Abstract
Small interfering RNA (siRNA) induces sequence-specific post-transcriptional gene silencing in mammalian cells. Different efficacy of each siRNA is considered to result from sequence preference by protein components in RNAi. To obtain mechanistic insight into siRNA functionality, here we describe a complete data set of siRNA activities targeting all possible position of a single mRNA in human cells. Seven hundred and two siRNAs covering open reading frame of enhanced green fluorescent protein mRNA ( 720 bases) were examined with minimized error factors. The most important finding is that specific residues at every third position of siRNAs greatly influence its RNAi activity; the optimized base composition at positions 3n + 1 (4,7,10,13,16,19) in siRNAs have positive effects on the activity, which can explain the waving siRNA activity with 3 nucleotides (nt) periodicity in the sequential positions of mRNAs. Since there was an obvious correlation between siRNA activity and its binding affinity to TRBP, a partner protein of human Dicer, the 3-nt periodicity might correlate with the affinity to TRBP. As an algorithm (‘siExplorer’) developed by this observation successfully calculated the activities of siRNAs targeting endogenous human genes, the 3-nt periodicity provides a new aspect unveiling siRNA functionality.
INTRODUCTION
The double-stranded RNA (dsRNA) interference (RNAi) method results in sequence-specific gene silencing and as such is an invaluable tool for rapid genetic analysis in many eukaryotic systems, including plants, fungi, insects, protozoans and mammals (1–6). In mammalian cells, such sequence-specific gene silencing has been successfully performed by utilizing short 21–23 nucleotide (nt) dsRNAs known as small interfering RNAs (siRNAs) (7–10) that escape from the dsRNA-induced antiviral pathways (11–14). To date, many endogenous genes in mammalian cells have been characterized using the siRNA technique (1,2,15). Instant knockdown of mRNAs as well as non-coding RNAs by siRNAs have now become a general tool in many fields of life science. siRNA also has a great potential to be used in medical applications. Together with development of delivery technique in vivo, several examples of animal experiments using siRNAs have been reported (16,17).
Fundamental studies on RNAi pathway to explain siRNA functionality are supporting rapid development of siRNA technology and its application. The guide strand of siRNA complementary to the target mRNA is loaded into RNA-induced silencing complex (RISC) (7,18). RISC recognizes and determines the target site for cleavage by hybridizing the guide strand of siRNA. Argonaute 2 has been identified as a catalytic subunit of RISC for mRNA cleavage (19–26). In many organisms, RNAi machinery is involved in translational repression mediated by micro(mi)RNAs, which are natural substrates for RNAi machinery. miRNA is a single strand non-coding RNA that matures from precursor RNA with short hairpin structure (pre-miRNA). RNase III-nuclease Dicer processes pre-miRNA to produce double-stranded miRNA and selects one strand to be a guide strand serving as a translational suppressor in RISC-like miRNP (27–29). Thus, the two processed strands are destined to distinct pathways. According to biochemical studies in Drosophila melanogaster, RISC assembly is initiated with siRNA recognition by Dicer2 and its partner protein R2D2 to form RISC-loading complex (RLC) (30). During this complex formation, it is known that relative thermodynamic instability at the 5′ end of siRNA results in asymmetric recognition of siRNA by Dicer-2–R2D2 complex. This asymmetric recognition is believed to determine which strand to serve as a guide strand in both miRNA and siRNA (31,32). In fact, many knockdown experiments using large number of siRNAs show that efficacious siRNAs have unstable 5′ end of their guide strands (33). On the basis of this observation, algorithms that design efficacious siRNAs were developed by several groups (34–39). In addition, assuming dicing is coupled with RISC formation, two groups independently succeeded in designing siRNAs to be good substrates for Dicer with potent efficacy at sub-nanomolar level (40,41).
RNAi pathway consists of many steps, including siRNA recognition by Dicer with its partner protein, unwinding siRNA, RISC assembly, target selection, target cleavage by Argonaute protein, product release and RISC recycling (27,42). Efficacy of siRNA is supposed to be determined by total efficiency of each step. In the initial step of RISC formation, strand selection of siRNA by asymmetric recognition of Dicer and its partner protein is a dominant role. In human, transactivating response (TAR) RNA-binding protein (TRBP) was identified as a partner protein for human Dicer (43,44). TRBP has three double-stranded RNA-binding domains (dsRBDs), two of which have a pivotal role in recognizing dsRNA (45) and the third domain at the C-terminus is required for interaction with human Dicer. Molecular mechanism on how Dicer–TRBP complex recognizes siRNA asymmetry is not fully understood. Recently, another dsRBD protein, PACT, was identified as a partner protein for human Dicer (46).
To obtain general rules embedded in siRNA sequences that provide a clue to understand molecular mechanism in RNAi pathway, we precisely determined RNAi activities of 702 siRNAs with complete double-stranded form targeting all possible position of ORF of EGFP mRNA (720 bases) in HeLa cells and analyzed relationship between siRNA sequences and their activities. The data clearly showed that efficacy of siRNAs is associated with the thermodynamic instability of the guide strand 5′ end as proposed. In addition, we found total base composition and specific residues at every third position of siRNAs to be involved in the RNAi activity. We have developed an algorithm on the basis of this observation that successfully predicts the siRNA activities targeting endogenous genes, suggesting that the three nucleotides periodic efficacy of siRNA was shown to be a universal phenomenon in human RNAi pathway.
MATERIALS AND METHODS
siRNA preparation
shRNA was transcribed by T7 RNA polymerase and then converted into siRNA by limited digestion with ribonuclease T1 (47) (see supporting information). We prepared 702 species of siRNAs that target all positions of the 720 nt EGFP coding region. The siRNA sequences targeting glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (1008 nt) start from nucleotide positions 8, 23, 33, 43, 53, 65, 93, 109, 120, 132, 145, 167, 198, 231, 241, 304, 337, 349, 368, 383, 402, 409, 448, 473, 491, 519, 539, 587, 595, 613, 636, 655, 676, 710, 734, 746, 761, 777, 789, 798, 802, 817, 845, 863, 889, 916, 941, 950, 974 and 989 for the experiment shown in Figure 7A, and sequential nucleotide positions 950–974 for the experiment shown in Figure 7B. The siRNA sequences targeting β-catenin mRNA start from nucleotide positions 579, 598, 654, 1224, 1694, 1681, 1898, 2101, 2219 and 2254 (Figure S4). For fluorescence correlation spectroscopy (FCS), 3′-TAMRA (6-carboxy-tetramethylrodamine)-labeled passenger strands for EGFP416 and EGFP64 were chemically synthesized and purified by HPLC (Hokkaido System Science, Inc.). Four nanomoles of TAMRA-labeled RNA was annealed with equal amount of its non-labeled guide strand at 37°C for 30 min in 20 μl of 30 mM HEPES-KOH (pH 7.8), 100 mM potassium acetate and 2 mM magnesium acetate, which was then purified by non-denaturing gel.
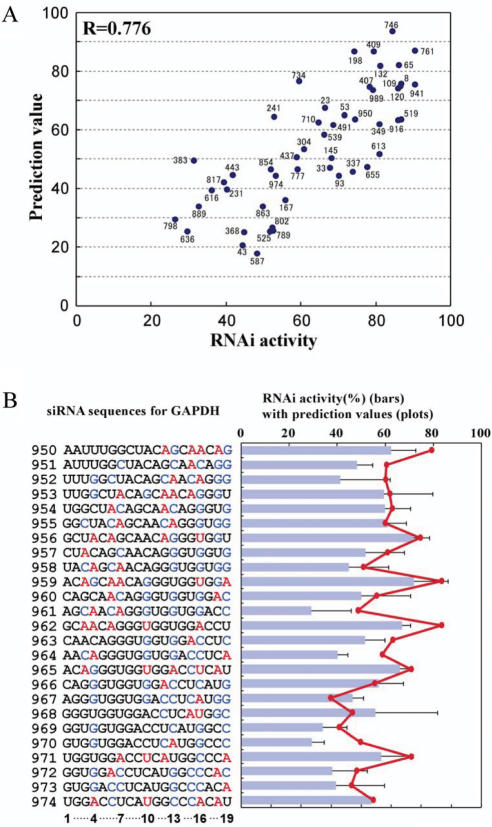
Figure 7.
Prediction of siRNA activities for endogenous gene. (A) Scatter plot for 50 GAPDH–siRNA activities versus their prediction values. The correlation coefficient is 0.776 (P = 3.4 × 10−11). Number near the spot corresponds to the passenger strand 5′ end position of each siRNA in GAPDH mRNA. (B) Prediction of sequential siRNA activities for GAPDH. The nt sequences show the sense strand of GAPDH–siRNAs for positions 950–974. The color code for bases represent bases that have positive or negative effects on the activity as described in the legend of Figure 3B. siRNA activities (%) for GAPDH are shown in bars. The prediction value calculated for each siRNA is plotted in red.
Expression and purification of human TRBP
The plasmid pGSX–TRBP (48) was kindly provided by Dr Joo Yong Lee (The Catholic University of Korea). The coding region of human TRBP in pGSX–TRBP was amplified using primers 5′-cgcggatcccatgagtgaagaggagcaaggct-3′ and 5′-gcagatcgtcagtcagtcacgatg-3′. The product was cloned into EcoRI/HindIII site of pET-21b. The recombinant TRBP with C-terminal his-tag was induced by 100 μM IPTG and expressed at 25°C for overnight. Then, it was purified by Ni-charged Hi-trap chelating column according to the manufacturer's instruction (GE healthcare).
Cell culture and transfection
HeLa cells stably expressing Hyg/EGFP were kindly provided by Dr M. Miyagishi (University of Tokyo). Hyg/EGFP gene was derived from pHygEGFP (Clontech), which expresses EGFP fused with a hygromycin-resistance gene. The cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS), 100 unit/ml penicillin, 100 μg/ml streptomycin and 100 μg/ml hygromycin. About 7.5 × 104 cells were inoculated in 6-well plates and cultivated for 48 h. The medium was then changed to 1 ml OPTI-MEM I (Gibco) without hygromycin. The cells were transfected with 10 μmol siRNAs for EGFP (f.c. 10 nM) by using 10 μl Oligofectamine (Invitrogen) and then incubated for 4 h. Thereafter, 500 μl Dulbecco's modified Eagle's medium with 30% FBS was added. In the case of GAPDH, normal HeLa cells were transfected with 25 μmol siRNAs (f.c. 25 nM).
Flow cytometry and immunofluorescence
Seventy-two hours after the transfection, the cells were trypsinized and washed twice with cold PBS, and resuspended in Hanks’ buffer containing 0.1% sodium azide and 0.2% BSA. EGFP fluorescence was measured by flow cytometry using FACScalibur (Becton Dickinson). GAPDH was measured by fixing the washed cells with 3% paraformaldehyde for 1 h, permeabilizing with 0.2% Triton X-100 in PBS for 2 min, followed by the addition of mouse anti-GAPDH or anti-β-catenin monoclonal antibodies (Chemicon). After a 30 min incubation at room temperature, the cells were washed three times with PBS containing 1% BSA and then FITC-conjugated anti-mouse antibodies (Zymed laboratories) were added and incubated for 30 min. The cells were washed three times with PBS containing 1% BSA and resuspended in Hanks' buffer/0.2% BSA. The FITC fluorescence was measured by flow cytometry.
Fluorescence correlation spectroscopy (FCS)
FCS measurements were performed using a MF20 single molecule fluorescence detection system (Olympus). Helium–Neon laser (543 nm) was used for the detection of TAMRA-labeled siRNA. TAMRA-labeled siRNA or single-stranded (ss)RNA (1 nM) was mixed with different concentrations (0–200 nM) of recombinant human TRBP in 25 μl mixture consisting of 50 mM HEPES-KOH (pH 7.8), 100 mM KCl, 5 mM MgCl2 and 7mM 2-mercaptoethanol. For the competition analysis, non-labeled competitor siRNA (5 nM) was added to the mixture in the same buffer and the concentration of TRBP was fixed at 150 nM. Competitor siRNA sequences targeting EGFP mRNA start from nt positions 8, 25, 37, 50, 64, 99, 123, 166, 176, 242, 264, 299, 359, 371, 383, 416, 417, 418, 450, 476, 485, 532, 591, 617, 626, 662 and 701. Data acquisition time was 3 or 5 s and measurements were repeated 5 or 6 times per well.
Thermodynamic energy calculation of 5′ ends of siRNAs
Thermodynamic parameters for the second 3′ dangle base, the first 3′ dangle base and RNA duplexes were obtained from the literatures (49–51). For each of 702 siRNAs targeting EGFP mRNA, thermodynamic stability (kcal/mol) of four base pairs in the passenger or guide strand 5′ end with 2-nt 3′-overhang was calculated. Stability differences were obtained by the value of the guide strand minus the value of the passenger strand. All calculated values for 702 siRNAs are shown in Table S1.
RESULTS
Activity distribution of 702 siRNAs that target EGFP mRNA
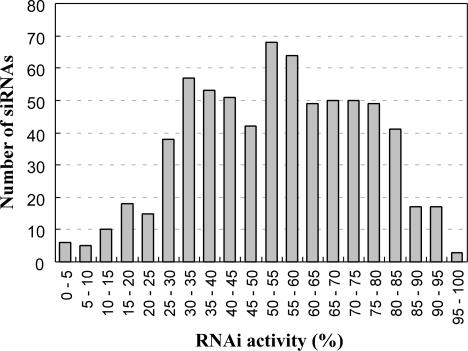
We have recently developed a method for synthesizing siRNAs derived from in vitro transcribed short hairpin (sh)RNA (47) (supplementary information, Figure S1). Seven hundred and two different species of siRNAs that target all the nucleotide positions of EGFP mRNA (720 nt) were prepared with high quality (Figure S2). If siRNA has an inverted repeat in its sequence, it is difficult to obtain dsRNA by simple annealing procedure of both strands that are synthesized chemically, because each strand tends to form intra-molecular hairpin-like structure. In this method, since siRNAs are generated from shRNA, all siRNAs are prepared as complete double-stranded form (47) (Figure S2). The prepared siRNAs consist of a 19-nt double-stranded region corresponding to the target sequence of mRNA with 2-nt overhangs at both 3′ ends. The quality of 702 siRNAs was confirmed by mass spectrometry and polyacrylamide gel electrophoresis (Figure S2). Each siRNA (10 nM) was then introduced into HeLa cells that stably express EGFP. The fluorescence of EGFP was measured by flow cytometry 72 h after the siRNA transfection. The rate with which EGFP fluorescence was decreased by siRNA knockdown was quantified as the RNAi activity (the best and the worst activities are defined as 100 and 0%, respectively). The data obtained in this experiment were highly reproducible. The RNAi activities of the 702 siRNAs show nearly normal probability distribution (Figure 1). Large majority of siRNAs show middle activities and fewer ones show very high or very low activities. IC50 value of the most efficacious siRNA was estimated as about 0.1 nM (data now shown). The average activity of 702 siRNAs is 53.75% and the standard deviation is 20.6. This is the first demonstration of a complete set of siRNA activities targeting all possible positions in ORF of a certain mRNA.
Figure 1.
Distribution histogram of 702 siRNA activities that target EGFP mRNA. RNAi activities are described as a percentage of decreased EGFP fluorescence by siRNA knockdown. The average activity of 702 siRNAs is 53.75% and the standard deviation is 20.6.
Thermodynamic instability of the guide strand 5′ end of siRNA involved in its activity
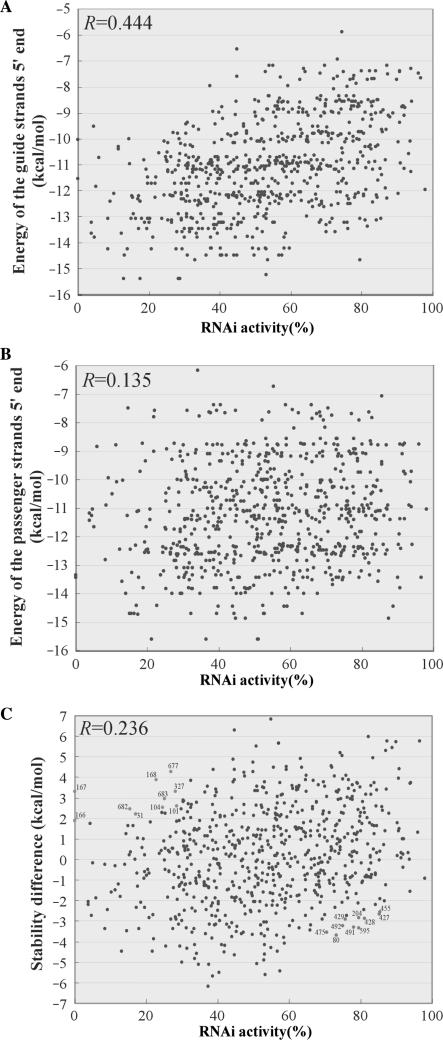
It is known that Drosophila Dicer-2 with its partner protein recognizes siRNA asymmetrically by sensing relative thermodynamic instability at the 5′ ends of both strands. This asymmetric recognition is supposed to be the fundamental principle of the strand selection for siRNAs (31,32). To investigate whether the complete set of siRNA activities in this study actually correlate with the instability of the guide strand 5′ end, we calculated thermodynamic energy of both 5′ ends for 702 siRNAs (see Materials and Methods and Table S1). The thermodynamic stability of the guide or passenger strand 5′ end for each siRNA is plotted with its RNAi activity (Figures 2A,B). As expected, there is an apparent correlation between instability of the guide strand 5′ end and its RNAi activity with a correlation coefficient (R value) 0.444 (P = 2.6 × 10−35) (Figure 2A). Meanwhile, as shown in Figure 2B, there is little correlation between the instability of passenger strand 5′ end and its RNAi activity (R value is 0.135, P = 3.4 × 10−4). If human Dicer with its partner (TRBP) senses the stability of the passenger strand 5′ end in a positive manner, the correlation coefficient should be a negative value in this plot. In addition, to examine relative stability of both strands, we made another plot (Figure 2C) for the RNAi activity against the energy difference for each siRNA which was calculated by the differential energy of the guide and the passenger strand 5′ ends (Table S1). Although it is a positive correlation, the plots are still dispersed in the graph with R factor 0.236 (P = 2.4 × 10−10). These results clearly illustrated that the instability of the guide strand 5′ end is a major factor contributing to the asymmetrical strand selection.
Figure 2.
Scatter plots for the knockdown activities of the 702 EGFP siRNAs versus thermodynamic stability of both ends in siRNAs. (A) Scatter plot for the activities of 702 siRNAs versus thermodynamic energy of the guide strand 5′ end. The correlation coefficient (R) is 0.444 (P = 2.6 × 10−35). (B) Scatter plot for the activities of 702 siRNAs versus thermodynamic energy of the passenger strand 5′ end. The correlation coefficient (R) is 0.135 (P = 3.4 × 10−4). (C) Scatter plot for the activities of 702 siRNAs versus stability difference (g.s. − p.s.) of both strands. The correlation coefficient (R) is 0.236 (P = 2.4 × 10−10). Blue spots represent 10 siRNAs that exhibit less stable guide strand 5′ end but show lower activity (less than 30). Red spots represent 10 siRNAs that exhibit more stable guide strand 5′ end but show higher activity (more than 70). Number near the spot corresponds to the passenger strand 5′ end position of each siRNA in EGFP mRNA.
Base composition of siRNA and its activity
To elucidate the other factors in siRNA sequence to be involved in its activity, we next examined base composition of each siRNA. The base composition of each siRNA was represented as the value B (base composition value), which was calculated by Equation (1) given below, where NX stands for the number of specific bases (X = A, U, G or C) in the siRNA and PX stands for the coefficient given to each base (X = A, U, G or C). Summation of NX and PX must be 19 and 1, respectively.
 |
1 |
The correlation between the B values obtained with various PX values for each base and the RNAi activities of all the siRNAs were then analyzed to calculate R values. This analysis was completely carried out by examining all possible combination of PX values changing at 0.05 steps, showing the R values ranging from −0.601 to 0.601 (Figures 3A,B and S3A–S in supplementary information). To obtain a higher R value between the B value and the RNAi activity, we found that PA + PU has to be greater than PG + PC, PA has to be greater than PU, and PG has to be greater than PC (Figures 3A,B and S3A–S). The best coefficients to achieve the highest R factor (0.601) for each base was found to be PA:PU:PG:PC = 0.4:0.35:0.15:0.1 (Figure 3A). The worst coefficients to yield the lowest R factor (−0.601) was found to be PA:PU:PG:PC = 0.1:0.15:0.35:0.4 (Figure 3B). The results showed that the RNAi activity turned out to correlate closely with the total base composition of siRNA sequence.
Figure 3.
Effect of the total base composition of 702 siRNAs on their ability to silence EGFP expression. Correlation between the total base composition of the 702 EGFP siRNAs and their activities. The correlation coefficients are plotted against the different B values that were obtained by changing the coefficients for each base in Equation (1). (A) Coefficients for A + U = 0.75, G + C = 0.25. (B) Coefficients for A + U = 0.25, G + C = 0.75. Other graphs with different coefficients are shown in Figure S3A–S.
Periodical effect of every third position in siRNA involved in RNAi activity
We next analyzed the effect of the residues at certain positions in the siRNA on its RNAi activity. At each position (1–19) of the 702 siRNAs, the R values between the best PX values (PA, PU, PG and PC are 0.4, 0.35, 0.15 and 0.1) and their activities (1-nt analysis) are plotted in Figure 4A. For example, if the position 1 of a certain siRNA has A-base, its PX value is 0.4. The result shows that Px values for positions 4, 7, 10, 13, 16 and 19 (i.e. the 3n + 1 positions, where n = counting number) correlate relatively well with the activity to yield positive R factor, while the Px values for positions 2, 5, 8, 11, 14 and 17 (i.e. the 3n + 2 positions) do not correlate with the activity (R factors are nearly 0). Positions 3, 6, 9, 12, 15 and 18 (i.e. the 3n positions) show R factors that fall between the R factors of the 3n + 1 and 3n + 2 positions. In contrast, we also examined the correlation between the B values calculated from 18-nt positions for 702 siRNAs in which each specific position had been systematically excluded and their activity (18-nt analysis) (Figure 4A, bars). The resulting R factors arising from this 18-nt analysis showed a clear negative relationship with the R factors of the 1-nt analysis at each position (Figure 4A, plots). Thus, the R factors of the 18-nt analysis for 702 siRNAs that lack a residue at position 3n + 1 show a significant reduction, the R factors from the data omitting position 3n show a mild reduction, and the R factors from the data omitting position 3n + 2 show no reduction. Position 1 does not strongly correlate with the activity.
Figure 4.
Effect of the base position of 702 siRNAs on their ability to silence EGFP expression. (A) Effect of certain positions in the 702 siRNAs on their activities. The correlation coefficients between the B values obtained from 1-nt at each position of 702 siRNAs and their activities are plotted in red. The correlation coefficients arising from the correlation analysis of the B values obtained from 18-nt of siRNAs in which each specific position is systematically excluded and the activities are shown in bars. Nucleotide positions represent the passenger strand of the siRNA. (B) Effect of different bases at each specific nucleotide position in the 702 siRNAs on their activities. The base-specific values (one or zero) were obtained as described in the text. The correlation coefficients between the base-specific values of 702 siRNAs and their activities are plotted for each base (A in red, U in pink, G in green and C in blue). Bases on the top of the graph that have positive effects on the activities are shown in red (beyond 0.2) or in orange (beyond 0.1) at the top of the graph. Bases on the bottom of the graph with negative effects on the activities are shown in blue (under −0.2) or in light blue (under −0.1) at the bottom of the graph.
The effect of particular bases at each nucleotide position on the RNAi activity was further assessed by correlation analysis (Figure 4B). For example, in the case of analysis for A-base, if the position 1 of a certain siRNA has A-base, its base-specific value is counted as 1. If it is a base other than A-base (U, C or G), its value is counted as 0. The base-specific values (one or zero) for each base against each specific position of 702 siRNAs were obtained. Then, we examined correlation analysis of the base-specific value (one or zero) for each siRNA against its activity (Figure 4B). A-base was found to be the most effective base for the RNAi activity, whereas G- and C-bases confer the negative effect. A-base at positions 3n and 3n + 1 shows the positive effect. U-base at positions 3n + 1 shows the positive effect, whereas U-base at positions 3n + 2 shows the negative effect. Namely, A3, A4, A6, A7, U10, A12, A13, A15, W(A&U)16, A18 and A19, have positive effects on the RNAi activity. In particular, A19 has the strongest effect on the activity. G- and C-bases at positions 3n and 3n + 1, namely, C3, G4, C6, C7, G10, C12, S(G&C)13, C15, S16, G18 and S19, are associated with a negative effect on the RNAi activity. No strong correlation with the activity was found for any of the bases in positions 3n + 2, which indicates that any of the bases can be present at positions 3n + 2 without influencing the activity of the siRNA. These results are consistent with the periodical effect of every third position of siRNA found in Figure 4A. As a result of this experimentation, the ideal siRNA sequence that would yield the most efficient activity was found to be 5′-NNAANAANNUNAANAWNAA-3′, although total base composition should be considered as another factor.
As shown in Figure 5C, the RNAi activities of siRNAs targeting sequential positions (1–50) in EGFP mRNA clearly showed periodical fluctuation at every third position. This phenomenon can be explained by the periodic efficacy of every third position in siRNAs. The 3-nt periodicity of waving RNAi activities is well observed in mRNA regions where A and U frequently appear at every third position, such as in positions 1–50, 110–160, 270–300 and 450–550 (Table S1). This periodical effect of every third position of siRNA implies the molecular mechanism of siRNA functionality in RNAi pathway.
Figure 5.
Comparison of the sequential knockdown activities of the 702 siRNAs targeting EGFP with the prediction values. (A) Correlation between siRNA activities and the B values (macro effect). siRNA activities (%), which are the rate of EGFP fluorescence reduction, are shown by bars. The B values for 702 siRNAs are plotted in blue. The correlation coefficient of the RNAi activity against the B values is 0.601 (P = 2.6 × 10−70). (B) The prediction values calculated by the algorithm are plotted in red. The correlation coefficient of the RNAi activities against the prediction values is 0.726 (P = 5.9 × 10−116). (C) Predicting periodical fluctuation of sequential siRNA activities. The nucleotide sequences show the passenger strand of EGFP siRNAs for positions 1–50. The colored bases represent bases that have positive or negative effects on the activity according to the analysis shown in Figure 3B. The RNAi activities (%) of the siRNAs for EGFP are shown in bars. The B value and the prediction value calculated for each siRNA are plotted in blue and red, respectively. The correlation coefficient of the RNAi activities against the prediction values in this region is 0.805 (P = 1.9 × 10−12).
Development of an algorithm that calculates siRNA activity
Based on these results, we tried to calculate the activity of a certain siRNA from its sequence information. To simplify the calculation, the B value associated with the optimal base composition of the 19-nt siRNA (which we termed the ‘macro effect’) and the positional effect of the individual base at each position (which we termed the ‘micro effect’) were considered separately. These effects were parameterized so that the concrete value that represented the prediction of the siRNA activity could be calculated. First, the best coefficients PX to yield the highest R factor were standardized from PA:PU:PG:PC = 0.4:0.35:0.15:0.1 to 6.073:5.111:1.265:0.304 by a linear expression 19.23 × PX + 1.6193 to calculate the transformed B values (macro effect) that could be parallelized to the actual RNAi activity (0–100%) (Equation (2)). For example, the macro effect value of the siRNA that targets position 5 of EGFP mRNA (Figure 2C), whose passenger strand is 5′-UGAGCAAGGGCGAGGAGCU-3′, is calculated as 6.073 × 5 + 5.111 × 2 + 1.265 × 9 + 0.304 × 3 (=52.884).
 |
2 |
The macro effect values for 702 siRNAs were calculated and plotted with their actual activities in Figure 5A. The line chart of the macro effects fits well with a large undulating curve formed by the sequential RNAi activities of the 702 siRNAs, the R factor of which results in 0.601 (P = 2.6 × 10−70). However, as shown in Figure 5C, the macro effects do not correspond well with sequential activities of siRNAs fluctuating at every third position. To elevate higher R factor by predicting such fluctuating activity, the micro effect values associated with periodical effect of every third position of siRNA are required.
In contrast, the parameters for the micro effect cannot be calculated directly from the data shown in Figure 4A and B because they contain both the macro and micro effects. Thus, the parameters for the micro effect were calculated from the difference values between the siRNA activities and the macro effect values. The difference values can be assumed to be comprised mainly of the micro effect that is derived from the positional effect of the individual bases without the macro effect. To calculate the micro effect value, the micro parameter QXi given to each base (X = A, U, G or C) at each position (i = position 1–19) for each siRNA is defined. Thus, the micro effect value is obtained by summing up the QXi parameters for each siRNA (Equation (3)). The optimized parameter sets of QXi for the micro effect were determined computationally to maximize the correlation between the micro effect value and the difference value (Table 1). For example, the micro effect value of the siRNA that targets position 5 of EGFP mRNA (5′-UGAGCAAGGGCGAGGAGCU-3′) (Figure 5C) is calculated as − 3.155 + 0.364 + 1.190 − 1.362 + 0.698 + 3.034 + 3.038 − 0.735 − 0.938 − 0.737 − 1.119 − 0.177 + 2.601 + 1.037 + 1.962 + 2.758 − 1.090 − 0.530 + 1.354 ( =8.143).
 |
3 |
Finally, the prediction value was obtained by adding the micro effect value to the macro effect value. For example, the prediction value of the siRNA that targets position 5 is 52.884 + 8.143(=61.027), which nicely correlates with its actual activity (63 ± 1.5). The prediction values against the 702 siRNAs were calculated accordingly (Table S1).
Table 1.
Optimized parameters for the micro effect.
| Bases/Positions | A | U | G | C |
|---|---|---|---|---|
| 1 | −1.555 | −3.155 | 3.492 | 3.136 |
| 2 | −2.237 | −1.856 | 0.364 | −0.067 |
| 3 | 1.190 | −0.341 | −1.226 | −0.808 |
| 4 | −0.179 | 0.070 | −1.362 | −0.136 |
| 5 | −0.154 | −2.258 | −0.454 | 0.698 |
| 6 | 3.034 | −1.407 | 0.387 | −4.081 |
| 7 | 3.038 | −1.440 | 0.637 | −2.676 |
| 8 | 2.341 | −2.697 | −0.735 | −0.317 |
| 9 | −2.028 | 2.113 | −0.938 | 1.935 |
| 10 | −1.455 | 2.521 | −0.787 | −0.126 |
| 11 | −0.938 | −0.136 | 0.614 | −1.119 |
| 12 | 1.112 | −1.519 | −0.177 | −1.279 |
| 13 | 2.601 | 0.200 | −2.095 | −1.296 |
| 14 | 2.092 | −2.258 | 1.037 | −1.388 |
| 15 | 1.797 | −1.356 | 1.962 | −1.801 |
| 16 | 2.758 | 2.128 | −2.188 | −1.491 |
| 17 | 0.915 | −1.295 | −1.090 | −1.119 |
| 18 | 1.971 | 2.482 | −2.853 | −0.530 |
| 19 | 5.310 | 1.354 | −2.785 | −2.627 |
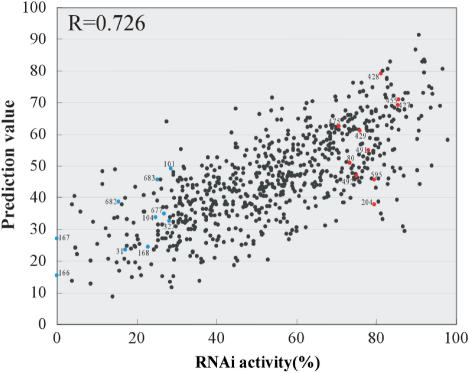
The correlation coefficient of the prediction values against the 702 siRNA activities increased to 0.726 (P = 5.9 × 10−116). The line chart of the prediction values nicely fits with the sequential RNAi activities of the 702 siRNAs (Figure 5B). In positions 1–50 (Figure 5C), the periodic fluctuation of the activity at every third position can be estimated completely by the prediction values as a correlation coefficient 0.805 (P = 1.9 × 10−12). As shown in the scatter plots (Figure 6), actual RNAi activities of 702 siRNAs aligned well with their prediction values ranging from high to low values, so that the plots appear to assemble in a diagonal line. Nine of the 702 siRNAs with prediction values beyond 80 also showed RNAi activities of over 80%. Thus, it is possible to design highly active siRNAs based on this algorithm. This algorithm is available as ‘siExplorer’ (http://rna.chem.t.u-tokyo.ac.jp/siexplorer.htm).
Figure 6.
Scatter plots for the knockdown activities of the 702 EGFP siRNAs versus the prediction values. Prediction value for each siRNA is listed in Table S1. Blue and red spots correspond to those in Figure 2C. The correlation coefficient (R) is 0.726 (P = 5.9 × 10−116).
Calculation of the siRNA activities targeting endogenous genes
To validate this calculation method in terms of its general applicability for endogenous genes, we prepared 50 randomly selected designs of siRNAs that target GAPDH mRNA. The knockdown efficiency measured by immunofluorescence using flow cytometry was quantified as the RNAi activity (%). The correlation coefficient of the prediction values against the activities was found to be 0.776 (P = 3.4 × 10−11) (Figure 7A). To see whether the periodical effect of every third position of siRNAs in sequential RNAi activity occurred with this gene as well, we prepared 25 siRNAs that target sequential positions within a 3′ end region of GAPDH mRNA (positions 950–974) where A and U residues appear frequently at every third position. As shown in Figure 7B, the siRNAs bearing A or U residues at 3n + 1 positions showed higher RNAi activity than ones targeting the next positions. Thus, the waving RNAi activity with 3-nt periodicity of siRNAs was clearly found in positions 956, 959, 962, 965, 968, 971 and 974. The line chart of the prediction values fits well with the sequential RNAi activities. This result demonstrated that the micro effect associated with the 3-nt periodicity is also important for calculating activities of siRNAs against GAPDH mRNA (Figure 7B). Furthermore, we designed 10 siRNAs that target β-catenin mRNA; five siRNAs with high prediction values (75–95) and five siRNAs with low prediction values (22–42). This algorithm can be applicable to design efficacious siRNAs for β-catenin mRNA (Figure S4). These results support the validity of our method for calculating siRNA activity that effectively silence endogenous genes. In addition, the 3-nt periodical fluctuation of siRNA activity was supposed to be a general phenomenon in human RNAi.
Examination of siRNA–TRBP interactions by fluorescence correlation spectroscopy (FCS)
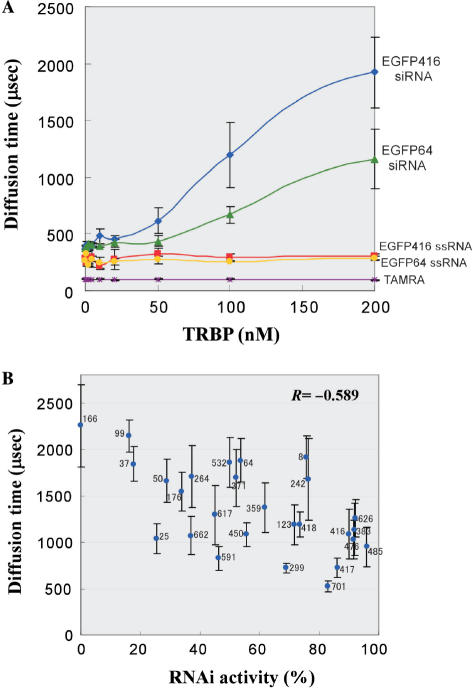
The periodical RNAi efficacy of every third position in siRNA sequences indicates the molecular interaction of siRNA with a protein component in RNAi machinery. In the cell, siRNA is initially recognized by Dicer with its partner protein to form RLC. We suppose that a part (not all) of siRNA functionality should be determined in this step. According to the biochemical data (43), it was reported that human Dicer itself has a weak affinity to dsRNA, while its partner protein TRBP has a dominant role for dsRNA recognition in Dicer–TRBP complex. We have examined binding affinity of recombinant human TRBP against EGFP siRNAs with various sequences. To measure siRNA–TRBP interaction precisely, we employed a new technique to study molecular interaction by FCS (52). FCS is a high-sensitive technique to observe a single fluorescence (F)-labeled molecule in a micro spot (1 femto-L) in a solution, which is illuminated by a confocal laser. FCS exactly determines the diffusion time of F-labeled molecules passing by the laser spot, which reflects the molecular weight of the labeled molecules. If the F-labeled siRNA interacts with TRBP, its increased molecular weight can be observed by its slow diffusion time. Another merit of this analysis is that FCS can analyze the complex formation under the equilibrium condition in a solution mixture. Different concentrations of recombinant human TRBP were incubated with TAMRA-labeled siRNA or ssRNA, then the diffusion time of the complex was measured (Figure 8A). We prepared two F-labeled siRNAs having equal GC content, which target positions 416 and 64 in EGFP mRNA. EGFP416 (GGCACAAGCUGGAGUACAA) was used as a representative siRNA with the 3-nt periodicity showing high activity (89.95); A or U at every 3n + 1 position (except position 1) and G or C at every 3n + 2 position. EGFP64 (GACGUAAACGGCCACAAGU) is a less active siRNA (53.64) with no 3-nt periodicity. In the absence of TRBP, diffusion time of both TAMRA-labeled siRNAs showed ∼400 μs. When TRBP was titrated, a dose-dependent prolongation of the diffusion time was observed in both EGFP416 and EGFP64 siRNAs, whereas no increase of the diffusion time was seen in their ssRNAs, demonstrating that TRBP specifically recognized double-stranded form of siRNAs. In the presence of 200 nM TRBP, the diffusion time of EGFP416 and EGFP64 siRNAs was found to be ∼1900 and ∼1200 μs, respectively. The result revealed that TRBP prefers to bind to efficacious siRNA having the 3-nt periodicity. To confirm this result, we carried out a competition experiment using 27 EGFP siRNAs having various RNAi activities (Figure 8B). Each non-labeled siRNA (5 nM) as a competitor was mixed with TAMRA-labeled EGFP416 siRNA (1 nM) in the presence of 150 nM TRBP. The FCS analysis monitoring diffusion time of the labeled siRNA revealed that efficacious siRNAs are more likely to compete with the labeled siRNA. The negative correlation between the diffusion time of the labeled siRNA and the RNAi activity of each competitor siRNA was observed with R factor −0.589 (P = 1.2 × 10−3), indicating that functionality of siRNA is associated with the binding affinity to TRBP. The 3-nt periodicity in siRNAs is supposed to originate from siRNA–TRBP interaction in the initial step of RNAi.
Figure 8.
FCS measurement of binding affinity of TRBP against various siRNAs. (A) FCS analysis measured translational diffusion velocity of TRBP–siRNA complex passing through a confocal laser spot in a mixture. Human TRBP was titrated in a range of 0–200 nM. Concentration of TAMRA-labeled siRNA, ssRNA or free TAMRA is fixed to 1 nM. (B) Competition analysis using non-labeled siRNAs. Competitor siRNA activities versus diffusion times are plotted with error bars. Competitor siRNA sequences targeting EGFP mRNA start from nt positions 8, 25, 37, 50, 64, 99, 123, 166, 176, 242, 264, 299, 359, 371, 383, 416, 417, 418, 450, 476, 485, 532, 591, 617, 626, 662 and 701. Concentration of competitor siRNA and TAMRA-labeled EGFP 416 siRNA are 5 and 1 nM, respectively. TRBP is fixed to 150 nM. The correlation coefficient (R) is −0.589 (P = 1.2 × 10−3).
DISCUSSION
Wealth of data set of siRNA activities tells us general rules that explain molecular mechanism of RNAi. We have described the first demonstration of a complete set of siRNA activities targeting all possible positions in ORF of single mRNA, while several groups have developed algorithms based on knockdown experiments in non-uniform condition using siRNAs against plural targets, various cell lines and different measurement method. To obtain comparable data set with the highest quality, we have designed a systematic RNAi experiment with minimized error factor. We employed HeLa cells that stably express EGFP as host cells to measure the RNAi activity accurately, because transient expression of EGFP results in scattering data. In addition, we have prepared gel-purified siRNAs with complete double-stranded form for quantitative knockdown experiment. RNAi activity was quantified by decreased fluorescence of EGFP that was measured by flow cytometry. We also confirmed with a quantitative RT-PCR that the decreased EGFP fluorescence by siRNA introduction was actually derived from RNAi-directed mRNA degradation (data not shown). This system allowed us to obtain reproducible data sets with limited error values. The correlation coefficient of two independent experiments for systematic knockdown achieved 0.851 (data not shown).
Most of the siRNAs showed medium activities and fewer ones showed very high or very low activities (Figure 1). This result indicates that it is rather difficult to design efficacious siRNAs from long mRNA sequence. It is possible to consider that RNAi activity depends on the accessibility of siRNA to the target mRNA. However, there was no evident correlation between the secondary structure of EGFP mRNA predicted and the full set of siRNA activities (data not shown), which suggests that the accessibility of siRNA to the mRNA may not be so important to the RNAi activity. The effects of mRNA-binding proteins, such as the initiation complex or UTR-binding proteins, on mRNA accessibility to siRNA still remain to be elucidated. However, the siRNAs targeting initiation region of GAPDH mRNA (positions 8, 23 and 33) exhibited good activities (79, 61 and 62%, respectively; Figure 5C). Thus, it can be assumed that the sequence of the target site is the most important factor that determines the activity.
It has been reported that thermodynamic property of siRNA plays an important role in overall efficiency of RNAi. Asymmetrical internal stabilities of both strands in siRNA is required for specifically introducing the guide strand into RISC to exert efficient silencing (31,33). Actually, it is known that functional siRNAs display lower internal stability at the 5′-guide strand as compared with the 5′-passenger strand (33). As shown in this study using complete set of siRNA activities, we have found clear correlation between instability of the guide strand 5′ end of each siRNA and its activity (R = 0.444). However, activity of each siRNA does not correlate with the stability of the passenger strand 5′ end. The relative stability of both strand for each siRNA showed a positive correlation (R = 0.236), but the plots are widely dispersed in the graph (Figure 2C) and the activities of many siRNAs are not predicted correctly. For instance, we picked 10 siRNAs (blue plots in Figure 2C and Table 2) that exhibit less stable guide strand 5′ end but show lower activity (less than 30), and 10 siRNAs (red plots in Figure 2C and Table 2) that exhibit more stable guide strand 5′ end but show higher activity (more than 70). As shown in Figure 6 and Table 2, the functionality of these siRNAs was nicely fitted with our prediction values calculated in consideration of the 3-nt periodicity. These results demonstrate that the instability of the guide strand 5′ end is a dominant factor to determine the asymmetric recognition of siRNA in the process of RISC assembly, and siRNA functionality is never determined only by the relative stability of 5′-ends of both strands.
Table 2.
Representative siRNAs whose activities can be predicted by the 3nt-periodicity.
A. Ten siRNAs that exhibit less stable guide strand 5′ end but show lower activity (less than 30).
| Position# | Sequence of the passenger strand | p.s. 5′-end (kcal/mol) | g.s. 5′-end (kcal/mol) | g.s. − p.s. (kcal/mol) | Activity (relative) | Our prediction values |
|---|---|---|---|---|---|---|
| 31 | GGGGUGGUGCCCAUCCUGG | −14.42 | −12.21 | 2.21 | 17.03 | 23.42 |
| 101 | GCGAGGGCGAUGCCACCUA | −12.61 | −10.01 | 2.6 | 28.52 | 49.06 |
| 104 | AGGGCGAUGCCACCUACGG | −13.22 | −10.69 | 2.53 | 24.66 | 33.73 |
| 166 | GUGCCCUGGCCCACCCUCG | −13.43 | −11.55 | 1.88 | 0.00 | 15.34 |
| 167 | UGCCCUGGCCCACCCUCGU | −13.35 | −10.03 | 3.32 | 0.01 | 26.90 |
| 168 | GCCCUGGCCCACCCUCGUG | −14.42 | −10.56 | 3.86 | 22.78 | 24.37 |
| 327 | CCGCGCCGAGGUGAAGUUC | −13.1 | −9.8 | 3.3 | 28.13 | 32.45 |
| 677 | CCGCCGCCGGGAUCACUCU | −14 | −9.75 | 4.25 | 26.93 | 34.99 |
| 682 | GCCGGGAUCACUCUCGGCA | −14.7 | −12.25 | 2.45 | 15.45 | 38.79 |
| 683 | CCGGGAUCACUCUCGGCAU | −13.84 | −10.89 | 2.95 | 25.13 | 45.64 |
| B. Ten siRNAs that exhibit more stable guide strand 5′ end but show higher activity (more than 70). | ||||||
| 80 | AGUUCAGCGUGUCCGGCGA | −8.8 | −12.49 | −3.69 | 73.34 | 50.73 |
| 204 | CGUGCAGUGCUUCAGCCGC | −11.83 | −14.66 | −2.83 | 79.54 | 37.74 |
| 427 | GAGUACAACUACAACAGCC | −10.4 | −13.07 | −2.67 | 85.32 | 69.32 |
| 428 | AGUACAACUACAACAGCCA | −9.09 | −11.97 | −2.88 | 81.16 | 79.11 |
| 429 | GUACAACUACAACAGCCAC | −10.32 | −13.23 | −2.91 | 75.82 | 61.18 |
| 455 | AUAUCAUGGCCGACAAGCA | −7.08 | −9.64 | −2.56 | 85.53 | 70.93 |
| 475 | AAGAACGGCAUCAAGGUGA | −7.49 | −11.06 | −3.57 | 70.62 | 62.47 |
| 491 | UGAACUUCAAGAUCCGCCA | −8.93 | −12.25 | −3.32 | 78.09 | 54.98 |
| 492 | GAACUUCAAGAUCCGCCAC | −10 | −13.23 | −3.23 | 75.05 | 46.75 |
| 595 | AACCACUACCUGAGCACCC | −9.74 | −13.07 | −3.33 | 79.56 | 45.68 |
Total base composition of siRNA was found to be an important factor that influence RNAi activity. The best coefficients to achieve the highest correlation coefficient (0.601) was found to be PA:PU:PG:PC = 0.4:0.35:0.15:0.1. As the B value obtained by Equation (1) correlates with the activity, efficacious siRNA should have AU-rich composition. However, as the GC content of the siRNAs for EGFP mRNA ranges from 36.8 to 84.2%, this rule can be applied to siRNAs bearing a similar range of GC contents. In fact, we observed that siRNAs with 10–20% GC contents represented lower RNAi activities than the expected values (data not shown). We do not have clear answer for the reason why low GC content of siRNA composition contributes the activity. One explanation is that siRNA with lower melting temperature due to AU-rich composition may be advantageous in the unwinding step of dsRNA or the recycling step of the guide strand.
The most important finding in this study is that the RNAi activities of siRNAs targeting sequential positions clearly showed periodical fluctuation at every third position in EGFP and GAPDH mRNAs. This waving RNAi activity with 3-nt periodicity is not observed in every sequential region, but observed in regions where A and U frequently appear at every third position. By analyzing positional effect of siRNA on the RNAi activity, this phenomenon can be explained by the periodical efficacy at positions 3n + 1 (4,7,10,13,16 and 19) in siRNA, which implies the molecular mechanism of siRNA functionality in RNAi pathway. The optimized base composition at positions 3n + 1 in siRNAs have positive effects on the RNAi activity, whereas no strong correlation with the activity was found for any of the bases at positions 3n + 2 (2,5,8,11,14 and 17). The positions 3n (3,6,9,12,15,18) also have positive effects on the activity but less efficient than those of 3n + 1. Reynolds et al. (34) reported siRNA-specific features that contribute to overall efficiency of RNAi by a systematic analysis of 180 siRNAs targeting two different genes, and found the passenger strand-specific base preferences; A-base at position 3 and 19, U-base at position 10, the absence of G at position 13 and the absence of G or C at position 19. This result completely falls within our observation that A and U at positions 3n + 1 are involved in efficient RNAi activity. The strongest effect of A19 on RNAi efficacy is a common observation with other groups (34–39). Basically, the effect of A-base at positions 3n + 1 show more positive than that of U-base (Figure 4B). Exceptionally, the effect of U-base goes beyond that of A-base at position 10. U10 seems to correlate with the cleavage reaction catalyzed by RISC, which cleaves the target mRNA between positions 10 and 11 (9,21,22). Similar tendency was also reported by Jagla et al. (53). They performed knockdown experiments using 601 siRNAs targeting one exogenous and three endogenous genes, and observed A/U preference at positions 10 and 19, and G/C preference at position 1, which is consistent with our results. However, none of the previous studies found any positive effects of A- or U-base at positions 3, 4, 6, 7, 12, 13, 15, 16 and 18, which clearly shape 3n and 3n + 1 periodicity in our study. The common problem in the previous studies is that the source data used include considerable error values by merging data obtained from different knockdown experiments for various targets with different measurement methods. On the other hand, as we performed RNAi experiments using the largest set of siRNAs (702 designs) targeting a single mRNA, we got a comparable data set with minimized error factor.
In order to derive a fundamental principle of RNAi mechanism from the correlation between siRNA sequences and their activities, we have developed an algorithm that calculates the activity of siRNA based on the analysis of the complete data set. A simple algorithm calculating the prediction value of siRNA functionality was developed by combining the macro effect associated with the optimal base composition and the micro effect associated with the positional effect. In fact, the prediction values of 702 siRNAs for EGFP mRNA obtained by this algorithm showed very good match with their actual activities with a high correlation coefficient (0.726). Actually, the algorithm successfully predicted activities of endogenous genes; 75 siRNAs for GAPDH and 10 siRNAs for β-catenin mRNAs. Recently, we have reported two examples for successfully knocking down human mitochondrial enzymes by siRNAs designed by this algorithm (54,55).
Several groups have already reported the algorithms or guidelines for the selection of efficacious siRNAs based on the activities of siRNAs that target different genes (34–39,53). Functional siRNAs are selected through several criteria, such as GC content, base preference and bias of internal stability of strand termini. Meanwhile, here we suggest that siRNAs targeting only one gene are needed for such statistical analysis because it is difficult to normalize siRNA activities against different target mRNAs that are measured by means of different methods. Our algorithm does not incorporate any criteria to select functional siRNAs, but just calculates the predicted activity from their sequence information. Thus, we could successfully predict wide variety of siRNAs whose activity range from low to high. As shown in Figures 5B and 6, the prediction values extensively fit well with actual activities of most of the siRNAs. This algorithm can design siRNAs with lower activity (such as 50 or 30%), which might be applicable to knockdown essential genes or to control gene function by changing siRNA activity.
RNAi pathway consists of multiple RNA–protein interactions characterized by many steps. Although our experiment observed overall efficiency of RNAi and cannot differentiate efficacy of each step, the periodical effect at every third position and base specificity of positional effect may provide a new aspect in RNAi mechanism. At the initial step of RNAi pathway in human, siRNA is recognized by Dicer with its partner protein TRBP or PACT. Two dsRBDs in TRBP are considered to have a dominant role for siRNA recognition. FCS measurement of TRBP–siRNA interaction revealed that TRBP preferentially interacts with efficacious siRNAs having A/U at 3n + 1 positions, although the correlation does not appear to be so significant. Thus, strong affinity of siRNA–TRBP interaction may contribute siRNA functionality. In addition, it is supposed that the 3-nt periodicity is derived from binding affinity of siRNA–TRBP interaction. According to the structural studies of dsRBD–dsRNA interaction (56–58), dsRBD binds to only one face of the A-form helix of dsRNA and spans 16 bp (Figure S5). The regions 1 and 2 of dsRBD mainly interacts with two adjacent minor grooves so as to form hydrogen bonds with base pairs in the dsRNA which correspond to positions 3, 4 and 16 in the passenger strand of siRNA. The region 3 interacts with phosphate backbone in the major groove (positions 7 and 13 in siRNA). Since base pairs at every third position of both strands align as straight lines on the two opposite faces of A-form helix of siRNA (Figure S5), it can be speculated that one dsRBD in TRBP binds to one of the two faces of the siRNA helix by mainly recognizing base pairs at positions 4 and 16 (3n + 1) in two adjacent minor grooves. Additional contact between phosphate backbone in the major groove at positions 7 and 13 (3n + 1) and region 3 of dsRBD may support this interaction. Another face of siRNA helix might be recognized by the other dsRBD in TRBP. A- or U-base preference at position 3n + 1 for siRNA functionality might be explained by sequence-specific recognition of dsRBD in TRBP. Further study will be necessary to test sequence preference of PACT which is another partner for human Dicer. Otherwise, considering that Dicer has a single dsRBD to be required for its function, one face of siRNA helix might be recognized by the dsRBD of Dicer. Alternative interpretation of the 3-nt periodicity is involved in the last step of RNAi. After unwinding siRNA, the guide strand paired with the target mRNA is recognized by Argonaute 2 protein in the RISC complex. Argonaute 2 has a highly conserved Piwi domain which is a slicer for the target cleavage (19). The crystal structure of Archaeoglobus fulgidus Piwi protein bound to dsRNA have shown that the guide–target duplex forms a short A-form helix (19,24), one face of which is bound to a positive surface of Piwi where putative catalytic site is located. In this interaction, if Piwi has a sequence preference of base pairs at every third position which aligns on the one face of the RNA helix, it is another candidate for the 3-nt periodicity. Further studies are still necessary to elucidate the molecular mechanism of this issue.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Suzuki lab members especially, Takeo Suzuki, N. Umeda, S. Kurata and A. Nagao for their technical assistance and fruitful discussion. We thank M. Susa, S.Y. Yang, J. Hida and T. Sugita (Hokkaido System Science, Inc.) for preparing siRNAs, F. Sekiya (Maze, Inc.) for computational analysis of siRNA activity, J.Y. Lee (Catholic University of Korea) for kind gift of pGST–TRBP. Special thanks are due to Drs P. Zamore and Y. Tomari (Massachusetts Medical School) for critical reading of the manuscript and many productive comments. This work was supported by grants from the New Energy and Industrial Technology Development Organization (NEDO), grants-in-aid for scientific research on priority areas from Japan Ministry of Education, Science, Sports and Culture (to T.S.). Funding to pay the Open Access publication charge was provided by Japan Ministry of Education, Science, Sports and Culture.
REFERENCES
- 1.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 2.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 3.Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 4.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 5.Zamore PD. Ancient pathways programmed by small RNAs. Science. 2002;296:1265–1269. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- 6.Tuschl T. Functional genomics: RNA sets the standard. Nature. 2003;421:220–221. doi: 10.1038/421220a. [DOI] [PubMed] [Google Scholar]
- 7.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minks MA, West DK, Benvin S, Baglioni C. Structural requirements of double-stranded RNA for the activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J. Biol. Chem. 1979;254:10180–10183. [PubMed] [Google Scholar]
- 13.Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 14.Macrae IJ, Zhou K, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 15.Truss M, Swat M, Kielbasa SM, Schafer R, Herzel H, Hagemeier C. HuSiDa–the human siRNA database: an open-access database for published functional siRNA sequences and technical details of efficient transfer into recipient cells. Nucleic Acids Res. 2005;33:D108–111. doi: 10.1093/nar/gki131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 17.Yano J, Hirabayashi K, Nakagawa S, et al. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clin. Cancer Res. 2004;10:7721–7726. doi: 10.1158/1078-0432.CCR-04-1049. [DOI] [PubMed] [Google Scholar]
- 18.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 19.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. Epub 2005 Mar 2030. [DOI] [PubMed] [Google Scholar]
- 20.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. Epub 2004 Jul 1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. Epub 2004 Jul 1429. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. Epub 2004 Jul 1429. [DOI] [PubMed] [Google Scholar]
- 25.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 26.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 29.Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 32.Preall JB, He Z, Gorra JM, Sontheimer EJ. Short interfering RNA strand selection is independent of dsRNA processing polarity during RNAi in Drosophila. Curr. Biol. 2006;16:530–535. doi: 10.1016/j.cub.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 33.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. Epub 2004 Feb 2001. [DOI] [PubMed] [Google Scholar]
- 35.Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 36.Ui-Tei K, Naito Y, Takahashi F, et al. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 2004;32:W130–134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huesken D, Lange J, Mickanin C, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 39.Holen T. Efficient prediction of siRNAs with siRNArules 1.0: An open-source JAVA approach to siRNA algorithms. RNA. 2006;12:1–6. doi: 10.1261/rna.81006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. Epub 2004 Dec 2026. [DOI] [PubMed] [Google Scholar]
- 41.Siolas D, Lerner C, Burchard J, et al. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. Epub 2004 Dec 2026. [DOI] [PubMed] [Google Scholar]
- 42.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 43.Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haase AD, Jaskiewicz L, Zhang H, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Reports. 2005;10:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 46.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katoh T, Susa M, Suzuki T, Umeda N, Watanabe K, Suzuki T. Simple and rapid synthesis of siRNA derived from in vitro transcribed shRNA. Nucleic Acids Res. Supplement. 2003;3:249–250. doi: 10.1093/nass/3.1.249. [DOI] [PubMed] [Google Scholar]
- 48.Lee JY, Kim H, Ryu CH, et al. Merlin, a tumor suppressor, interacts with transactivation-responsive RNA-binding protein and inhibits its oncogenic activity. J. Biol. Chem. 2004;279:30265–30273. doi: 10.1074/jbc.M312083200. [DOI] [PubMed] [Google Scholar]
- 49.Xia T, SantaLucia J, Jr, Burkard ME, et al. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 50.Sugimoto N, Kierzek R, Turner DH. Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry. 1987;26:4554–4558. doi: 10.1021/bi00388a058. [DOI] [PubMed] [Google Scholar]
- 51.O’Toole AS, Miller S, Serra MJ. Stability of 3′ double nucleotide overhangs that model the 3′ ends of siRNA. RNA. 2005;11:512–516. doi: 10.1261/rna.7254905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maiti S, Haupts U, Webb WW. Fluorescence correlation spectroscopy: diagnostics for sparse molecules. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11753–11757. doi: 10.1073/pnas.94.22.11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jagla B, Aulner N, Kelly PD, et al. Sequence characteristics of functional siRNAs. RNA. 2005;11:864–872. doi: 10.1261/rna.7275905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Umeda N, Suzuki T, Yukawa M, Ohya Y, Shindo H, Watanabe K. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005;280:1613–1624. doi: 10.1074/jbc.M409306200. Epub 2004 Oct 1626. [DOI] [PubMed] [Google Scholar]
- 55.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;14:14. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- 56.Blaszczyk J, Gan J, Tropea JE, Court DL, Waugh DS, Ji X. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure (Camb) 2004;12:457–466. doi: 10.1016/j.str.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 57.Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 58.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985;13:3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.