 2602.49 |
| GpppAC7 | dAC4/48 h | 36.4 | 91 | 1 | 2 | 34.9 | 2906.83 | 2907.67 |
| 7MeGpppAC | dAC4/48 h | 26.7 | 99 | 2 | 3 | 13.1 | 1092.33 | 1090.61 |
| 7MeGpppAC2 | dAC2/15 h | 28.7 | 100 | 31 | 61 | 16.8 | 1397.25 | 1395.79 |
| 7MeGpppAC3 | dAC3/21 h | 30.7 | 99 | 52 | 103 | 20.4 | 1701.58 | 1700.97 |
| 7MeGpppAC4 | dAC4/48 h | 31.9 | 100 | 13 | 26 | 24.1 | 2006.91 | 2006.15 |
| 7MeGpppAC5 | dAC4/48 h | 32.9 | 100 | 34 | 68 | 27.7 | 2311.68 | 2311.33 |
| 7MeGpppAC6 | dAC4/48 h | 34.5 | 100 | 17 | 33 | 31.4 | 2616.77 | 2616.51 |
| 7MeGpppAC7 | dAC4/48 h | 36.0 | 98 | 2 | 3 | 35.1 | 2921.68 | 2921.69 |
Methyltransferase experiments
Dengue virus MTase domain
The reaction was set up in 50 µl reaction volume containing 40 mM Tris, pH 7.5, 5 mM DTT, 500 nM NS5MTaseDV, 2 μM GpppAC3 and 20 μM AdoMet, incubated for 60 min at 30°C and stopped by immediate freezing. For HPLC analysis, the sample was mixed with 210 μl of TEAB (0.05 M) and analyzed. The gradient started after 5 min at 100% eluent A with an increase to 10% eluent B after 25 min, and to 30% eluent B after 45 min. The product peak was purified and analyzed by mass spectrometry.
Human N7 MTase
Reactions were set up in 40 µl reaction volume containing 40 mM Tris, pH 7.5, 5 mM DTT, 500 nM hMTase, 2 μM capped RNA oligonucleotide and 10 μM AdoMet. Reactions were incubated for 30 min at 30°C and stopped by immediate freezing. For HPLC analysis, 40 µl of sample were mixed with 210 µl of TEAB (0.05 M) and analyzed. The gradient started after 5 min at 100% eluent A with an increase to 10% eluent B after 25 min, and to 30% eluent B after 45 min. The product peak was purified and analyzed by mass spectrometry.
Analysis of GpppA2′OMeC3 by enzymatic digestion and HPLC
A preparative reaction (3 × 250 µl) was set up using the same conditions as described above for Dengue virus MTase except that 1 μM NS5MTaseDV was used and the reaction stopped after 8 h. Samples of 250 μl reaction mixture were mixed with 750 µl of TEAB (0.05 M) and injected into the HPLC system. The gradient started after 5 min at 100% eluent A with an increase to 10% eluent B after 25 min and to 20% after 35 min. After peak separation and collection, the product was lyophilized three times. Product yield was around 1 nmol of pure, lyophilized GpppA2′OMeC3. Then 0.5 nmol GpppA2′OMeC3 was digested in 20 µl reaction volume containing 50 mM Tris, pH 8.5, 5 mM MgCl2, 0.05 units of nucleotide pyrophosphatase type II from Crotalus adamanteus (Sigma, enzyme preparation contains side activity phosphodiesterase I, 0.005 units in 20 µl reaction volume) and 10 units of calf intestine phosphatase (New England Biolabs). Reaction was allowed to proceed for 11 min at 37°C and stopped by heating to 95°C for 5 min. The crude reaction mixture was then diluted with water (120 µl), filtered on Nanosep 3K omega (Pall Corporation) and centrifuged at 16 000 × g for 20 min. Washing of the filter was done by adding 60 µl of water followed by centrifugation at 16 000 × g for 10 min. The combined filtrates were mixed with 200 µl of TEAB (0.05 M) and analyzed by HPLC without using the on-line cleaning procedure. The gradient started after 5 min at 100% eluent A with an increase to 10% eluent B after 25 min and to 30% after 45 min.
RESULTS AND DISCUSSION
Synthesis of (7Me)GpppACn by T7 DNA primase
Bacteriophage T7 DNA primase-helicase is able to synthesize small RNA primers with the sequence pppA(C)n (n = 3,4) from DNA templates containing the recognition sequence 5′GTC3′ in which the cytidine is cryptic (22). The initiation site being flexible in terms of the phosphate moiety of the initiating adenosine (23) can accommodate GpppA or 7MeGpppA allowing thus the production of capped RNA oligonucleotides (17). For this study, we used the N-terminal fragment of the protein (residues 1 to 271), which contains the N-terminal Zn-binding domain (residues 1 to 56) and the catalytic RNA polymerase domain (residues 71–245) connected by a flexible linker (18). The DNA primase fragment was efficiently expressed in E. coli, soluble and easily purified (see Material and Methods).
We started with a DNA template, CCCCGGGTCT25 (transcribed sequence underlined) designed for the synthesis of AC3. Instead of the initiating ATP, we used GpppA and 7MeGpppA in order to produce (7Me)GpppAC3. Product formation was analyzed by reverse-phase chromatography on a HPLC system. An ‘on-line cleaning’ process (24,25) was used to remove unwanted proteic material from the sample. This technique allowed direct analysis of crude enzymatic reaction mixtures, with no significant loss of material. The sample was loaded onto the pre-column, thus eliminating proteic material while retaining the oligonucleotide mixture. After 5 min, the flow path was inverted and the substrates and produced capped RNAs were delivered onto the separating column by starting an acetonitrile gradient. Figure 1 shows the profiles after 6 h of primase reaction. Single peak material was purified and the molecular masses determined by mass spectrometry (see Material and Methods). Substrates GpppA and 7MeGpppA were efficiently incorporated by T7 DNA primase into small-capped RNAs (7Me)GpppACn (1 ≤ n ≤ 4). Apart from the main product (7Me)GpppAC3, smaller products were generated, (7Me)GpppAC and to a lower extent (7Me)GpppAC2 (not labeled in Figure 1), as well as a longer product, (7Me)GpppAC4. The long DNA template size which was chosen albeit the primase fragment works equally on short templates (26), allowed its easy separation from the reaction products because of its longer retention time (38.3 min).
Figure 1.
HPLC profiles of T7 DNA primase reaction mixtures using DNA template CCCCGGGTCT25. Crude enzymatic mixtures were analyzed directly. Reaction conditions were as follows: T7 DNA primase reaction buffer, DTT and BSA as given in Material and Methods, 5 mM CTP, 1 mM GpppA (A) or 7MeGpppA (B), 10 µM DNA template CCCCGGGTCT25 (named dAC3) and 4 µM T7 DNA primase. Reactions were incubated at 37°C for 6 h. The first section (marked in gray) denotes the removal of proteic material and remaining CTP by an on-line cleaning procedure using a pre-column. The produced capped RNAs were then separated on the reverse-phase C18 column. The gradient started after 5 min at 100% eluent A with an increase to 10% eluent B after 25 min, reaching 30% after 35 min and 50% after 40 min.
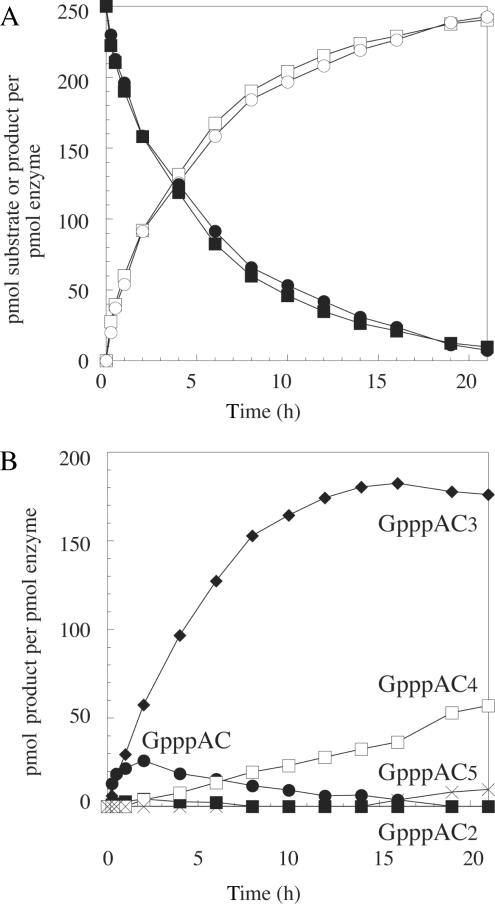
Subsequently, we followed product formation over time in order to reach optimal substrate conversion (Figure 2). Figure 2A shows the overall yield when GpppA or 7MeGpppA were used as cap analogs. T7 DNA primase incorporates both to same extent. Reactions were completed to 96% after 21 h. Figure 2B shows accumulation of individual products GpppAC to GpppAC5 of one of the reactions using GpppA as cap analog. GpppAC was produced during the early phase of reaction and completely converted into longer products after 16 h. GpppAC2 was equally only present during the early phase of the reaction but to very low extent. After 1 h, GpppAC3 was the main product and remained so throughout the reaction. GpppAC4 appeared almost immediately and its yield grew linearly, whereas GpppAC5 appeared after 15 h. Thus leaving the reaction longer opens the possibility to produce higher amounts of longer substrates (see below). The product profiles were similar when 7MeGpppA was used as cap analog (not shown).
Figure 2.
Time course of capped RNA synthesis by T7 DNA primase on DNA template dAC3 (CCCCGGGTCT25). Reaction conditions are given in the legend of Figure 1. Samples were taken at the given time points and analyzed by HPLC. The determination of yields (pmol product per pmol enzyme) is detailed in Material and Methods. (A) global analysis of substrate depletion (▪ GpppA, • 7MeGpppA) and product generation (□ GpppACn, ◯ 7MeGpppACn) with time, (B) formation of individual products in the GpppA-capped RNA product mixture (• GpppAC, ▪ GpppAC2, ♦ GpppAC3, □ GpppAC4, ×GpppAC5).
Higher accumulation of AC than of AC2 products has been observed repeatedly (27,28). Recently, it was proposed that the accumulation of the abortive AC product results from a change in conformation, namely in the relative position of the Zn-binding and RNA polymerase domains, occurring after the synthesis of the first phosphodiester bond so that the AC product is released more readily (29). Once the elongation-mode conformation is in place, longer products are formed. As it has been described before for non-capped RNA oligonucleotide synthesis by T7 DNA primase (22,27,28), the primase added extra non-templated cytidines and thus produced (7Me)GpppAC4 and (7Me)GpppAC5. This phenomenon might be due to either the ability of the primase to mis-incorporate CMP opposite cytidine (27), the addition of a non-templated overhang (27) or pseudo-templated transcription where the primase reiteratively transcribes a small homopolymeric template stretch (30).
Variation of DNA template, CTP, cap analog and enzyme concentration
The initially used conditions for the production of capped RNA oligonucleotides were 5 mM CTP, 1 mM cap analog, 10 μM DNA template CCCCGGGTCT25 (named dAC3) and 4 μM T7 DNA primase. Routinely, we obtained overall conversions of 81–96% after 21 h. In view of the possibility to obtain a higher percentage of longer products maintaining overall yields approaching 100%, the influence of DNA template, CTP, cap analog and enzyme concentration was studied.
The DNA template concentration was varied between 5 and 50 μM. CTP and cap analog concentrations were not varied in total independence because a higher proportion of CTP than cap analog will always be incorporated into the products. The ratio GpppA to cytidine in product GpppAC5 for instance is 5 to 1. We kept this ratio as a minimum and increased CTP and GpppA concentration together from the starting values 5 and 1 mM to 20 and 4 mM, respectively. Furthermore, we kept the cap analog concentration at 1 mM raising only the CTP concentration. Table 1 shows the results of optimization using cap analog GpppA, which were similar for 7MeGpppA reactions (not shown). Note that there is a difference in substrate conversion (81.2% versus 90.4%) between two identical reactions (0.01 mM DNA template and 5 mM CTP/1 mM GpppA, respectively), which reflect the variations we obtained between independent reaction series. Nevertheless product distribution was always very similar.
Table 1.
Influence of DNA oligonucleotide dAC3 (CCCCGGGTCT25), CTP and GpppA concentrations on T7 DNA primase reaction yield and product profile. Standard reaction conditions were used as given in the legend to Figure 1 corresponding to the presence of 250 pmol of cap analog substrate per pmol T7 DNA primase. The concentrations of DNA template and CTP/cap analog were then changed as given. Reactions were incubated for 21 h and analyzed by HPLC. Determination of yields of total product (pmol per pmol enzyme and percentage of substrate conversion) and of single products (percentage of total product) is explained in Material and Methods
| Variable | Concentration | Total product GpppACn | GpppAC | GpppAC2 | GpppAC3 | GpppAC4 | GpppAC5 | |
|---|---|---|---|---|---|---|---|---|
| (mM) | (pmol/pmol enzyme) | (%) | (% of total product) | |||||
| DNA template | 0.005 | 150.9 | 60.3 | 8.7 | 1.7 | 62.6 | 21.6 | 5.4 |
| 0.01 | 203.0 | 81.2 | 3.4 | 0.6 | 61.9 | 27.4 | 6.7 | |
| 0.02 | 239.8 | 95.9 | 0.6 | 0.5 | 53.1 | 37.1 | 8.6 | |
| 0.05 | 249.0 | 99.6 | 0 | 4.2 | 38.9 | 47.4 | 9.4 | |
| CTP/GpppA | 5/1 | 222.9 | 90.4 | 1.8 | 0 | 62.4 | 28.8 | 7.1 |
| 10/2 | 130.5 | 26.1 | 30.3 | 4.8 | 54.9 | 10.0 | 0 | |
| 20/4 | 47.5 | 4.7 | 77.7 | 8.7 | 13.6 | 0 | 0 | |
| 10/1 | 65.3 | 26.2 | 31.3 | 5.1 | 52.2 | 10.0 | 1.4 | |
| 20/1 | 25.2 | 10.1 | 58.2 | 7.7 | 29.3 | 4.7 | 0 | |
The increase in DNA template concentration up to 50 μM led to an increase in overall yield and product length. Thus 50 μM DNA template seems to be the concentration of choice. On the other hand, it has to be considered that template usage is much more efficient when lower concentrations are used. At 50 µM DNA template concentration, ca. 20 pmol RNA oligonucleotide are produced from one pmol DNA template, i.e. the template is recycled 20 times supposing that all T7 DNA primase molecules are active. This can be calculated from the amount of total product generated (249.1 pmol per pmol primase) and the amount of DNA of template used in the reaction (12.5 pmol per pmol primase). When 5 and 10 μM DNA template is used, initiation happens 126 and 80 times, respectively, at each template molecule.
Increasing CTP and cap analog concentration at the same time maintaining a ratio of 5 to 1 resulted in a decrease in total yield and product length, thus both the initiation and elongation part of the primase reaction seem to be affected drastically. When we increased the CTP concentration alone, inhibition was stronger regarding product formation in absolute terms (pmol product per pmol enzyme) whereas product distribution essentially stayed the same. Inhibition by CTP may be explained by the formation of a dead-end complex by non-productive binding between the enzyme and CTP (31) that prevents productive binding of the cap analog. Activity seems to be partially rescued by higher concentrations of cap analog. However, higher cap analog concentrations result in lower overall substrate conversion. In conclusion, CTP and cap analog concentrations were kept at 5 mM and 1 mM, respectively.
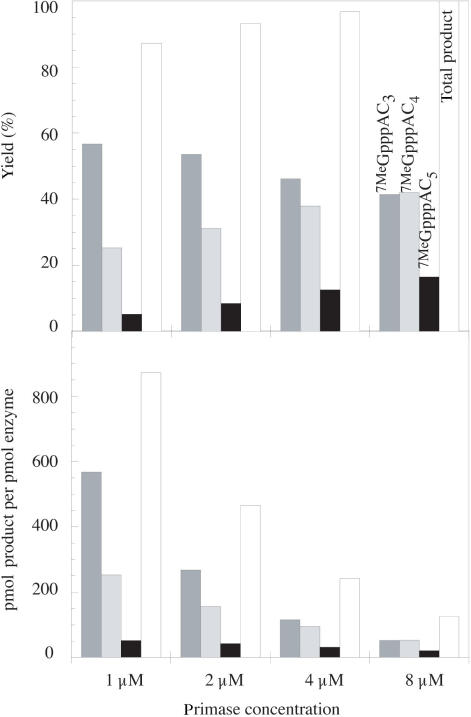
Enzyme concentration was varied between 1 and 8 μM. As shown in Figure 3, the overall yield and the length of formed products increase with enzyme concentration within the tested range (upper panel). Nevertheless, the efficiency of the enzymatic reaction decreases from 867.5 pmol per pmol enzyme at 1 μM to 125.0 pmol per pmol enzyme at 8 μM enzyme concentration (lower panel). In conclusion, 4 µM T7 DNA primase was kept as enzyme concentration for oligonucleotide production on a preparative scale and further optimization studies.
Figure 3.
Influence of T7 DNA primase concentration on reaction yield and product profile. Standard reaction conditions were used as given in the legend of Figure 1 except for T7 DNA primase concentration which was varied between 1 and 8 μM corresponding to the presence of 1000 and 125 pmol of cap analog substrate (7MeGpppA) per pmol T7 DNA primase. DNA template was dAC3 (CCCCGGGTCT25). Reactions were incubated for 21 h and analyzed by HPLC as described in Material and Methods. Yields in percentage conversion of cap analog into each single product (upper panel) and pmol product per pmol enzyme (lower panel) were determined as detailed in Material and Methods. Total product represents the sum of conversion into GpppAC3, GpppAC4 and GpppAC5. Amounts of GpppAC and GpppAC2 products were negligible under the tested reaction conditions.
Variation of DNA template length and reaction time
In order to produce a series of capped RNA oligonucleotides of different length, several DNA templates (C(6−n)GnGTCT25, 1 ≤ n ≤ 4) were used and the reaction time was varied. Table 2 shows that the use of templates dAC2 (C5GGTCT25) to dAC5 (C3GGGGGTCT25) varying the reaction time between 21 and 71 h allowed the production of capped oligonucleotides 7MeGpppAC to 7MeGpppAC9. In accordance with Figure 2, almost identical results were obtained with GpppA as cap analog (not shown). Highest yields (100 and 96.1%, respectively) were obtained when DNA templates dAC2 and dAC3 were used. The main products after 21 h corresponded to 7MeGpppAC2 (75.2%) and 7MeGpppAC3 (72.1%), respectively. As observed before, considerable amount of (n + 1) products (21.2 and 23.6%, respectively) and small amounts of (n + 2) products (3.6% and 4.2%, respectively) were formed. With DNA templates dAC4 and dAC5, the overall yield of the reactions were lower (78.1 and 55.7%, respectively). Yields could be increased to a certain limit (93.4 and 70.6%, respectively) by extending the reaction time to 48 h. When dAC4 was used, the main product after 21 h was 7MeGpppAC4 (44.7%) but the (n + 1) product 7MeGpppAC5 after 48 h (50.5%). The (n + 1) product 7MeGpppAC6 was the main product in the case of dAC5 after 21 h (30.4%) and remained so after 48 h (33.9%). Allowing the reaction using dAC5 to proceed further to 71 h led to a negligible increase in overall product yield and chain length. In conclusion, an adequate DNA template and reaction time of up to 48 h can be chosen to produce a desired spectrum of (7Me)GpppACn (1 ≤ n ≤ 9). The chain length distribution of the capped RNA oligonucleotides is wider when DNA templates with longer guanine sequences are used (dAC4 and dAC5).
Table 2.
Influence of DNA template length and reaction time on T7 DNA primase reaction yield and product profile. Standard reaction conditions were those given in the legend of Figure 1 corresponding to the presence of 250 pmol of cap analog substrate per pmol T7 DNA primase. DNA templates dAC2 to dAC5 correspond to CCCCCGGTCT25 to CCGGGGGTCT25, respectively. Reactions were incubated at 37°C and analyzed by HPLC as described in Material and Methods. Products are listed with the sequence following the cap 7MeG, e.g. AC stands for 7MeGpppAC. Yields of total product (pmol per pmol enzyme and percentage conversion of substrate) and of single products (percentage of total product) were determined as explained in Material and Methods
| DNA template | Reaction Time | Total product 7MeGpppACn | AC | AC2 | AC3 | AC4 | AC5 | AC6 | AC7 | AC8 | AC9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (h) | (pmol/pmol enzyme) | (%) | (% of total product 7MeGpppACn) | |||||||||
| dAC2 | 21 | 250.0 | 100 | 0 | 75.2 | 21.2 | 3.6 | 0 | 0 | 0 | 0 | 0 |
| dAC3 | 21 | 240.3 | 96.1 | 0 | 0 | 72.1 | 23.6 | 4.2 | 0 | 0 | 0 | 0 |
| dAC4 | 21 | 195.3 | 78.1 | 4.6 | 0 | 31.8 | 44.7 | 15.8 | 3.1 | 0 | 0 | 0 |
| 48 | 233.6 | 93.4 | 1.9 | 0.5 | 0 | 27.2 | 50.5 | 18.0 | 1.8 | 0 | 0 | |
| dAC5 | 21 | 139.2 | 55.7 | 17.8 | 6.5 | 4.9 | 0.4 | 21.1 | 30.4 | 15.7 | 3.1 | 0 |
| 48 | 176.6 | 70.6 | 9.9 | 3.5 | 2.7 | 0.3 | 19.3 | 33.9 | 22.7 | 6.5 | 1.2 | |
| 71 | 179.3 | 71.7 | 9.1 | 3.1 | 1.6 | 0 | 13.5 | 35.3 | 27.3 | 8.2 | 1.9 | |
Addition of fresh T7 DNA primase and CTP
Using dAC5, it was tested if yield and product chain length could be increased by adding fresh enzyme after 48 h and leaving the reaction to 71 h. Additionally, CTP was added to replenish free CTP thus trying to allow synthesis of longer products but to avoid inhibition by high CTP concentrations (see above). The addition of enzyme alone increased the overall yield of the reaction and product chain length only to a limited extend (76.3% substrate conversion versus 73.7% without addition of enzyme). When enzyme and CTP were added at the same time (the resulting CTP concentration was adjusted to 2 mM supposing that CTP had been consumed completely), the overall yield decreased to 68.0% substrate conversion but the product chain length was shifted to longer products. After 71 h, 7MeGpppAC8 represented 18% of total product instead of 8.2% without addition of enzyme and CTP. When the amount of added CTP was further increased (adjusted to 5 mM concentration), the reaction yield decreased further without increasing the product length in comparison to 2 mM. Therefore if longer products are desired, the addition of enzyme and CTP up to 2 mM is beneficial.
Synthesis of (7Me)GpppACn on a preparative scale
Large-scale reactions (200 μl reaction volume) were set up using the optimized conditions described above, but trying to use reasonable concentrations of enzyme and HPLC grade DNA template (4 and 10 μM, respectively). Globally, no significant differences in activity and product distribution were observed between small (20 μl) and large (200 μl) scale reactions (not shown). Parallel reactions were set up to further increase production scale. We used the DNA templates dAC2 and dAC3 for the production of (7Me)GpppAC2 and (7Me)GpppAC3, respectively, whereas dAC4 was used as the template for the production of (7Me)GpppAC4 to (7Me)GpppAC7. (7Me)GpppAC was purified as a minor product of the dAC4 reaction. The reaction times were chosen to ensure the best conversion rate of the starting material while avoiding excessive distribution of the products, especially for the shorter capped RNAs. After purification, products were lyophilized three times to remove the volatile buffer solution. Each compound was characterized by mass spectrometry and its purity confirmed by HPLC (Table 3).
The isolated yields of each product after lyophilization are given in Table 3. The best yields (42 and 52%) were obtained for the dAC3 reactions where also a good balance between conversion rate of the starting material and low dispersion of the products could be obtained. Slightly lower yields were obtained for the dAC2 reactions. As expected the global dispersion of the products for the dAC4 series was the widest with the advantage that this reaction afforded reasonable amounts of (7Me)GpppAC and (7Me)GpppAC4 to (7Me)GpppAC7 to be produced. Thus we were able to produce a range of short GpppA- or 7MeGpppA-capped RNA molecules with yields up to 52% corresponding to ca. 100 nmol (corresponding to ca. 20 μg) of product 7MeGpppAC3 starting from 200 nmol cap analog. The obtained yields may still be increased when higher DNA template and enzyme concentrations are used and, concerning longer products, when enzyme and CTP are added after 48 h (see above).
Chemical approaches (the advantage being that the sequence can be varied) for the synthesis of short-capped RNAs described in the literature gave isolated yields from 2 to 37%, as summarized by Koukhareva (8). Similar or superior results have been obtained, at least for the main products, using our enzymatic approach. Other strategies to produce GpppA-capped RNA oligonucleotides, which combine chemical synthesis of diphosphorylated RNA followed by enzymatic addition of the cap by commercially available guanylyltransferase (9), or use two enzymatic reactions (synthesis of triphosphate poly(rA) (32) followed by capping with guanylyltransferase (10)), produced radiolabeled products for which no product yields were given. Concerning one-pot enzymatic reactions, E. coli as well as bacteriophage T7, SP6 and T3 DdRps using their promoter sequences were reported to have produced GpppA-capped RNA in microgram range (12,13,16). Nevertheless 1 μg of one these long transcripts of ca. 2200 nucleotides (12) correspond to 65 fmol of capped RNA and thus they are useful for cellular transfection assays but not as purified substrates in enzymatic assays. Very recently, the T7 class II ϕ2.5 promoter (14) was successfully used with T7 DdRp to produce semi-purified GpppA-capped RNA transcripts of ca. 180 nucleotides. No product yields were given. The capped RNAs served as substrates in multiple-step, radioactive Flavivirus MTase assays (15). This approach offers the advantage that sequence-specific RNA molecules can be generated. It will be interesting to see if it allows production of sufficient amounts for wider biochemical, inhibition and crystallographic studies.
GpppA- and 7MeGpppA-capped RNAs as methyltransferase substrates
The purified capped RNA oligonucleotides were tested as substrates of Dengue 2′OMTase, expressed as recombinant N-terminal domain of protein NS5 (NS5MTaseDV) in E. coli (6). In contrast to the usual set-up of methyltransfer assays (15), we used non-radioactive AdoMet and RNA substrate. The reaction mixture was analyzed by reverse-phase HPLC including an on-line cleaning procedure without any additional sample treatment. Figure 4A shows the resulting profile after a 30-min reaction using GpppAC3 as substrate. The retention time of substrate GpppAC3 was 30.6 min. An additional peak was observed at 35.4 min. Its molecular mass was determined by mass spectrometry as being 1702.44 (m/z, [M−H]−) compared to 1688.24 (m/z, [M−H]−, see Table 3) of GpppAC3, thus one methyl group was transferred. In order to identify the receiving position, we digested the product at 35.4 min by a mix of nucleotide pyrophosphatase, phosphodiesterase I and calf intestine phosphatase rendering the corresponding nucleosides. The resulting mixture was analyzed by HPLC (Figure 4B, lower chromatogram). The comparison with standard compounds (upper chromatogram), which was verified by co-injection (not shown), allowed the identification of 2′O position of the adenosine as the methylated position. Thus, in accordance with earlier results using non-purified substrates (6), we found that the product of methyltransfer by NS5MTaseDV using substrate GpppAC3 corresponds to GpppA2′OMeC3. Note that the important delay in elution of GpppA2′OMeC3 in comparison to substrate GpppAC3 is caused by an increase in hydrophobicity due to the methylation of the 2′OH group combined with the use of a very shallow gradient (see figure legend). In order to see if methylation could also be achieved at the guanine-N7 position of GpppAC3, we used recombinant human mRNA cap N7MTase (33,34). The HPLC analysis of the reaction products (Figure 4C, lower chromatogram) in comparison to a control reaction using 7MeGpppAC3 (upper chromatogram) shows that GpppAC3 is indeed efficiently methylated at the guanine-N7 position. Note that in this case, a positive charge is generated upon methylation leading to a shorter elution time of the product (29.3 min) in comparison to the non-methylated substrate (30.7 min). Thus both methyltransfer reactions were readily detectable by our HPLC separation method. They can be easily analyzed in a quantitative way in terms of picomoles methyl group transferred by one picomole enzyme.
Figure 4.
Methyltransferase reactions using GpppAC3 as substrate. (A): HPLC profile of the NS5MTaseDV reaction mixture using GpppAC3 as substrate. Reaction conditions are given in Material and Methods. The crude enzymatic mixture was analyzed without sample treatment. The first section (in gray) indicates the removal of proteic material and remaining AdoMet by on-line cleaning on the pre-column (see Material and Methods). The gradient started after 5 min at 100% eluent A with an increase to 10% eluent B after 25 min reaching 30% after 45 min. (B): HPLC profile (lower chromatogram) of nucleoside product mixture after enzymatic digestion of the product generated by methylation of GpppAC3 by NS5MTaseDV (see panel A at 35.4 min). Enzymatic digestion was done using a mix of nucleotide pyrophosphatase, phosphodiesterase I and calf intestine phosphatase. The upper chromatogram shows a mixture of standard compounds. No on-line cleaning was used. The gradient started after 5 min at 100% eluent A with an increase to 10% eluent B after 25 min and to 30% after 45 min. (C): HPLC profile of the human mRNA cap N7MTase reaction mixture using GpppAC3 as substrate (lower chromatogram) in comparison to 7MeGpppAC3 being used as a control substrate (upper chromatogram). Reaction conditions are given in Material and Methods. The crude enzymatic mixtures were analyzed without sample treatment as described in the legend of panel A.
CONCLUSION
We were able to produce and purify a series of GpppA-capped RNA oligonucleotides of different length, (7Me)GpppACn (1 ≤ n ≤ 9), in a simple one-step enzymatic reaction using a recombinant bacteriophage T7 DNA primase fragment. Our yields (Table 2) correspond well to the yield of 75% substrate conversion obtained upon production of 7MeGpppACC using the full-length T7 DNA primase-helicase (17). In our study, we went further in optimizing the production of longer molecules using just the T7 DNA primase fragment. The isolated yields of products (7Me)GpppACn (1 ≤ n ≤ 7) obtained after large-scale synthesis and purification, in average 99% (Table 3), are superior or comparable to other methods, the additional advantage of the method presented here being the simplicity at least for biochemical laboratories with protein expression facilities. Moreover, this method allows the production of highly purified and easily quantified capped RNAs of different lengths for (1) non-radioactive activity assays, (2) studies of methyltransferase specificity in relation to substrate length and (3) co-crystallization studies of a variety of eukaryotic and viral methyltransferases and guanylyltransferases.
We showed that our purified capped RNA oligonucleotides serve as substrate of a mRNA cap N7MTase and a 2′OMTases of human origin and Dengue virus, respectively. Both methyltransfer reactions were monitored by a non-radioactive, quantitative HPLC assay.
In addition, our optimized method allows also the production of di- or triphosphate RNA oligonucleotides (using ATP or ADP instead of GpppA), which are valuable tools for the biochemical and structural studies of RNA triphosphatases and guanylyltransferases. Besides, preliminary tests have shown that the production of GpppACUCn and possibly other RNA oligonucleotides varying at positions 3 and further are attainable (not shown).
ACKNOWLEDGEMENTS
We wish to thank Charles C. Richardson (Harvard Medical School, Boston, USA) and Aaron J. Shatkin (Center of Advanced Biotechnology and Medicine, Piscataway, USA) for kindly providing the T7 DNA primase expression vector and the cDNA of human mRNA cap N7MTase, respectively. We thank David Karlin for the production of the T7 DNA primase as well as Marie-Pierre Egloff for purified NS5MTaseDV. This study was supported, in part, by the integrated project VIZIER (LSHG-CT-2004-511960) of the European Union 6th PCRDT. F.P. was supported by a postdoctoral fellowship from the Centre National de la Recherche Scientifique (CNRS), France. Funding to pay the Open Access publication charge was provided by CNRS.
REFERENCES
- 1.Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 3.Gould EA, de Lamballerie X, Zanotto PM, Holmes EC. Origins, evolution, and vector/host coadaptations within the genus Flavivirus. Adv. Virus Res. 2003;59:277–314. doi: 10.1016/s0065-3527(03)59008-x. [DOI] [PubMed] [Google Scholar]
- 4.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 2004;10:S98–109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 5.Benarroch D, Egloff M-P, Mulard L, Guerreiro C, Romette J-L, Canard B. A structural basis for the inhibition of the NS5 Dengue virus mRNA 2′-O-methyltransferase domain by ribavirin 5′-triphosphate. J. Biol. Chem. 2004;279:35368–35643. doi: 10.1074/jbc.M400460200. [DOI] [PubMed] [Google Scholar]
- 6.Egloff M-P, Benarroch D, Selisko B, Romette J-L, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jemielity J, Heinonen P, Lonnberg H, Darzynkiewicz E. A novel approach to solid phase chemical synthesis of oligonucleotide mRNA cap analogs. Nucleosides, Nucleotides & Nucleic Acids. 2005;24:601–605. doi: 10.1081/ncn-200061922. [DOI] [PubMed] [Google Scholar]
- 8.Koukhareva II, Lebedev AV. Chemical route to the capped RNAs. Nucleosides, Nucleotides & Nucleic Acids. 2004;23:1667–1680. doi: 10.1081/NCN-200031492. [DOI] [PubMed] [Google Scholar]
- 9.Brownlee GG, Fodor E, Pritlove DC, Gould KG, Dalluge JJ. Solid phase synthesis of 5′-diphosphorylated oligoribonucleotides and their conversion to capped m7Gppp-oligoribonucleotides for use as primers for influenza A virus RNA polymerase in vitro. Nucleic Acids Res. 1995;23:2641–2647. doi: 10.1093/nar/23.14.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuman S. Functional domains of vaccinia virus mRNA capping enzyme. Analysis by limited tryptic digestion. J. Biol. Chem. 1989;264:9690–9695. [PubMed] [Google Scholar]
- 11.Lockless SW, Cheng H-T, Hodel AE, Quiocho FA, Gershon PD. Recognition of capped RNA substrates by VP39, the vaccinia virus-encoded mRNA cap-specific 2′-O-methyltransferase. Biochemistry. 1998;37:8564–8574. doi: 10.1021/bi980178m. [DOI] [PubMed] [Google Scholar]
- 12.Holden KL, Harris E. Enhancement of dengue virus translation: role of the 3′ untranslated region and the terminal 3′ stem-loop domain. Virology. 2004;329:119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Yun SI, Kim SY, Rice CM, Lee YM. Development and application of a reverse genetics system for Japanese encephalitis virus. J. Virol. 2003;77:6450–6465. doi: 10.1128/JVI.77.11.6450-6465.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman TM, Wang G, Huang F. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 phi 2.5 promoter. Nucleic Acids Res. 2004;32:e14. doi: 10.1093/nar/gnh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West nile virus 5′-cap structure is formed by sequential Guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contreras R, Cheroutre H, Degrave W, Fiers W. Simple, efficient in vitro synthesis of capped RNA useful for direct expression of cloned eukaryotic genes. Nucleic Acids Res. 1982;10:6353–6362. doi: 10.1093/nar/10.20.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuo H, Moriguchi T, Takagi T, Kusakabe T, Buratowski S, Sekine M, Kyogoku Y, Wagner G. Efficient synthesis of 13C,15N-labeled RNA containing the cap structure m7GpppA. J. Am. Chem. Soc. 2000;122:2417–2421. [Google Scholar]
- 18.Kato M, Ito T, Wagner G, Richardson CC, Ellenberger T. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol. Cell. 2003;11:1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- 19.Frick DN, Baradaran K, Richardson CC. An N-terminal fragment of the gene 4 helicase/primase of bacteriophage T7 retains primase activity in the absence of helicase activity. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7957–7962. doi: 10.1073/pnas.95.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawson RMC, Elliott DC, Elliott WH, Jones KM. Data for Biochemical Research. Oxford: Oxford University Press; 1986. [Google Scholar]
- 21.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 22.Mendelman LV, Richardson CC. Requirements for primer synthesis by bacteriophages T7 63-kDa gene 4 protein. J. Biol. Chem. 1991;266:23240–23250. [PubMed] [Google Scholar]
- 23.Kusakabe T, Richardson CC. Gene 4 DNA primase of bacteriophage T7 mediates the annealing and extension of ribo-oligonucleotides at primase recognition sites. J. Biol. Chem. 1997;272:12446–12453. doi: 10.1074/jbc.272.19.12446. [DOI] [PubMed] [Google Scholar]
- 24.Pompon A, Lefebvre I, Imbach J-L, Kahn S, Farquhar D. Decomposition pathways of the mono- and bis(pivaloyloxy methyl) esters of azidothymidine 5′-monophosphate in cell extract and in tissue culture medium: an application of the ‘on-line IRSP-cleaning’ HPLC technique. Antivir. Chem. Chemother. 1994;5:91–98. [Google Scholar]
- 25.Pompon A, Lefebvre I, Imbach JL. ‘On-line internal surface reversed-phase cleaning’: the direct HPLC analysis of crude biological samples. Application to the kinetics of degradation of oligonucleotides in cell culture medium. Biochem. Pharmacol. 1992;43:1769–1775. doi: 10.1016/0006-2952(92)90709-r. [DOI] [PubMed] [Google Scholar]
- 26.Frick DN, Richardson CC. Interaction of bacteriophage T7 gene 4 primase with its template recognition site. J. Biol. Chem. 1999;274:35889–35898. doi: 10.1074/jbc.274.50.35889. [DOI] [PubMed] [Google Scholar]
- 27.Kusakabe T, Richardson CC. Template recognition and ribonucleotide specificity of the DNA primase of bacteriophage T7. J. Biol. Chem. 1997;272:5943–5951. doi: 10.1074/jbc.272.9.5943. [DOI] [PubMed] [Google Scholar]
- 28.Frick DN, Kumar S, Richardson CC. Interaction of ribonucleotide triphosphates with the gene 4 primase of bacteriophage T7. J. Biol. Chem. 1999;274:35899–35907. doi: 10.1074/jbc.274.50.35899. [DOI] [PubMed] [Google Scholar]
- 29.Qimron U, Lee SJ, Hamdan SM, Richardson CC. Primer initiation and extension by T7 DNA primase. EMBO J. 2006;25:2199–2208. doi: 10.1038/sj.emboj.7601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacques JP, Kolakofsky D. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev. 1991;5:707–713. doi: 10.1101/gad.5.5.707. [DOI] [PubMed] [Google Scholar]
- 31.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. 3rd edn. Oxford: Portland Press; 2004. [Google Scholar]
- 32.Venkatesan S, Moss B. Donor and acceptor specificities of HeLa cell mRNA guanylyltransferase. J. Biol. Chem. 1980;255:2835–2842. [PubMed] [Google Scholar]
- 33.Pillutla RC, Yue Z, Maldonado E, Shatkin AJ. Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J. Biol. Chem. 1998;273:21443–21446. doi: 10.1074/jbc.273.34.21443. [DOI] [PubMed] [Google Scholar]
- 34.Saha N, Schwer B, Shuman S. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 1999;274:16553–16562. doi: 10.1074/jbc.274.23.16553. [DOI] [PubMed] [Google Scholar]