Abstract
In Caenorhabditis elegans, the Mex-3 protein is a translational regulator that specifies the posterior blastomere identity in the early embryo and contributes to the maintenance of the germline totipotency. We have now identified a family of four homologous human Mex-3 genes, called hMex-3A to -3D that encode proteins containing two heterogeneous nuclear ribonucleoprotein K homology (KH) domains and one carboxy-terminal RING finger module. The hMex-3 are phosphoproteins that bind RNA through their KH domains and shuttle between the nucleus and the cytoplasm via the CRM1-dependent export pathway. Our analysis further revealed that hMex-3A and hMex-3B, but not hMex-3C, colocalize with both the hDcp1a decapping factor and Argonaute (Ago) proteins in processing bodies (P bodies), recently characterized as centers of mRNA turnover. Taken together, these findings indicate that hMex-3 proteins constitute a novel family of evolutionarily conserved RNA-binding proteins, differentially recruited to P bodies and potentially involved in post-transcriptional regulatory mechanisms.
INTRODUCTION
Cell polarization and asymmetric division are fundamental processes for generating cell diversity. Studies initially developed in Caenorhabditis elegans have led to the identification of six genes, named par for partitioning defective, that are conserved across evolution and encode proteins that act as key effectors of cell polarity (1). However, the mechanisms by which polarity information is connected to cell-fate specification remain elusive (2). PAR molecules are known to regulate both the localization and the activity of several maternal factors which transduce polarity cues in the C. elegans embryo and the protein Mex-3 is one of these regulators (3).
Mex-3 contains two K homology (KH) domains, initially characterized in heterogeneous nuclear K ribonucleoproteins as a conserved region of 65–70 amino acids which interacts with RNA (4). Mutations that disrupt the Mex-3 locus are embryonic lethal and cause defects in the AB anterior blastomere descendants which inappropriately produce body wall muscle, hence the name Mex-3 for muscle excess 3 (3). The Mex-3 protein is distributed uniformly in oocytes but becomes asymmetrically enriched in the two anterior blastomeres at the four-cell stage. Furthermore, both Mex-3 mRNA and protein are components of the P granules in germ cells (3). Previous reports have established that Mex-3 localization is complementary to that of PAL-1 (3,5,6), the C. elegans ortholog of the Caudal homeoprotein in Drosophila. PAL-1 is expressed in posterior blastomeres but is abnormally present in all blastomeres at the four-cell stage in Mex-3 mutants. PAL-1 is required to specify the identity of the posterior blastomere and Mex-3 prevents translation of the PAL-1 mRNA in the anterior blastomeres presumably through its binding to the PAL-1 3′UTR (5,6). In addition, it has been suggested that Mex-3 controls the spatial repression of PAL-1 and relays polarity cues of PAR proteins through the specific interaction with two RNA-binding proteins, Mex-6 and Spn-4 (6).
Besides its role in the specification of posterior blastomeres, Mex-3 is also involved in the correct segregation of P granules at the eight-cell stage (3). P granules contain maternally contributed mRNAs and these structures are thought to be involved in the determination of the germline, thus suggesting a potential role of Mex-3 in the specification of this cell lineage. Consistent with this idea, disruption of mex-3 together with gld-1, a gene coding a member of the STAR/KH family of RNA-binding protein, results in the transdifferentiation of germ cells into somatic cells (7).
In the search for human homologs of the C. elegans cell-fate determinants, we have identified and characterized a family of four human gene homologs to Mex-3 (hMex-3) that map to distinct chromosomes. Here, we report that the hMex-3 proteins characterized are related RNA-binding phosphoproteins that shuttle between the nucleus and the cytoplasm and that they show differential localization to cytosolic foci recently identified as sites of mRNA storage, translation regulation and degradation (8). Furthermore, the hMex-3 proteins display distinct modes of association with Ago molecules which are members of a large ribonucleoprotein complex involved in RNA interference (RNAi) and microRNA (miRNA) pathways. These results establish that hMex-3 proteins constitute a novel subfamily of evolutionarily conserved RNA-binding proteins potentially involved in the regulation of post-transcriptional events.
MATERIALS AND METHODS
Bioinformatics
Existence of human and mouse Mex-3 gene homologs was determined by comparison of ceMex-3 protein sequence (Genbank accession number AAK73873) with human or mouse genome using Blast (http://www.ncbi.nlm.nih.gov/BLAST/) (9). Gene structures were predicted by GenScan (http://bioweb.pasteur.fr/seqanal/interfaces/genscan.html) (10). Potential structural protein domains were predicted by SMART (http://dylan.embl-heidelberg.de/) (11) and PSORT (http://psort.nibb.ac.jp/) (12). Consensus sequence L-x(2,3)-[LIVFM-x(2,3)-L-x-[LI] from NESbase (http://www.cbs.dtu.dk/databases/NESbase-1.0/) (13) was used to predict NES sequence localization. Amino acid sequences were aligned by ClustalW (http://www.ebi.ac.uk/clustalw/) (14) and T-coffee (http://igs-server.cnrs-mrs.fr/Tcoffee/) (15). A tree was done with PhyML Method (model JTT) (16) on the multiple alignment obtained by T-coffee.
RNA extraction and RT-PCR analysis
Total cellular RNA were extracted from various human cell lines using the RNeasy Mini kit (Qiagen) and incubated in DNAse I (RNAse-free DNAse set) according to the manufacturer's instructions. Total RNA (1 μg) from the cell lines or from 20 human tissues (Multiple Tissue Total RNA panel, BD Biosciences) were reverse transcribed to cDNA by the Omniscript Reverse Transcription kit (Qiagen). To check expression level of the four hMex-3 genes, Cdx2 gene or GAPDH (glyceraldehyde 3-phosphate dehydrogenase) and β2M (β2-microglobulin) genes as controls, 40 cycles (35 for Cdx2, 30 for GAPDH and β2M) of PCR were performed according to the manufacturer's instructions on cDNA with Taq DNA Polymerase (Invitrogen) for GAPDH and β2M, High Fidelity PCR system (Roche), for hMex-3C or GC-rich PCR system (Roche) for hMex-3A, hMex-3B, hMex-3D and Cdx2 in presence of 200 nM internal primers specific for each cDNA:
hM3AiF:5′-AGGGCTGCTGGAAGACGAG-3′,
hM3AiR:5′-TGAGATGATTTCCCGCCG-3′,
hM3BiF:5′-CCTGCTGGGGCTGGACA-3′,
hM3BiR:5′-TGGAAGTCGTTCTCGTCTGT-3′,
hM3CiF:5′-TGAACGGGGAGCAGGCG-3′,
hM3CiR:5′-TGACTTGGACGGTGGTTTGA-3′,
hM3DiF:5′-CCGACCAGATGAGCGTGAT-3′,
hM3DiR:5′-CGAGCAGGTCCAGGCAGA-3′,
Cdx2Fi:5′-GAACCTGTGCGAGTGGATG-3′,
Cdx2Ri:5′-TCTCAGAGGACCTGGCTGAG-3′,
GAPDHiF:5′-TCCCATCACCATCTTCCAGG-3′,
GAPDHiR:5′-ATGAGTCCTTCCACGATACC-3′,
β2MF:5′-TCGCGCTACTCTCTCTTTCTG-3′,
β2MR:5′-AACTTCAATGTCGGATGGATG-3′.
For cloning of complete hMex-3 coding sequence, 30 cycles of PCR were performed on BOSC cell cDNA with GC-rich PCR system (Roche) in the presence of 200 nM of specific external primers extended with a restriction enzyme site recognition sequence:
hM3AeF:5′-CTAGGGATCCACCATGCCTAGTCTAGTGGTATCTGG-3′,
hM3AeR:5′-CTAGGAATTCTTAGGAGAATATTCGGATGGC-3′,
hM3BeF:5′-CTAGGGATCCACCATGCCCAGCTCGCTGTTC-3′,
hM3BeR:5′-CTAGGAATTCTTAAGAAAAGATGCGGATGGC-3′,
hM3CeF:5′-CTAGGGATCCACCATGCCCAGCGGCAGCTC-3′,
hM3CeR:5′-CTAGGTCGACTTAAGAGTGAATTTGGATTGCCTGAG-3′.
Constructs, mutagenesis and antibodies
Amplified human coding sequences for hMex-3A, hMex-3B and hMex-3C were inserted into pCMV-Tag 3B after BamH1/EcoRI (for hMex-3A and hMex-3B) or BamH1/XhoI (for hMex-3C) digestion. Fragments of cDNAs coding for Bpag1 and the p62-sequestosome were cloned in the same vector and were used as controls. The pCDNA-PABP plasmid was generated by PCR amplification using the pGEM1-PABP construct as a template (17). hMex-3B and hMex-3C mutants of predicted nuclear export signal (NES) sequence, KH domain and RING domain were generated by directed mutagenesis by using QuikChange XL Site-Directed Mutagenesis kit (Stratagene), according to the manufacturer's instructions. The mutated amino acids in each domain are indicated by an arrow on Figure 1. hMex-3B/KH: G177D, hMex-3B/RG: C521G, hMex-3C/NES: L52A + L57A, hMex-3C/KH: G343D, hMex-3C/RG: C608G + C611G. GFP-hDcp1a (18) and pEGFP-Rev-NES (19) constructs were also used. pIRESneo-FLAG/HA Ago1 and pIRESneo-FLAG/HA Ago2 constructs (Addgene plasmid 10820 and 10822) were obtained from the laboratory of T. Tuschl (20).
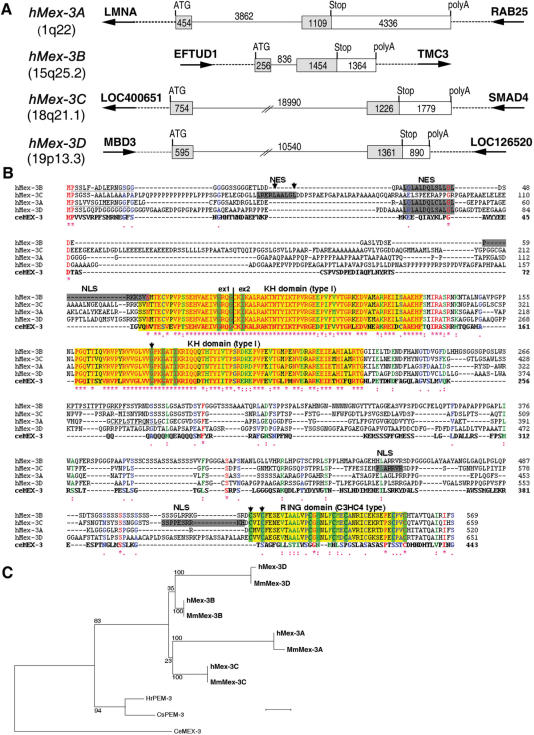
Figure 1.
Human family of Mex-3 proteins. (A) Structure and chromosomal localization of the four human Mex-3 genes. Exons are depicted as boxes, with coding sequences as grey boxes and 3’-UTR as white boxes. Genes lying downstream and upstream of hMex-3A, hMex-3B, hMex-3C and hMex-3D genes are displayed as large arrows. (B) Sequence alignment of the hMex-3 proteins and their Caenorhabditis elegans ortholog. C. elegans Mex-3 and hMex-3 amino acid sequences were aligned with ClustalW. Identical residues (red) are annotated by an asterisk, whereas similar residues (green) and lightly similar residues (blue) are denoted with two or one dot, respectively. Junctions between the two exons (vertical line) lie in the same position for the four hMex-3 mRNA. One NES is predicted in each hMex-3 protein, and one NLS is also predicted for hMex-3B and hMex-3C. Two strongly conserved KH domains of type I were predicted in hMex-3 proteins. One conserved RING domain of C3H4 type was also predicted in C-terminus of the four human proteins. The highly conserved amino acids of KH and RING domains are shown as green boxes. Arrows indicate the amino acids which were mutated in this study. (C) Phylogenetic tree of the Mex-3 gene family. The 180 most conserved amino acids around the KH domains of human Mex-3, mouse Mex-3, ceMex-3 (Genbank accession number AAK73873) and two Ascidian Mex-3 homologous sequences from Ciona savignyi and Halocynthia roretzi (Genbank accession number BAB03404 and BAC10968) were aligned by T-coffee. A tree was done with PhyML method on this alignment. Bootstrap values are indicated as percentages (500 replicates). Scale bar: 0.05.
Mouse monoclonal anti-myc (9E10) (Roche), anti-flag (M2) (Sigma) and anti-actin (ICN) antibodies were used in western blot at 1:5000, 1:1300 and 1:25 000 dilutions, respectively. Mouse monoclonal anti-myc and anti-HA 11 (Covance) antibodies were used in immunofluorescence at 1:300 dilution. Mouse monoclonal antibody against human PABP (clone 10E10) was a kind gift from G. Dreyfuss (University of Pennsylvania, USA) and was used in western blot at 1:2000. Rabbit polyclonal anti-hDcp1a serum (18) was used in western blot at 1:1000. Rabbit polyclonal anti-hMex-3A, hMex-3B and hMex-3C antibodies anti-M3Aα, Anti-M3Aβ, Anti-M3Bα, Anti-M3Bβ and Anti-M3Cα were produced and immunopurified by Eurogentec. They were raised against the peptides PSLVVSGIMERNGGF, CKPLSTFRQNSLG, PSSLFADLERNGSGG, KPTPSITPTPGRKPF and CNPVPPSRARMISNYR, respectively. They were used in western blots and for immunofluorescence at 1:1000 and 1:100 dilutions respectively (both anti-M3A) or 1:20 000 and 1:150, respectively (both anti-M3B). For immunohistochemistry experiments, primary antibodies used were anti-M3Bβ antibody (dilution 1:200), rabbit anti-MUC2 (dilution 1:1000, kindly provided by I Van Seuningen, Inserm U560, Lille, France), rabbit anti-Chromogranin-A (dilution 1:300, DiaSorin) and rabbit anti-Lyzosyme (dilution 1:100, Dako).
Cell culture and transfections
MCF7 and BOSC cells were cultivated in DMEM medium containing FBS 10%, penicillin 100 U/ml and streptomycin 0.1 mg/ml, and incubated at 37°C in 5% CO2. Cells were plated 24 h before transfections. Plasmids encoding different cDNAs were transfected into cells using ExGen 500 (Upstate Biotechnologies) as a transfection reagent, according to the manufacturer's instructions.
Protein analyses, immunoprecipitation and kinase assay
Cells were lysed 48 h post-transfection and western blot analyses were performed as described earlier (21). In order to test for the phosphorylation of proteins, protein extracts prepared in lysis buffer without phosphatase inhibitors were treated for 1 h at 37°C with 8 U λ-Phosphatase (Biolabs) per μg of protein. For immunoprecipitation, Myc-tagged proteins were immunoprecipitated from protein extracts (1 mg) with the anti-myc antibody (5 μg) as described earlier (21) and western blot analyses were performed as described on the immunocomplexes obtained. Protein extracts were treated with or without RNAseA (0.2 mg/ml) prior to immunoprecipitation. For kinase assay, Myc-tagged hMex3-A, hMex-3B and Bpag1 proteins were immunoprecipitated with the anti-myc antibody and the immunocomplexes were incubated in kinase buffer as described earlier (21).
Ribonucleotide polymers binding assays
Proteins encoded by expression vectors were in vitro transcribed/translated with TNT Coupled Reticulocyte Lysate system (Promega) in the presence of [35S] methionine, according to the manufacturer's instructions. RNA homopolymer-conjugated agarose beads poly(A) (Sigma) (20 μl) were washed five times with RNA-binding buffer (10 mM Tris-HCl, pH 8, 2.5 mM MgCl2, 0.5% Triton X-100, 100 mM NaCl) and incubated with 5 μl (1/10) of in vitro translated proteins for 30 min at 4°C in 500 μl of the same buffer. After five washes with RNA-binding buffer containing 200 mM NaCl, bound proteins were eluted with SDS sample buffer and analysed by SDS-PAGE and autoradiography.
RNA immunoprecipitation
Cells were lysed 48 h post-transfection as described earlier, except that RNAsin (Promega) was added to the lysis buffer (10 U/ml) and lysis time was extended to 45 min. Myc-tagged proteins were immunoprecipitated from protein extracts (750 μg) with the anti-myc antibody (5 μg). After three washes in lysis buffer containing RNAsin and one wash containing DNAse I (50 U/ml), RNA were extracted from the sepharose-protein A beads (Amersham) by TRI Reagent (Sigma) and resuspended in 30 μl H2O. RT-PCR analysis was directly performed on the RNA, as described earlier.
Immunofluorescence
MCF7 or HeLa cells were plated on glass cover slips and were transiently transfected as described. After 48 h, cells were fixed 20 min in 4% paraformaldehyde, permeabilized 5 min in 0.5% Triton X-100 and blocked 20 min in PBS containing BSA 0.3%. Cells were then incubated overnight with primary antibody diluted in PBS–BSA and were incubated 1 h with fluorescent-labeled secondary antibody. Cells were mounted with Vectashield mounting medium containing DAPI (Vector Laboratories) and were observed by a confocal microscope TSC SP2 (Leica). To study a potential nucleocytoplasmic transport of hMex-3 proteins, cells were incubated for 1 h with 10 μg/ml Cycloheximide (Sigma), and then 20 ng/ml Leptomycin B (LMB) (Sigma) was added and left for 5 h before fixation. In experiments of colocalization with hDcp1a, cells were treated with or without 5 μg/ml Cycloheximide (Sigma) and left for 2 h before fixation.
Immunohistochemistry
The human intestinal samples used in this study were from the collection of surgical specimens of the Department of Visceral Surgery stored at the Department of Histopathology (Hôpitaux Universitaires de Strasbourg, France) under the guidelines approved by the Institutional review board. Access to these samples was approved by the local ethical committee. These samples corresponded either to biopsies of histologically normal colon mucosa taken during colorectal tumour resection, or to specimens of resected Meckel's diverticulum. Tissue samples were fixed in 4% paraformaldehyde and embedded in paraffin. Serial sections (6 μm) were deparaffinized and antigens were retrieved by heating twice for 5 min in 10 mM citrate (pH 6) using a microwave (700 W). The sections were blocked in PBS, 0.1% Triton X100 and 5% normal goat serum and incubated overnight at 4°C with the appropriate primary antibodies. Secondary biotinylated anti-rabbit or anti-mouse antibodies (dilution 1:200, Vector Laboratories) were revealed using the Vectastain ABC Elite kit according to the recommendations of the supplier (Vector Laboratories). 129sv mouse duodenum samples were fixed in 4% paraformaldehyde and embedded in paraffin. Serial sections were deparaffinized and antigens were retrieved as follows: 15 min in a 0.1% trypsin (Gibco) solution in CaCl2 0.1% at 37°, and 15 min microwave treatment using Dako's Antigen Unmasking Solution. Sections were blocked in Dako's Antibody Diluent, then primary antibody was incubated overnight at 4°C diluted 1:100 in Dako's Antibody Diluent. Secondary goat anti-rabbit alexafluor555 conjugated antibody (Invitrogen) was incubated 1 h at room temperature diluted 1:1000 in Dako's Antibody Diluent. Tissues were mounted with Vectashield mounting medium containing DAPI (Vector Laboratories).
RESULTS
Identification and characterization of four human Mex-3 homologous genes
Using Blast search and GenScan prediction, we have identified four potential human genes homologous to the C. elegans Mex-3 gene. These genes, designated as hMex-3A, -3B, -3C and -3D, are located on distinct chromosomes at positions 1q22, 15q25.2, 18q21.1 and 19p13.3, respectively (Figure 1A). The four hMex-3 genes are composed of two exons and one intron which vary in size from 836 bp for hMex-3B to 18 990 bp for hMex-3C. The sequences containing the entire open reading frame were amplified from RNA by RT-PCR and fully sequenced (Genbank accession numbers: AY950677, AY950678, AY950679, AY950680) with the results confirming the in silico predictions. The 3′-UTR of the hMex-3 genes was also predicted by GenScan and then confirmed after a Blast search of the hMex-3 genomic regions in a human EST library. A variant form of hMex-3D, called TINO, has been recently cloned (22). Compared to hMex-3D, TINO is truncated in its N-terminal region, beginning at the first KH domain. This form of hMex-3D also differed at its C-terminal end, with 19 amino acids, encoded by a potential alternative exon, replacing the four last terminal amino acids of hMex-3D.
Sequence alignment of the hMex-3 proteins with the C. elegans Mex-3 protein shows that all proteins contain two highly conserved KH domains presenting 79–81% of sequence identity (Figure 1B). Database searches revealed a RING finger domain in the carboxy-terminal region of the four hMex-3 proteins. This RING finger is lacking in the C. elegans Mex-3 but was identified in the Ascidian ortholog pem-3 (23) and was also predicted in the orthologous Drosophila sequence based on in silico comparisons (data not shown).
We have also identified four mouse Mex-3 (mMex-3) genes on chromosomes syntenic to the human chromosomes containing the hMex-3 genes (data not shown). The four putative mMex-3 proteins display a strong similarity to their human counterpart, being approximately 95% identical at the amino acid level. A phylogenetic tree was constructed using the PhyML method (Figure. 1C) which suggests that a unique ancestral Mex-3 gene was duplicated after the time of divergence between urochordates and vertebrates.
Tissue-specific expression of the hMex-3 genes
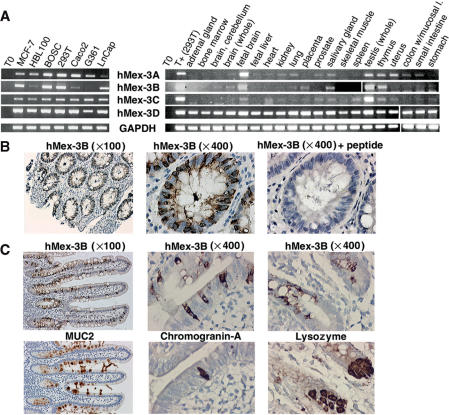
To examine the expression pattern of the hMex-3 genes, RT-PCR analyses were performed on 8 human cell lines and 20 different human tissues with oligonucleotide specific to each hMex-3 transcript (Figure 2A). hMex-3D mRNA was found ubiquitously expressed in all the cell lines and tissues tested. The other three hMex-3 genes were expressed at varying levels in different tissues, with the highest expression being found in fetal brain and testis. hMex-3A, -3B and -3C genes were also expressed in thymus, salivary gland and uterus (Figure 2A). A weak expression of these three hMex-3 genes was also detected in the intestine.
Figure 2.
Expression profile of hMex-3 genes and hMex-3B protein. (A) Human Mex-3 gene expression levels (panels 1–4) were examined by RT-PCR with specific internal primers and were compared with expression level of ubiquitously expressed GAPDH gene (panel 5). RNA were extracted from 7 human cell lines (left) and from 20 human tissues (Multiple Tissue Total RNA panel, BD Biosciences) (right). (B) Human colon sections (magnification: ×100 or ×400) were stained with anti-hMex-3Bβ antibody (panels 1 and 2), anti-hMex-3Bβ antibody + hMex-3B peptide, as control (panel 3). (C) Serial sections of human Meckel's diverticulum (magnification: ×100 or ×400) were stained with anti-hMex-3Bβ (top left) and anti-MUC2 (bottom left), anti-hMex-3Bβ (top middle) and anti-Chromogranin-A (bottom middle), anti-hMex-3Bβ (top right) and anti-Lysozyme (bottom right).
In order to study the expression of hMex-3 proteins in human tissue, we have analyzed histologically normal colon mucosa and Meckel's diverticulum which gave a weak, but nevertheless reproducible signal in RT-PCR. Using a polyclonal antibody raised in the laboratory, hMex-3B expression was found to be restricted to a subpopulation of intestinal epithelial cells in the colon (Figure 2B) and in the small intestine (Figure 2C), whereas we failed to detect any expression in the lamina propria and the intestinal smooth muscles. The signal was specific for hMex-3B since pre-incubation of the antibody with the corresponding immunogenic peptide eliminated the labeling (Figure 2B). We noticed that hMex-3B was expressed in a peculiar repeated pattern reminiscent of the distribution of the epithelial cells belonging to the secretory lineage, whereas cells of the absorptive lineage remained unlabeled (Figure 2B and C). Consistent with this observation, staining of serial sections with an anti-MUC2 antibody, which recognizes a specific marker of goblet cells, showed a distribution similar to that of hMex-3B (Figure 2C). As shown in Figure 2C, staining with an anti-hMex3B antibody revealed no overlap between hMex-3B-containing cells and either enteroendocrine cells or Paneth cells (distinguished by staining for Chromogranin-A and lysozyme, respectively), thus confirming that the hMex-3B protein is specifically expressed in intestinal mucin-secreting cells. Using sections of mouse testis, we have also obtained evidence that hMex-3B protein is expressed in the epididymal epithelial cells (data not shown). Overall, these results are consistent with the mRNA expression profile and confirm that the endogenous hMex-3B protein is expressed.
hMex-3 proteins are RNA-binding proteins
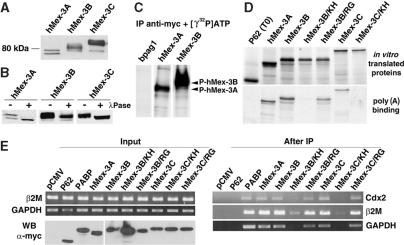
During the course of this study, we cloned the RT-PCR amplified hMex-3A, -3B and -3C coding sequences into an eukaryotic expression vector. The three hMex-3 proteins were tagged N-terminally with the c-myc epitope and these constructs transfected into BOSC cells (derived from the human embryonic kidney epithelial cell line 293T). Two days post-transfection, cells were lysed and protein samples were analysed by western blot with an anti-myc monoclonal antibody. As shown in Figure 3A, the myc-hMex-3 fusion proteins range between 60 and 90 kDa. The anti-myc monoclonal antibody reacted with several bands from the different protein extracts originating from the transfected cells and these same bands were detected using specific polyclonal sera raised against hMex-3A or hMex-3B (data not shown). The observation of an electrophoretic mobility shift of the three hMex-3 proteins suggests that these proteins could be post-translationally modified. Treatment of protein lysates with λ-phosphatase (Figure 3B) followed by western blotting with the anti-myc antibody resulted in the disappearance of the higher molecular weight bands, thus indicating that the hMex-3 proteins are phosphorylated. To confirm that hMex-3A and hMex-3B are phosphoproteins, we performed an immunokinase assay. hMex-3A and hMex-3B were immunoprecipitated with the anti-myc monoclonal antibody and the resulting immunocomplexes were incubated in the presence of [γ-32P]ATP (Figure 3C). Autoradiography revealed that hMex-3A and hMex-3B, but not the control protein Bpag1, were phosphorylated under these conditions.
Figure 3.
Biochemical characterization of hMex-3 proteins. (A and B) BOSC cells were transiently transfected with vectors expressing myc-tagged forms of hMex-3A, -3B and -3C. Western blot analysis was performed with anti-myc antibody. In (B) treatment of protein extracts with (+) or without (−) λ-Phosphatase. (C) Kinase assay. hMex-3A and -3B proteins or a control protein (Bpag1) expressed in BOSC cells were immunoprecipitated with the anti-myc antibody and incubated with kinase buffer and [γ32P] ATP. Labeled proteins were revealed by autoradiography. (D) RNA homopolymer binding assay. Proteins from indicated expression vectors were in vitro translated in the presence of [35S] methionine (top). Binding to agarose beads coupled to poly(A) (bottom) RNA homopolymers is shown for in vitro translated proteins. As a negative control, a fragment of P62-sequestosome protein was incubated with RNA homopolymers in the same conditions. One-tenth of the initial translation reactions and all the bound proteins were analysed by SDS-PAGE and autoradiography. (E) In vivo hMex-3 binding to mRNA. BOSC cells were transiently transfected with vectors expressing myc-tagged proteins, as indicated. RT-PCR amplification was performed on total RNA extracted from those cells (top left). Western blot analysis performed with anti-myc antibody (bottom left). RT-PCR amplification performed on total RNA extracted from sepharose-protein A beads after immunoprecipitation by an anti-myc antibody (right).
Next, we explored whether the hMex-3 proteins interact with RNA by measuring the ability of hMex-3A, -3B and -3C to bind agarose beads on which ribonucleotide polymers of poly(A) had been immobilized. hMex-3 proteins, in vitro translated in the presence of [35S]-methionine, were incubated with the beads and the bound proteins eluted and detected by autoradiography. As shown in Figure 3D, hMex-3A, -3B and -3C are able to bind poly(A) in vitro. They most likely bind RNA via their KH domains because a specific mutation in the second KH domain prevents hMex-3B and hMex-3C binding, whereas mutations at conserved residues of the RING finger motif has no effect. Similar results were obtained with ribonucleotide poly(U) (data not shown).
To confirm that hMex-3 proteins interact with RNA in vivo, we used a protocol which relied on an immunopurification step followed by the identification of the RNA co-precipitated by RT-PCR. BOSC cells were transfected with vectors expressing hMex-3A, -3B, -3C as well as various mutants, and 48 h post-transfection, cells were lysed and hMex-3 protein–RNA complexes were immunoprecipitated with the anti-myc monoclonal antibody. As a positive control, the poly(A)-binding protein 1 (PABP) was similarly expressed in BOSC cells. The RNA recovered by immunoprecipitation was analysed by RT-PCR using primers specific for the genes coding for the transcription factor Cdx2, β2-microglobulin and the housekeeping enzyme GAPDH. As shown in Figure 3E, those mRNAs were detected in hMex-3 and PABP immunoprecipitates, whereas no mRNA was purified with the myc-tagged p62 protein. Furthermore, these mRNAs were not found in association with the KH mutants, whereas no difference was observed with the RING finger mutant. Interpretation of immunoprecipitation analysis must be taken with caution since it is known that RNA-binding proteins may interact with mRNAs subsequent to cell lysis (24), and additional experiments will sort out what the genuine hMex-3 mRNA targets are. However, the above experiments are consistent with the in vitro results and confirm that hMex-3 proteins interact with mRNAs via their KH domains.
Nucleocytoplasmic shuttling of hMex-3 proteins
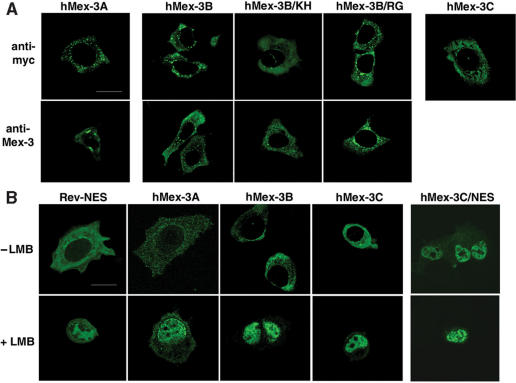
To examine the subcellular distribution of hMex-3 proteins, we transiently expressed hMex-3A, -3B and -3C into human breast cancer MCF7 cells and carried out indirect immunofluorescence analysis with the anti-myc tag antibody. Confocal microscopy analysis revealed that hMex-3 proteins are located in the cytoplasm and excluded from the nucleus. However, we noticed a clear, specific labeling pattern, with hMex-3A and -3B showing a punctuated staining whereas hMex-3C was distributed more homogenously throughout the cytoplasm (Figure 4A). Strikingly, a mutation in the KH domain eliminated the spotted localization of hMex-3B, while a mutation in the RING finger domain did not modify the topology of the staining (Figure 4A). Using our polyclonal antibodies directed against hMex-3A and -3B we observed an expression pattern similar to the one observed with the anti-myc antibody (Figure 4A), thus further confirming the specificity of these anti-hMex-3 sera. However, we were unable to detect the endogenous hMex-3 proteins, probably due to their weak expression in MCF7 cells (see Figure 2A).
Figure 4.
Subcellular localization of hMex-3 proteins. (A) MCF7 cells expressing hMex-3A, -3B and -3C were stained with anti-myc antibody (top) or with specific anti-hM3Aβ or anti-hM3Bβ antibodies (bottom) and revealed by FITC-conjugated secondary antibodies. (B) MCF7 cells expressing hMex-3A, -3B, -3C proteins or hMex-3C mutated within the nuclear export signal (NES) sequence were treated with or without 20 ng/ml of Leptomycin B (LMB) and were stained as above. pEGFP-Rev-NES construct was used as a positive control. Scale bar: 20 μm.
The hMex-3 proteins contain a potential NES consensus sequence in their N-terminal region (Figure 1B). To determine whether hMex-3 proteins are nucleocytoplasmic shuttling proteins, we expressed them in MCF7 cells in the presence or absence of Leptomycin B (LMB), a specific inhibitor of the CRM1-mediated nuclear export of NES-containing proteins (25) (Figure 4B). As a positive control, we observed that upon LMB treatment, a protein containing the enhanced green fluorescent protein (EGFP) fused to the NES of the protein Rev concentrated in the nucleus (19) (Figure 4B). LMB treatment of the cells expressing the hMex-3 proteins resulted in the massive accumulation of these proteins in the nucleus, thus indicating that they shuttle between the nucleus and the cytoplasm through the CRM1 export pathway. To confirm these results, we introduced two point mutations at conserved residues within the NES sequence of hMex-3C and transiently expressed this mutant form of hMex-3C (hMex-3C/NES) into MCF7 cells. As shown in Figure 4B, hMex-3C/NES was mainly detected in the nucleus, thus confirming that hMex-3 proteins are nucleocytoplasmic shuttling proteins dependent on the N-terminally located NES sequences of the hMex-3 proteins.
hMex-3A and hMex-3B proteins localize to Dcp-containing bodies
The recent demonstration that factors involved in mRNA degradation pathways are enriched in cytoplasmic granules, prompted us to study whether the hMex-3A and hMex-3B foci correspond to these structures. Granules known as processing bodies (P bodies) in yeast (26), and GW-182 bodies (27) or Dcp-containing bodies in mammals (18) are the cellular sites of mRNA turnover (we will refer these structures as P bodies in the rest of the text).
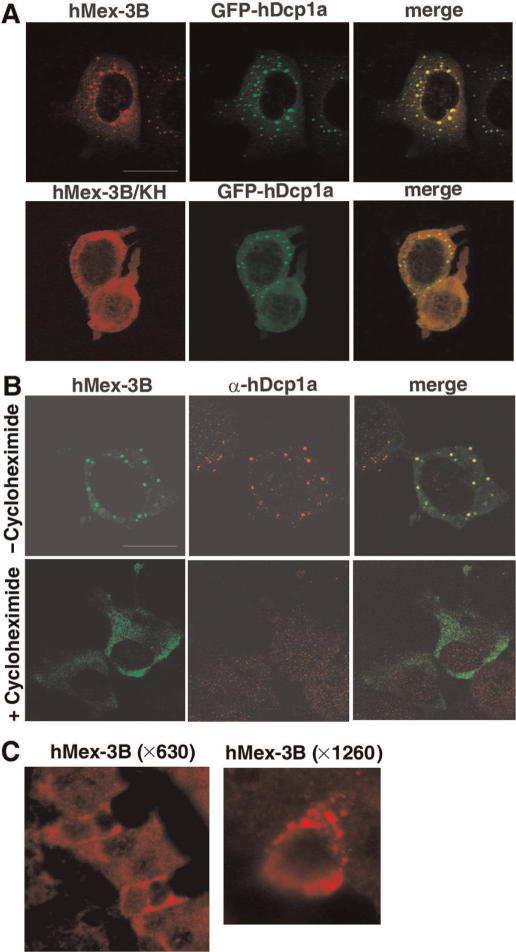
The human mRNA decapping enzyme, hDcp1a, is concentrated in P bodies (28). MCF7 cells were co-transfected with vectors expressing hMex-3B and a GFP-hDcp1a fusion protein (Figure 5A). Indirect immunofluorescence and confocal analyses revealed that hMex-3B colocalized with hDcp1 in cytoplasmic foci, although we repeatedly observed that some of the hMex-3B-containing granules were devoid of hDcp1. hMex-3A, but not hMex-3C, was also found in Dcp-containing granules (data not shown). As suggested by the results displayed in Figure 5A, the hMex-3B KH mutant was no longer localized in P bodies and this was not due to the disruption of the P bodies’ integrity, since the punctuate distribution of Dcp1a was not affected by the expression of the hMex-3B KH mutant (Figure 5A).
Figure 5.
Localization of hMex-3B in Dcp-containing bodies. (A) MCF7 cells expressing hMex-3B or hMex-3B/KH and GFP-hDcp1a proteins were stained with anti-hM3Bα antibody and revealed with ALEXA555-conjugated secondary antibody (left). Central panel shows GFP signal. Right panel shows overlay of the two signals. (B) MCF7 cells expressing hMex-3B protein treated with or without 5 μg/ml of cycloheximide were stained with anti-myc antibody (left) and anti-hDcp1a antibody (middle) and revealed with ALEXA488 and ALEXA555-conjugated secondary antibodies. Right panels show overlay of the two signals. Scale bar: 20 μm. (C) Sections of mouse duodenum (magnification: ×630 or ×1260) were stained with anti-hMex-3Bβ and revealed with an ALEXA555-conjugated secondary antibody. Tissues were mounted and observed by a confocal microscope, as described in the materials and methods section.
Cell treatment with a translational inhibitor, such as cycloheximide, is known to result in the disappearance of Dcp-containing-foci (18). MCF7 cells transiently expressing hMex-3B were treated with cycloheximide and stained by indirect immunofluorescence with an anti-myc tag antibody and anti-hDcp1a serum. As shown in Figure 5B, such treatment led to a near complete loss of P bodies and, concomitantly, to the alteration of the punctuate pattern of hMex-3B expression towards a more uniform cytoplasmic distribution. Consistent with these results, endogenous hMex-3B protein was found concentrated in cytoplasmic granules of duodenal goblet cells, which are morphologically similar to P bodies (Figure 5C). Overall, these data indicate that hMex-3A and hMex-3B, but not hMex-3C, should be considered as new components of P bodies.
Interaction of hMex-3 and Ago proteins
The human Argonautes (Ago) family comprises eight members, four of which (Ago1 to Ago4) constitute a subfamily of highly homologous proteins (29). Ago proteins are components of the RNA-induced silencing complex (RISC) and Ago2 is endowed with an endonuclease activity involved in the miRNA and siRNA-guided cleavage of target mRNAs (20). Consistent with their function in RNA-silencing mechanisms, Ago1 and Ago2 proteins have been localized in P bodies (30–32).
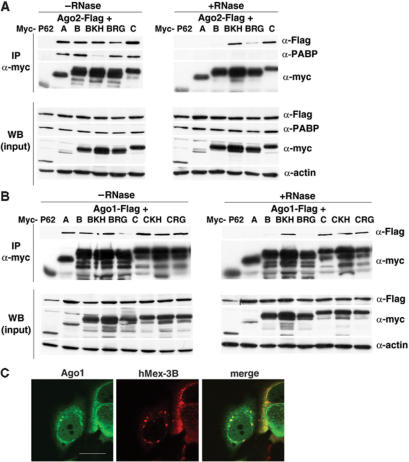
To determine whether hMex-3 proteins associate with Ago, BOSC cells were transiently transfected with constructs that encode Flag-tagged Ago1 or Ago2 together with vectors expressing hMex-3 proteins. hMex-3 proteins were immunoprecipitated with the anti-myc monoclonal antibody and the co-precipitated proteins were detected by western blot with an anti-Flag antibody. As shown in Figure 6A and B, Ago1 and Ago2 were co-precipitated with hMex-3A, -3B and -3C proteins as well as with KH and RING finger mutants but not with the myc-p62 control. The interaction between hMex-3A, -3B and Ago1, 2 proteins is most likely indirect and mediated by RNA because RNase treatment of cell lysates prevented co-precipitation of Ago1 or Ago2 by hMex-3 proteins (Figure 6A and B). In these conditions, the interactions between hMex-3 proteins and PABP were also completely disrupted (Figure 6A and B). Interestingly, the proteins Ago1 and Ago2 were efficiently co-precipitated by hMex-3C WT and hMex-3B bearing the KH mutation after RNAse treatment. These observations strongly suggest that the two proteins, hMex-3C WT and hMex-3B bearing the KH mutation, directly interact with Ago1 or Ago2 outside the P bodies. In contrast, hMex-3A and -3B WT localized inside the P bodies do not directly interact with Ago1 and Ago2. Accordingly, immunofluorescence analysis showed that hMex-3B and Ago1 colocalized in P bodies (Figure 6C).
Figure 6.
Interaction of hMex-3 proteins with Ago1 and Ago2. BOSC cells were transiently co-transfected with vectors expressing myc-tagged proteins, as indicated and Flag-HA-Ago2 (A) or Flag-HA-Ago1 (B). Protein extracts were treated with or without RNAseA (0.2 mg/ml) before immunoprecipitation (IP) with an anti-myc antibody followed by western blot analysis performed with indicated antibodies (top panels) or before direct western blot (WB) analysis performed with indicated antibodies (bottom panels). (C) MCF7 cells co-expressing myc–tagged hMex-3B and Flag-HA-Ago1 proteins were stained with the anti-hM3Bβ serum (middle) and a monoclonal anti-HA antibody (left) revealed with ALEXA488 and ALEXA555-conjugated secondary antibodies. Right panel shows overlay of the two signals. Scale bar: 20 μm.
DISCUSSION
hMex-3, a novel family of RNA-binding proteins
In this study, we have reported the characterization of the human homologs of the C. elegans Mex-3 gene. The four human genes (called hMex-3A, -3B, -3C, -3D) are located on distinct chromosomes and encode closely related proteins which contain two tandemly repeated KH domains. A carboxy-terminal RING finger domain, which is lacking in the C. elegans Mex-3, is present in an Ascidian ortholog of Mex-3, called pem-3 (23), as well as in Drosophila ortholog (data not shown). RING fingers function as ubiquitin–protein ligases (33), thus suggesting that hMex-3 proteins may mediate either self-ubiquitination or ubiquitination of target proteins and thus, regulate their stability and/or subcellular localization.
Using an RT-PCR approach, we showed that hMex-3D was ubiquitously expressed in human tissues whereas the other three hMex-3 genes showed a more heterogeneous expression with the highest levels being found in fetal brain and testis. At the protein level, using a polyclonal serum specific for hMex-3B, we detected this protein in the adult intestinal epithelium, specifically in goblet cells. These results raise the possibility that hMex-3B is either involved in the specification of intestinal progenitor in goblet cells or regulate their mucus ‘secretory’ phenotype, an observation which warrants further investigation. The characterisation of the protein expression profile of the other three hMex-3 proteins awaits the development of high specificity antibodies but it is possible that they too are present in distinct intestinal epithelial cell types, and this situation might similarly apply to other organs. Alternatively, hMex-3 may be coexpressed in the same cell type and play nonredundant biological functions, as suggested by the distinct cellular localization of hMex-3C and its distinct interaction with Ago proteins.
hMex-3 are RNA-binding phosphoproteins that shuttle between the nucleus and the cytoplasm
During this study, we found that hMex-3A, -3B and -3C are proteins which predominantly accumulate in the cytoplasm, and shuttle between the cytoplasm and the nucleus via their NES motif and the CRM1/exportin 1 machinery (25). We further obtained evidence that hMex-3 are phosphoproteins and several potential phosphorylation sites for known protein kinases are predicted on the hMex-3 protein sequences (data not shown). In agreement with these results, we have recently found that hMex-3B specifically interacts with 14-3-3 scaffold proteins in a phosphorylation-dependent manner (data not shown).
Consistent with the presence of the two KH domains, hMex-3A, -3B and -3C proteins interact in vitro with RNA homopolymers and with cellular mRNA. Introduction of a mutation, which results in the replacement of a conserved Gly by an Asp within the second KH domain, eliminates hMex-3 RNA binding. The same mutation was identified in C. elegans and found to disrupt the Mex-3 gene function (3). These results indicate that hMex-3 proteins bind RNA through their KH domains, although we cannot formally exclude that the binding is indirect and mediated by a protein that recognizes RNA and interacts with the KH domains of hMex-3 (34).
Donnini and coworkers recently reported that TINO, which corresponds to a shorter form of hMex-3D (see the results section), binds to a sequence containing the AU-rich elements (AREs) of the BCL-2 transcript (22). However, the target sequence of hMex-3D/TINO is not precisely defined yet and the recent resolution of the crystallographic structure of the third KH domain of hnRNP K suggests that the core sequence of the KH domain might be as short as a tetranucleotide (35). It is thus possible that each KH domain in the hMex-3 proteins interacts with distinct RNA sequences on the same transcript, or alternatively, that both KH domains may contribute to the high-affinity recognition of a single sequence on mRNA as previously exemplified for the Drosophila P-element somatic inhibitor (36).
hMex-3A and -3B are two novel components of P bodies
Analysis of the cellular distribution of hMex-3 proteins revealed that hMex-3A and -3B colocalize with both the decapping enzyme hDcp1a and Ago1 proteins in P bodies (28). These structures contain factors involved in the 5′–3′ degradative pathway of eukaryotic mRNA, such as the hCcr4 deadenylase and the Xrn1 exonuclease (18,26,28,37). Translation inhibition is known to block mRNA decay and to decrease drastically the number of foci (18). In agreement with this result, cycloheximide treatment similarly reduced the number of hMex-3 granules. Furthermore, mutation of the KH domain abrogated the accumulation of hMex-3B in cytoplasmic foci, indicating that recruitment to these structures depends on RNA binding. Taken together, our data provide evidence that hMex-3A and hMex-3B are two novel components of P bodies. Of note, hMex-3C did not show the same distribution pattern, an observation which supports the notion that hMex-3 proteins have intrinsically specific properties. Evidence is mounting that P bodies are both the sites of mRNA degradation and of sequestration of nontranslated transcripts (26,38). Therefore, the localization of hMex-3A and hMex-3B in P bodies is compatible either with a function in RNA decay, as proposed by Donnini and collaborators for TINO (22), or with a role in translational regulation as established for Mex-3 in the nematode (5,6). A role of hMex-3 in translation regulation is not incompatible with a function in the control of RNA stability, and there are several documented examples of RNA-binding proteins involved in both processes (39). Interestingly, we found that hMex-3A and hMex-3B do not form a physical complex with Ago proteins whereas hMex-3B KH mutant, which is located outside the P bodies, interact with Ago. These data suggest that, upon binding to their mRNA targets, Ago and hMex-3A or hMex-3B dissociate and the ribonucleoprotein complex is concomitantly ferried towards P bodies. This hypothesis is supported by the immunofluorescence analysis which showed that hMex-3B and Ago1 colocalized in P bodies.
Stress granules (SGs), which are cytoplasmic structures whose assembly is induced by the exposure of mammal cells to various environmental stresses (40,41) function as a triage center, exporting mRNA destined to be degraded to P bodies or allowing reinitiation of translation on polysomes (42,43). It is thus possible that hMex-3 proteins might relocate to these granules under certain conditions, a question which is currently under investigation.
Interestingly, in C. elegans, the Mex-3 protein is a constituent of P granules which are ribonucleoprotein particles associated with germ cells. Mex-3 appears to play a role in the correct segregation of these granules in the P4 blastomere which gives rise to the germline (3). Therefore, our data buttress the attracting idea that P granules and P bodies may constitute related structures with similar functions (40,44).
A conserved role for hMex-3 in the maintenance of stem cell pluripotency?
Previous studies have established that Mex-3 controls the spatial patterning of PAL-1 in the C. elegans early embryo (5,6,45,46). Homologs of pal-1 have been characterized in many organisms, including caudal in Drosophila (47) and Cdx in mammals (48,49). Interestingly, Cdx2 represses expression of Oct3/4, a transcription factor which is a critical determinant of the pluripotent stem cell phenotype (50). Furthermore, the Ascidian ortholog of Mex-3 is expressed in the blastomere supposed to give rise to the primordial germ cells (23). Collectively, these results suggest that the function of Mex-3 in the maintenance of cells with multiple morphogenetic potentialities, as recently demonstrated for the nematode germline (7), may have been conserved throughout evolution. The characterization of the human Mex-3 proteins reported here, will allow their contribution to these vital biological processes to be determined and to uncover their interplay with RNA metabolism.
ACKNOWLEDGEMENTS
We are grateful to I. Van Seuningen (Lille, France) and G. Dreyfuss (University of Pennsylvania, USA) for kindly providing the anti-MUC2 and anti-PABP antibodies, respectively. This work was supported by a grant from the French Ligue Nationale Contre le Cancer (équipe labelisée). KBP was the recipient of a grant from the French Fondation pour la Recherche Médicale, and from the Centre Régional de Lutte contre le Cancer Léon Bérard. JC was the recipient of a grant from the Centre National pour la Recherche Scientifique. Funding to pay the Open Access publication charge was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- 2.Labbe JC, Goldstein B. Embryonic development: a new SPN on cell fate specification. Curr. Biol. 2002;12:R396–398. doi: 10.1016/s0960-9822(02)00884-9. [DOI] [PubMed] [Google Scholar]
- 3.Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- 4.Grishin NV. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- 6.Huang NN, Mootz DE, Walhout AJ, Vidal M, Hunter CP. MEX-3 interacting proteins link cell polarity to asymmetric gene expression in Caenorhabditis elegans. Development. 2002;129:747–759. doi: 10.1242/dev.129.3.747. [DOI] [PubMed] [Google Scholar]
- 7.Ciosk R, DePalma M, Priess JR. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science. 2006;311:851–853. doi: 10.1126/science.1122491. [DOI] [PubMed] [Google Scholar]
- 8.Anderson P. A place for RNAi. Dev. Cell. 2005;9:311–312. doi: 10.1016/j.devcel.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 11.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S.A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 13.la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirot O, O'Toole E, Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 17.Grange T, de Sa CM, Oddos J, Pictet R. Human mRNA polyadenylate binding protein: evolutionary conservation of a nucleic acid binding motif. Nucleic Acids Res. 1987;15:4771–4787. doi: 10.1093/nar/15.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Degot S, Le Hir H, Alpy F, Kedinger V, Stoll I, Wendling C, Seraphin B, Rio MC, Tomasetto C. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J. Biol. Chem. 2004;279:33702–33715. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- 20.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Nony P, Gaude H, Rossel M, Fournier L, Rouault JP, Billaud M. Stability of the Peutz-Jeghers syndrome kinase LKB1 requires its binding to the molecular chaperones Hsp90/Cdc37. Oncogene. 2003;22:9165–9175. doi: 10.1038/sj.onc.1207179. [DOI] [PubMed] [Google Scholar]
- 22.Donnini M, Lapucci A, Papucci L, Witort E, Jacquier A, Brewer G, Nicolin A, Capaccioli S, Schiavone N. Identification of TINO: a new evolutionarily conserved BCL-2 AU-rich element RNA-binding protein. J. Biol. Chem. 2004;279:20154–20166. doi: 10.1074/jbc.M314071200. [DOI] [PubMed] [Google Scholar]
- 23.Satou Y. Posterior end mark 3 (pem-3), an ascidian maternally expressed gene with localized mRNA encodes a protein with Caenorhabditis elegans MEX-3-like KH domains. Dev. Biol. 1999;212:337–350. doi: 10.1006/dbio.1999.9336. [DOI] [PubMed] [Google Scholar]
- 24.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 26.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 32.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 33.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 34.Chen T, Damaj BB, Herrera C, Lasko P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol. Cell. Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Backe PH, Messias AC, Ravelli RB, Sattler M, Cusack S. X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure. 2005;13:1055–1067. doi: 10.1016/j.str.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 36.Chmiel NH, Rio DC, Doudna JA. Distinct contributions of KH domains to substrate binding affinity of Drosophila P-element somatic inhibitor protein. RNA. 2006;12:283–291. doi: 10.1261/rna.2175706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 38.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin. Cell Dev. Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fitzler MJ, Scheuner D, Kaufman RJ, Golan DE, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mootz D, Ho DM, Hunter CP. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development. 2004;131:3263–3272. doi: 10.1242/dev.01196. [DOI] [PubMed] [Google Scholar]
- 46.Bowerman B, Ingram MK, Hunter CP. The maternal par genes and the segregation of cell fate specification activities in early Caenorhabditis elegans embryos. Development. 1997;124:3815–3826. doi: 10.1242/dev.124.19.3815. [DOI] [PubMed] [Google Scholar]
- 47.Mlodzik M, Gehring WJ. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell. 1987;48:465–478. doi: 10.1016/0092-8674(87)90197-8. [DOI] [PubMed] [Google Scholar]
- 48.Gamer LW, Wright CV. Murine Cdx-4 bears striking similarities to the Drosophila caudal gene in its homeodomain sequence and early expression pattern. Mech. Dev. 1993;43:71–81. doi: 10.1016/0925-4773(93)90024-r. [DOI] [PubMed] [Google Scholar]
- 49.Suh E, Chen L, Taylor J, Traber PG. A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol. Cell. Biol. 1994;14:7340–7351. doi: 10.1128/mcb.14.11.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]