Abstract
We used a cDNA microarray approach to monitor the expression profile of rice (Oryza sativa) under cold stress and identified 328 cold-regulated genes. Thirteen such genes encoding MYB, homeodomain, and zinc finger proteins with unknown functions showed a significant change in expression under 72-h cold stress. Among them, OsMYB3R-2 was selected for further study. Unlike most plant R2R3 MYB transcription factors, OsMYB3R-2 has three imperfect repeats in the DNA-binding domain, the same as in animal c-MYB proteins. Expression of OsMYB3R-2 was induced by cold, drought, and salt stress. The Arabidopsis (Arabidopsis thaliana) transgenic plants overexpressing OsMYB3R-2 showed increased tolerance to cold, drought, and salt stress, and the seed germination of transgenic plants was more tolerant to abscisic acid or NaCl than that of wild type. The expression of some clod-related genes, such as dehydration-responsive element-binding protein 2A, COR15a, and RCI2A, was increased to a higher level in OsMYB3R-2-overexpressing plants than in wild type. These results suggest that OsMYB3R-2 acts as a master switch in stress tolerance.
Plants are exposed to environmental conditions that frequently impose constraints on growth and development. Among them, low temperature stress is one of the serious environmental stresses affecting plant growth and agricultural production. On exposure of plants to low temperature, a series of genes are induced, the products of which may either directly protect against stress or further control the expression of other target genes (Yamaguchi-Shinozaki and Shinozaki, 2006). In Arabidopsis (Arabidopsis thaliana), a major transcriptional regulatory system that controls abscisic acid (ABA)-independent gene expression in response to low temperatures has been identified (Stockinger et al., 1997; Liu et al., 1998). The system is based on the C-repeat (Baker et al., 1994)/dehydration-responsive element (Yamaguchi-Shinozaki and Shinozaki, 1994) that interacts with C-repeat-binding factors (CBFs). Under cold stress, CBF/dehydration-responsive element-binding protein 1 (DREB1) genes are rapidly and transiently induced and subsequently activate the expression of target genes (Gilmour et al., 1998). Several studies have reported that ectopic overexpression of some CBFs resulted in both activation of target genes and enhanced freezing, salt, or dehydration tolerance of transgenic plants (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999; Haake et al., 2002).
The CBF pathway is a central component of cold response, but CBF-independent pathways might also be necessary for the cold stress response (Zhu et al., 2004). Direct evidence exists for the activities of some cold-regulated transcription factors (TFs) not participating in the CBF cold-response pathway (Fowler and Thomashow, 2002), which suggests that TFs play a crucial role in controlling downstream gene expression as well as the regulation of cross talk between different signaling pathways. The key to understanding plant cold response lies in the identification of new components involved in those processes and the elucidation of the signaling pathways.
Rice (Oryza sativa) is a model monocot system and one of the most important food crops in Asia (Khush, 1997; Tyagi et al., 1999; Tyagi and Mohanty, 2000; Cantrell and Reeves, 2002). Unlike Arabidopsis and other crops such as wheat (Triticum aestivum), barley (Hordeum vulgare), and rye (Secale cereale), rice is adversely affected by cold, drought, and salt stress. Cold stress especially limits rice production. Minimizing the loss caused by low temperatures will not only help improve net product but will also extend rice cultivation in marginal lands not able to be cultivated (Khush, 1999; Tyagi and Mohanty, 2000). Rice exposed to cold stress showed marked changes in gene expression, biomembrane lipid composition, and small molecule accumulation (Iba, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). However, much less is known about the regulation mechanism of the rice response to cold stress. Therefore, identifying uncharacterized cold-related genes and defining their functions will enrich the understanding of stress-signaling networks in rice and be important for improving rice tolerance to cold stress.
Here, we report on the isolation and functional characterization of a nuclear-localized R1R2R3 MYB TF designated OsMYB3R-2 (O. sativa R1R2R3 MYB-2) in rice. The protein, like animal c-Myb proteins, contains three imperfect repeat sequences in the N-terminal DNA-binding domain (Jin and Martin, 1999). Overexpression of OsMYB3R-2 in Arabidopsis leads to increased tolerance to freezing, drought, and salt stress.
RESULTS
Isolation of Cold-Responsive MYB TFs from Cold-Tolerant Rice
Yuedongdao, a rice variety possessing characteristics of cold tolerance is a crossed progeny of cultivated rice and Dongxiang wild rice (Oryza rufipogon Griff.), which is a population of common wild rice with increased cold stress tolerance from Dongxiang in the Jiangxi province of China. The Dongxiang wild rice rhizome can survive at a freezing temperature to −12.8°C (Liu et al., 2003a). Our physiological analyses showed Yuedongdao and Dongxiang wild rice seedlings survived under 2°C cold treatment for 72 h, whereas cultivated rice (cold-sensitive rice varieties) did not survive (X. Dai, H. Liu, Y. Xu, and K. Chong, unpublished data). Expression profiles of Yuedongdao under cold stress with 2°C for 72 h were monitored by cDNA microarray (Biostar Genechip), which contains approximately 10,000 rice clones (Liu et al., 2003b). The probes were prepared from RNAs isolated from Yuedongdao seedlings under cold treatment for 72 h and nontreated controls. For hybridization, two replicates of a 2-d swap experiment were performed with RNAs extracted independently from a different batch of plants. Results of the two replicates were highly correlated (r = 0.86). We considered genes with an expression ratio (treatment to control) 2-fold greater or less than that of control genes (|Log2 ratio| ≥ 1) as cold-inducible or cold-repressive genes. A total of 328 genes showing reproducible 2-fold up- or down-regulation were selected. Among them, 157 genes were cold inducible and 171 cold repressive (Table I; Supplemental Table S1).
Table I.
Number of genes differentially expressed on microarray hybridization
Numbers in the table represent the number of genes detected as differentially expressed.
| Log 2 (Treated/Untreated) | 1–1.5 | 1.5–2.0 | 2.0–2.5 | 2.5–3.5 | 3.5–5.0 | Total |
|---|---|---|---|---|---|---|
| Up-regulated genes | 98 | 42 | 13 | 3 | 1 | 157 |
| Down-regulated genes | 93 | 55 | 20 | 2 | 1 | 171 |
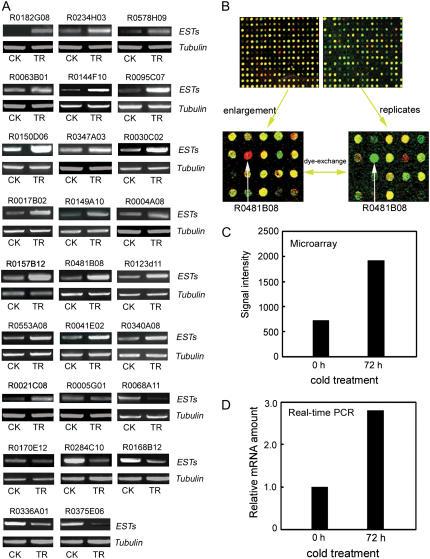
We performed semiquantitative reverse transcription (RT)-PCR to confirm the differentially expressed genes identified by microarray analysis. Twenty-six genes representing different expression profiles were analyzed, of which 25 exhibited expression patterns similar to that from microarray analysis (Fig. 1A); only one, R0005G01, showed no significant difference in gene expression between the treatment and control with RT-PCR amplification. Moreover, all randomly scattered expressed sequence tags that represent the same gene showed a similar differentially expressed pattern in the microarray analysis (Supplemental Table S1). Thus, our cDNA microarray hybridizations were stable and reliable.
Figure 1.
Isolation of cold-inducible MYB TF from microarray hybridization. A, Analysis of the reliability of microarray hybridization by semiquantitative RT-PCR. Expression pattern of 26 genes selected randomly from cDNA microarray shown in A. CK, Nontreatment control; TR, cold treatment for 72 h at 2°C. B, Microarray hybridization signal of R0481B08 in two dye-exchange replicates. C, Signal intensity of R0481B08 in microarray hybridization. D, Real-time PCR to validate R0481B08 microarray results presented in C.
Among the 328 cold-responsive genes, we identified three genes encoding cold-inducible TFs, including two MYB TFs and one homeodomain TF. One, an expressed sequence tag R0481B08 (accession no. BAD81765) encoding a putative R1R2R3 MYB TF was selected for further functional studies. In our microarray hybridization, R0481B08 showed a completely opposite hybridization signal in two dye-exchange replicates (Fig. 1B). The transcript level was increased 2.6-fold when the cold-treated sample was labeled with Cy5 and the untreated sample was labeled with Cy3, whereas in the dye-exchange hybridization, the transcript level was decreased 2.3-fold (Supplemental Table S1). The expression of R0481B08 in the microarray analysis was confirmed by semiquantitative RT-PCR (Fig. 1A). Moreover, real-time PCR used to further examine its expression in microarray hybridizations showed a similar amount of change as the microarray data (Fig. 1D), which strongly supports the validity of the cold-regulated expression pattern from the microarray analysis (Fig. 1, C and D).
Structural Features, Phylogenetic Tree, and Subcellular Localization of OsMYB3R-2
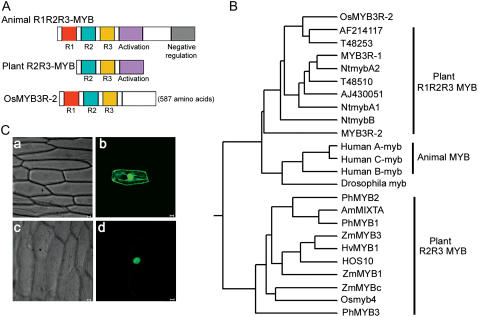
To investigate the function of R0481B08, we amplified its full-length cDNA by RT-PCR from rice seedlings treated for 72 h at 2°C. The full-length cDNA contains an open reading frame of 587 amino acids with a calculated molecular mass of 63.9 kD. Homological analysis showed that the gene shared the greatest sequence similarity with the MYB TFs from Arabidopsis, rice, Populus, and tobacco (Nicotiana tabacum) within the MYB domain. The MYB domain is composed of three imperfect repeat sequences (R1, R2, and R3) of 50 to 53 amino acids in the mammalian MYB protein c-MYB and related proteins A-MYB and B-MYB (R1R2R3-MYB; Carr and Mott, 1991), whereas most of the plant MYB proteins identified thus far contain only two repeats (R2R3-Myb; Jin and Martin, 1999; Stracke et al., 2001). Interestingly, it contains three repeat sequences (Fig. 2A), like animal c-MYB proteins, so this gene is an R1R2R3-type MYB TF and was designated OsMYB3R-2 (O. sativa R1R2R3 MYB-2). Other members of this group of MYB genes from plants (MYB3R-1, MYB3R-2, T48510, AF214117, T48253 from Arabidopsis; NtmybA1, NtmybA2, and NtmybB from tobacco) have been reported recently (Braun and Grotewold, 1999; Kranz et al., 2000), but the biological functions of the genes from Arabidopsis are unknown.
Figure 2.
Structure, localization, and homological analysis of OsMYB3R-2. A, Scheme showing structures of MYB proteins. Structure of OsMYB3R-2 shown together with functional domains of animal R1R2R3-Myb and typical plant R2R3-Myb proteins. B, Phylogenetic tree of Myb proteins. The tree was constructed with the DNAMAN tree program with amino acid sequences of MYB domains of OsMYB3R-2 and other members of the Myb family isolated from plants and animals, c-Myb, A-Myb, and B-Myb from humans, Drosophila melanogaster Myb, MYB3R-1, MYB3R-2, T48510, AF214117, T48253, and HOS10 from Arabidopsis, ZmMYB1, ZmMYB3, and ZmMYBC from maize (Zea mays), PhMYB1, PhMYB2, and PhMYB3 from petunia (Petunia hybrida), AmMIXTA from Antirrhinum majus, HvMYB1 from barley (Hordeum vulgare), NtmybA1, A2, and NtmybB from tobacco, and Osmyb4 and AJ430051 (MYB3R1) from rice. C, Localization of OsMYB3R-2-GFP protein. GFP alone (b) or OsMYB3R-2-GFP (d) in onion epidermal cells. Corresponding bright-field images (a and c). GFP or OsMYB3R-2-GFP fusion was driven by the control of the CaMV 35S promoter. Onion epidermal peels were bombarded with DNA-coated gold particles, and GFP expression was visualized 24 h later. Bars = 50 μm.
We constructed a phylogenetic tree based on the amino acid sequences of MYB domains of animal and plant Myb proteins (Fig. 2B). The OsMYB3R-2 protein is more similar to AF214117, T48253, MYB3R-1, and NtmybA2 proteins than to plant R2R3-Myb proteins. The OsMYB3R-2 protein showed 66.7% to 74.8% identity with human C-Myb and B-Myb but only 37.6% to 44.1% identity with plant R2R3-Myb proteins. However, OsMYB3R-2 protein did not group with any animal R1R2R3-type Myb proteins (A-Myb, B-Myb, and C-Myb) but, rather, formed a separate branch. OsMYB3R-2 and MYB3R-1 are more closely related to each other than to AJ430051, a putative R1R2R3-type MYB from rice.
To examine its subcellular localization, OsMYB3R-2 was fused in frame to the 5′ terminus of the green fluorescent protein (GFP) reporter gene under the control of the cauliflower mosaic virus 35S promoter (CaMV 35S). The recombinant constructs of the OsMYB3R-2-GFP fusion gene and GFP alone were introduced into onion (Allium cepa) epidermal cells by particle bombardment. As shown in Figure 2C, the OsMYB3R-2-GFP fusion protein accumulated mainly in the nucleus, whereas GFP alone was present throughout the whole cell. Thus, OsMYB3R-2 is a nuclear-localized protein, which is consistent with its predicted function as a TF (Fig. 2C).
Expression Pattern of OsMYB3R-2 in Response to Cold, Salt, and Drought Stress
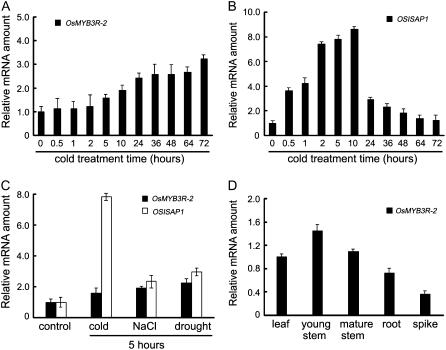
We performed real-time RT-PCR to examine the expression pattern of OsMYB3R-2 under different stress conditions. Under cold stress, the transcripts of OsMYB3R-2 began to increase after 5 h cold treatment and gradually accumulated up to 72 h of treatment (Fig. 3A), which was consistent with our microarray results. In the case of salt and dehydration stress, transcript levels of OsMYB3R-2 were also induced after 5 h treatment as compared with that of nontreated controls (Fig. 3C). To validate this experiment, we used the OSISAP1, a gene encoding zinc-finger protein from rice, as a positive control. OSISAP1 is induced under cold, salt, and drought stress. Under cold treatment, the transcript level of OSISAP1 was increased to a very high level during a 12-h cold treatment and declined thereafter (Mukhopadhyay et al., 2004). We confirmed that the expression of OSISAP1 was induced by cold, desiccation, and salt stress (Fig. 3, B and C), which was consistent with previous studies (Mukhopadhyay et al., 2004). The expression pattern of OsMYB3R-2 under cold stimulation was different from that of OSISAP1, although both were induced by cold stress in rice.
Figure 3.
Real-time PCR analysis for the expression of OsMYB3R-2 in rice. OSISAP1 was used as a positive control. A and B, Time course of OsMYB3R-2 and OSISAP1 expression during cold treatment. C, OsMYB3R-2 expression response to cold, salt, and drought stress. D, OsMYB3R-2 expression in various tissues. Actin was used as an internal control. Data represent means and ses of three replicates.
In addition, we examined tissue-specific expression of OsMYB3R-2 in rice using real-time RT-PCR. The OsMYB3R-2 transcripts were detected in all organs tested, but the highest level was in young stems and the lowest in spikes (Fig. 3D).
Taken together, these results suggest that OsMYB3R-2 is induced under cold, salt, and drought stimulation, which suggests that it functions during these stresses.
Overexpression of OsMYB3R-2 Increases Tolerance to Freezing, Drought, and Salt
To investigate the function of OsMYB3R-2 in plants, we overexpressed OsMYB3R-2 in transgenic Arabidopsis under control of a CaMV 35S promoter. Transformed lines of Arabidopsis were confirmed by hygromycin selection and Southern blotting. Southern blot was performed by using the DNA digested with HindIII or EcoRI and β-glucuronidase (GUS) gene as a probe. Three transgenic lines were randomly selected and showed different hybridized patterns to the GUS probe (Fig. 4A). In the wild type, however, no signals were detected under the same conditions. Therefore, the three transgenic lines could be independent. Furthermore, RNA gel-blot analysis showed that OsMYB3R-2 was expressed at the higher levels in transgenic Arabidopsis than in the wild type (Fig. 4B).
Figure 4.
Molecular characterization and phenotypes of OsMYB3R-2 transgenic Arabidopsis plants. A, Southern-blot assay for Arabidopsis transgenic plants. Genomic DNA isolated from the wild-type (WT) or transformed plants digested with EcoRI (E) or HindIII (H). The blot was hybridized with the open reading frame of the GUS gene labeled with α-32P-dCTP and α-32P-dATP as described in “Materials and Methods.” B, Expression of independent transgenic plant lines of Arabidopsis by RNA gel-blot analysis. Each lane was loaded with 10 μg total RNA isolated from 3-week-old seedlings of transgenic Arabidopsis. The RNA blot was hybridized with a 32P-labeled OsMYB3R-2 cDNA probe. Ethidium bromide-stained rRNA was used as a RNA-loading control. C, The phenotypes of the T3 generation of independent lines of overexpressed OsMYB3R-2 transgenic Arabidopsis. Wild-type and OsMYB3R-2-overexpressed lines after 3 weeks growth at 22°C. [See online article for color version of this figure.]
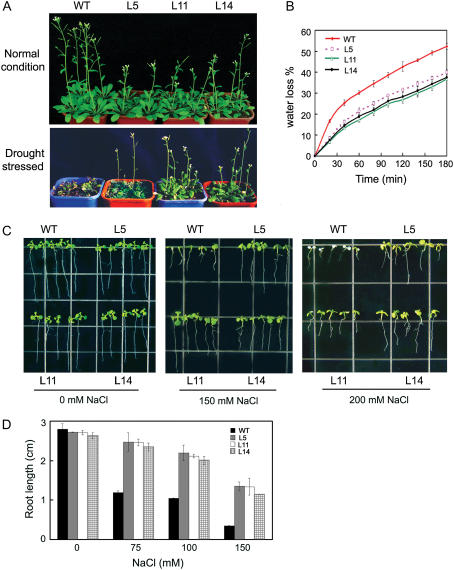
To examine the possible phenotypes of transgenic lines, T3 progeny of the OsMYB3R-2-overexpressed lines and the wild-type plants were grown in the greenhouse under identical conditions. Compared with wild-type plants, transgenic plants showed a little retarded growth under normal conditions (Fig. 4C).
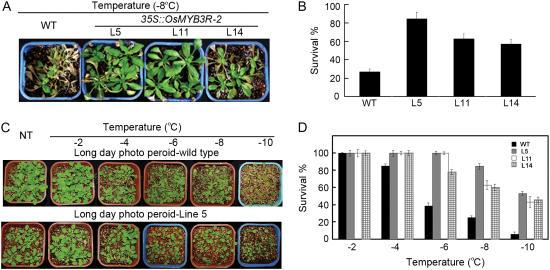
To investigate the effect of OsMYB3R-2 overexpression on freezing tolerance, T3 transgenic and wild-type seedlings were exposed to −8°C for 10 h. After 6 d recovery at normal conditions, survival was 26.8% for the wild-type and 84.5% for transgenic lines (Fig. 5, A and B). Phenotypically, most transgenic seedlings were green and could regrow as compared with the wild type, whereas most wild-type seedlings became white and did not regrow after removed to normal conditions. The survival percentage under different low temperatures also showed dramatic difference between the transgenic plants and wild-type plants (Fig. 5, C and D). At −10°C, the proportion of survived wild-type plants decreased to 5.6%, whereas more than 42.8% of transgenic plants survived. Thus, OsMYB3R-2-overexpression plants show high tolerance to freezing stress.
Figure 5.
Effect of OsMYB3R-2 expression on freezing tolerance in transgenic Arabidopsis plants. A, Four-week-old OsMYB3R-2-overexpressed and wild-type (WT) plants were cold stressed at −8°C for 10 h and then transferred back to the normal condition for recovery. Photographs of representative seedlings of WT and three transgenic lines were taken after 6 d of recovery. B, Quantitative analysis of the plant survival 6 d after the freezing treatment as shown in A. Error bars indicate sd. C, Tolerance of 3-week-old L5 plants at different temperatures below freezing for 10 h. Photographs were taken 6 d after freezing treatment. D, Survival percentage for L5, L11, L14, and wild-type plants was recorded on treatment at temperatures below freezing. Error bars indicate sd.
To determine the effect of OsMYB3R-2 overexpression on drought tolerance, 14-d-old plants grown on soil were not watered for 2 weeks and then watered and grown under normal conditions for 7 d (Fig. 6A). After watering was restarted, transgenic plants showed a stronger growth recovery phenotype than wild-type plants. Only 26.7% of the wild-type plants survived this treatment. In contrast, more than 85% of OsMYB3R-2-overexpressed plants survived (Table II), which suggests that the overexpression of OsMYB3R-2 in transgenic Arabidopsis results in greater tolerance to drought stress than in the wild type. The drought-tolerance phenotype of transgenic plants overexpressing OsMYB3R-2 was consistent with slower water loss in detached rosette leaves as compared with the wild type (Fig. 6B).
Figure 6.
Effect of OsMYB3R-2 expression on drought and salt tolerance in transgenic Arabidopsis plants. A, Drought tolerance of 35S∷OsMYB3R-2. Top, Wild-type and transgenic plants without drought stress treatment. Bottom, Wild-type and transgenic plants were grown for 2 weeks with normal watering, withheld from water for 2 weeks, and then rewatered for 7 d before photographs were taken. B, Water loss in wild-type and transgenic plants. Detached leaves from 25-d-old plants grown on soil were incubated on a bench, and the fresh weight (FW) was measured at the time intervals indicated. Water loss was calculated from the decrease in FW compared with time zero. Error bars indicate sd. C, Salt tolerance of 35S∷OsMYB3R-2. Wild-type and transgenic plants were germinated on MS agar plates, then transferred to a new MS agar plate supplemented with different concentrations of NaCl for 7 d. D, Dose response of transgenic and WT seedlings to NaCl. Data shown are root length. Error bars indicate sd.
Table II.
Survival rates of transgenic plants under drought stress conditions
Two-week-old soil-grown plants withheld from water for 2 weeks, rewatered, and scored 7 d later. Plants were considered dead if all the leaves were brown and there was no regrowth 7 d after rewatering.
| OsMYB3R-2-Overexpressed Lines | Survivala | Totalb | Survivalc |
|---|---|---|---|
| Wild-type | 16 | 60 | 26.7 |
| L5 | 51 | 60 | 85 |
| L11 | 60 | 60 | 100 |
| L14 | 58 | 60 | 96.7 |
Number of surviving plants.
Total plants used in drought assay.
Percentage of surviving plants.
To test the effect of OsMYB3R-2 overexpression on salt tolerance, transgenic and wild-type seedlings were grown as described in “Materials and Methods.” Seedlings of both genetic backgrounds grew normally in NaCl up to 150 mm, but the transgenic seedlings formed longer roots than the wild type when grown vertically under NaCl treatment (Fig. 6, C and D). When NaCl concentration was increased to 200 mm, the growth of the wild type was completely inhibited and the seedlings showed absence of greening, whereas transgenic seedlings were still green and continued to grow (Fig. 6C).
Germination of OsMYB3R-2-Overexpressed Seeds Is Insensitive to ABA and NaCl
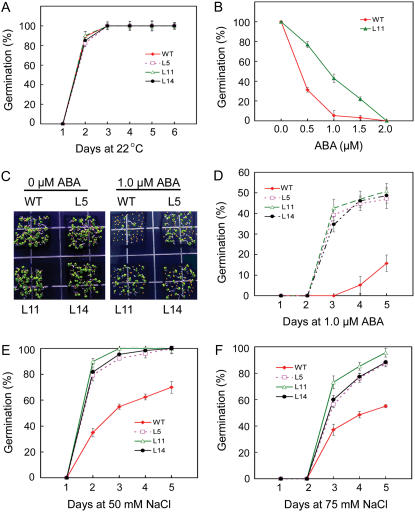
We tested the effect of ABA and NaCl on germination of OsMYB3R-2-overexpressed seeds. There was no difference in seed germination between the wild-type and transgenic plants under normal conditions (Fig. 7A). In the presence of exogenous ABA, the germination of both wild-type and OsMYB3R-2-overexpressed seeds was inhibited significantly, but transgenic seeds inhibited to a lesser extent (Fig. 7B). For example, at 0.5 μm ABA, approximately 80% of OsMYB3R-2-overexpressed seeds germinated comparing with only 30% seeds of the wild type. Under 1.0 μm ABA treatment, most seeds of the wild type did not germinate. In contrast, about one-half the seeds of the transgenic plants germinated and developed green cotyledons and true leaves (Fig. 7, C and D). At ABA levels higher than 2.0 μm, the germination of both wild-type and transgenic seeds was inhibited completely.
Figure 7.
Response of seed germination to ABA and NaCl in transgenic Arabidopsis plants. Seeds were incubated at 0°C for 48 h before being placed at 22°C for germination. Data are means of five replicates (each with 50 seeds for each line). A, Germination in the absence of ABA or NaCl (water only). B, Seed germination of L11 on MS agar plates with different concentrations of ABA. C, Seed germination on MS agar plates with or without 1.0 μm ABA. The picture was taken 10 d after imbibition. D, Germination in the presence of 1.0 μm ABA. E, Seed germination on MS agar plates saturated with 50 mm NaCl. F, Seed germination on MS agar plates saturated with 75 mm NaCl. [See online article for color version of this figure.]
We also observed that the germination of transgenic seeds was more tolerant to NaCl than that of wild type under different NaCl treatments (Fig. 7, E and F). At 50 mm NaCl, nearly 79% to 90% of transgenic seeds germinated at day 2 compared with only 35% seed germination for the wild type (Fig. 7E). At 75 mm NaCl, germination of both wild-type and OsMYB3R-2-overexpressed seeds was completely inhibited at day 2 after imbibition. At day 3, although seeds of both plants began to germinate, the germination in the transgenic plants was significantly higher than that of the wild type (Fig. 7F). Nevertheless, both wild-type and transgenic seeds were not observed to germinate at day 5 after imbibition when NaCl concentrations were at 100 mm (data not shown). Thus, overexpression of OsMYB3R-2 in Arabidopsis increased tolerance to NaCl and ABA during seed germination.
OsMYB3R-2 Activates the Expression of Cold-Responsive Genes
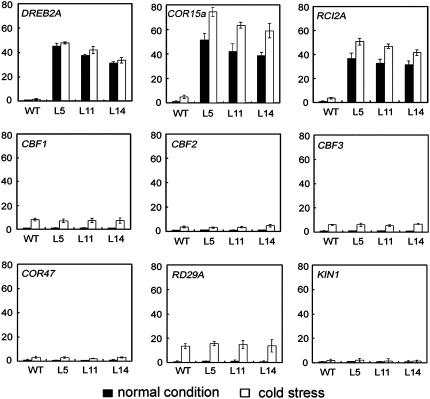
To elucidate the molecular mechanism of OsMYB3R-2 in the cold response, we monitored the expression of cold-responsive genes identified in the regulated pathways by real-time PCR analysis. Under 4°C cold treatment for 6 h, the tested marker genes, including RD29A, CBF1, CBF2, CBF3, KIN1, and COR47, showed slight induction in both wild-type and transgenic plants under cold-stress conditions, consistent with previous studies (Kurkela and Franck, 1990; Gilmour et al., 1992, 1998; Yamaguchi-Shinozaki and Shinozaki, 1993; Stockinger et al., 1997). However, under normal conditions (22°C), the expression of DREB2A, COR15a, and RCI2A in OsMYB3R-2-overexpressed transgenic plants was substantially higher than that in wild-type plants, whereas no significant induction in expression of RD29A, CBF1, CBF2, CBF3, KIN1, and COR47 in both transgenic and wild-type plants (Fig. 8). COR15a and DREB2A are involved in stress signaling by CBF/DREB1 pathways (Artus et al., 1996; Liu et al., 1998), but RCI2A by CBF/DREB1-independent pathways (Medina et al., 2005). Thus, overexpression of OsMYB3R-2 increases expression of DREB2A, COR15a, and RCI2A, which are involved in plant tolerance by different pathways.
Figure 8.
Expression patterns of stress-responsive genes in wild-type and transgenic Arabidopsis using real-time PCR. Total RNA was extracted from 14-d-old plants grown under normal or cold treatment for 6 h, respectively. Transcript levels measured by real-time RT-PCR of DREB2A, COR15a, RCI2A, CBF1, CBF2, CBF3, COR47, RD29A, and KIN1 under normal conditions (black bars) or 4°C treatment for 6 h (white bars). Actin was used as an internal control. Data represent means and ses of three replicates.
DISCUSSION
OsMYB3R-2 Encodes a Cold-Responsive R1R2R3 MYB TF
In plants, the transcripts of genes encoding several families of TFs, such as AP2/EREBP, bZIP/HD-ZIP, and several classes of zinc finger domains, are induced after exposure to various abiotic stresses (Shinozaki and Yamaguchi-Shinozaki, 2000; Seki et al., 2001). These TFs function in various pathways to confer stress tolerance in plants (Ingram and Bartels, 1996; Thomashow, 1999; Hasegawa et al., 2000; Zhu, 2002). MYB TFs are involved in numerous processes (Jin and Martin, 1999; Ito et al., 2001; Stracke et al., 2001). So far, only two R2R3-MYB TFs, HOS10 in Arabidopsis and Osmyb4 in rice, may play essential roles in cold stress by a possible CBF-independent pathway (Vannini et al., 2004; Zhu et al., 2005), but the role of R1R2R3 MYB involved in cold stress is poorly understood. In this study, we identified a cold-inducible R1R2R3 MYB TF, OsMYB3R-2, from cold-insensitive rice, a progeny of cultivated rice and common wild rice with cold-tolerance characteristics (Fig. 1).
R1R2R3-MYB genes seem to constitute a small gene family in plants. In Arabidopsis, five R1R2R3-type Myb genes have been described (Braun and Grotewold, 1999; Kranz et al., 2000), but little about their functions is known. Three R1R2R3-type genes from tobacco, NtmybA1, A2, and NtmybB, have been identified to be involved in M-specific activator (MSA)-mediated G2/M-phase-specific transcription through binding to the MSA element and modulating its activity (Ito et al., 2001). Apparently, unlike most plant R2R3-type MYB proteins, OsMYB3R-2 proteins are closer to the plant R1R2R3 MYB and the animal A-, B-, and C-MYB. The presence of the R1R2R3 MYB motif in the OsMYB3R-2 protein, as well as its nuclear localization, demonstrates that OsMYB3R-2 is an R1R2R3-type MYB TF (Fig. 2). R1R2R3-Myb genes occur in different plant evolutionary lineages, including mosses, ferns, and monocots (Kranz et al., 2000). Thus, in contrast to plant R2R3-Myb, the R1R2R3-type plant MYB proteins could have had a conserved function in eukaryotes. From this evidence, we suggest that OsMYB3R-2 plays a conserved role during stress tolerance in rice.
Usually transcriptional factors are induced rapidly during the early phase of the response to cold, drought, and salt stress, reach maximal induction at several hours, and then decrease in expression level (Thomashow, 2001; Yamaguchi-Shinozaki and Shinozaki, 2006). For example, CBF1 and CBF3 showed peak induction at 6 h in wild-type plants and CBF2 showed peak induction at 3 h during stress treatment (Gong et al., 2002). From the nature of early induction, CBF1/CBF2/CBF3 act early in the signal transduction pathway of the stress response. Expression-pattern analysis shows that the activation pattern of OsMYB3R-2 under cold stress differs from that of other stress-inducible TFs previously reported (Dubouzet et al., 2003; Vannini et al., 2004). The transcript level of genes such as OsDREBIA and OSISAP1, required early after stress, increase to a very high level within 1 h after cold treatment (Dubouzet et al., 2003; Mukhopadhyay et al., 2004), continue to increase until 5 or 3 h, remain at elevated levels until 10 or 12 h, and decline thereafter. However, the transcript level of OsMYB3R-2 increases after cold treatment for 5 h and gradually accumulates within 72 h (Fig. 3A). Furthermore, OsMYB3R-2 is induced by drought and salt stress (Fig. 3C), which is dissimilar to Osmyb4, another MYB TF involved in cold stress in rice that was induced only by cold stress for 4 h (Vannini et al., 2004). Thus, OsMYB3R-2 can be classified as a novel R1R2R3-type MYB TF, and this is the first report, to our knowledge, showing that an R1R2R3-type MYB is involved in cold, drought, and salt stress.
Overexpressed OsMYB3R-2 Increases Tolerance to Stress in Arabidopsis
Certain stress-induced proteins have been shown to impart stress tolerance. Overexpression of genes such as CBF/DREB1, OSISAP1, and HVA1 could confer stress tolerance in transgenic plants (Browse and Xin, 2001; Dubouzet et al., 2003; Mukhopadhyay et al., 2004), although their functions remain to be defined. These examples provide a target for improving stress tolerance of crop plants and give an opportunity to understand the function of previously uncharacterized genes. The expression of OsMYB3R-2 in rice is induced with exposure to cold, drought, and salt stress (Fig. 3). In Arabidopsis, the overexpression of OsMYB3R-2 led to increased tolerance to cold, dehydration, and salt stress (Figs. 5–7). Our data suggest that the overexpressed OsMYB3R-2 protein results in enhanced transduction of stress-response signals. Furthermore, the elevated stress tolerance of 35S∷OsMYB3R-2 plants coincides with up-regulated stress-responsive genes, including DREB2A, COR15a, and RCI2A. COR15a and DREB2A belong to the DRE/CRT class of stress-responsive genes. COR15a from Arabidopsis is induced after cold stress, and its overexpression in transgenic Arabidopsis leads to increased freezing tolerance (Artus et al., 1996; Steponkus et al., 1998). DREB1/CBFs are thought to function in cold-responsive gene expression, whereas DREB2s are involved in high salinity and drought-responsive gene expression (Liu et al., 1998). Thus, the enhanced stress tolerance in OsMYB3R-2 transgenic plants might depend in part on changes in the expression of those genes. However, several tested CBF class or CBF/DREB inducible marker genes, such as CBF1, CBF2, CBF3, RD29A, COR47, and KIN1, did not show increased expression in the 35S∷OsMYB3R-2 plants under normal conditions (Fig. 8), which suggests that some other stress pathways may be involved in OsMYB3R-2-mediated stress tolerance. The high transcription levels of RCI2A in 35S∷OsMYB3R-2 plants support this deduction. RCI2A protein is not a member of the CBF/DREB1 regulon and involvement of CBF/DREB1-independent pathways in modulating stress signaling (Medina et al., 2005). Hydrophilic RCI2A protein may contribute to increased stress tolerance in transgenic plants (Thomashow, 1998; Hasegawa et al., 2000). So the mechanism of OsMYB3R-2 may be to increase the expression of some hydrophilic proteins to enhance stress tolerance.
Several lines of evidence indicate that other signal pathways in addition to those mediated by CBF TFs are involved in cold stress (Fowler and Thomashow, 2002; Kreps et al., 2002). Cross talk between those signal transduction pathways is poorly understood. The high transcript level of DREB2A, COR15a, and RCI2A in 35S∷OsMYB3R-2 plants suggests that OsMYB3R-2 acts as a master switch in stress tolerance and is involved in the complex network controlling stress-responsive genes.
Usually, enhanced drought tolerance accompanies hypersensitivity to ABA treatments during seed germination and early seedling development (Hu et al., 2006; Ko et al., 2006). In contrast, in our system, enhanced tolerance to stress accompanies decreased sensitivity of germination to ABA in overexpressed OsMYB3R-2 transgenic plants. In fact, the phenotype exists in other genes previously reported, such as AtHD2C, CaXTH3, and AtTPS1 (Avonce et al., 2004; Cho et al., 2006; Sridha and Wu, 2006), although the precise mechanism is still unknown. It is possible that there are various ABA signal transduction pathways involved in both processes of tolerance and germination in Arabidopsis.
OsMYB3R-2 differs from HOS10 and Osmyb4. Importantly, the activated genes in 35S∷OsMYB3R-2 Arabidopsis also differ from those in Osmyb4 transgenic Arabidopsis. These results suggest that OsMYB3R-2 is a novel member of the R1R2R3-type MYB family in rice and is involved in stress response.
This study has characterized an R1R2R3-type MYB protein localized at the nucleus in rice and induced by cold, drought, and salt stress. The enhanced stress tolerance of 35S∷OsMYB3R-2 Arabidopsis plants reveals that OsMYB3R-2 could mediate signal transduction, regulating some stress-responsive genes involved in CBF-dependent or -independent pathways. Although the detailed mechanism of OsMYB3R-2 involvement in stress is not yet clear, the characterization of OsMYB3R-2 function will provide new insights into stress pathways. This report provides beneficial information for molecular breeding leading to improved stress tolerance of agricultural crops.
MATERIALS AND METHODS
Plant Materials
We used rice (Oryza sativa) L. cv Yuedongdao, which is insensitive to cold stress. Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used in gene transformation.
Rice seeds were surface sterilized for 5 min with ethanol (75% v/v) and 10 min with commercially diluted (1:3 v/v) NaOCl, followed by several rinses with sterile water. Germination was carried out for 72 h on sterile Murashige and Skoog (MS) medium in the dark at 28°C, then grown under 28°C-day/25°C-night temperatures, 12-h-light/12-h-dark cycle, and 50% humidity. After 2 weeks of germination, seedlings underwent several treatments: cold, 2°C for 0.5, 1, 2, 5, 10, 24, 36, 48, 64, and 72 h; drought, transferred to Whatman 3MM paper in a sterile petri dish for 5 h; and high salinity, 250 mm NaCl for 5 h. After all the treatments, seedlings were harvested, frozen in liquid nitrogen, and stored at −70°C for further analysis. Control plants were harvested at the same time as the treated plants.
Microarray Analysis
Rice seedlings were exposed to cold for 72 h; total RNAs from treated plants and nontreated plants were used for preparation of Cy5- and Cy3-labeled cDNA probes. P100S cDNA microarray (Biostar Genechip) were hybridized with Cy5- and Cy3-labeled probe pairs of cold-treated and nontreated plants. Labeling, hybridization, and washing were performed as described for the CyScribe Post-Labeling kit. Hybridized slides were scanned with use of a GenePix 4000B scanner (Axon Instruments) at 532 and 635 nm to capture the emission of Cy3 and Cy5, respectively. The intensity of each spot at the two wavelengths was transformed into a ratio value with use of the GenePix 4.0 software. Overall intensity of the hybridized slide was normalized by use of GenePix 4.0 software. With the removal of the spots automatically flagged Bad or Not Found by the software, the spots whose ([S − B]/B) < 3, referring to ([media of signal − media of background]/media of background) < 3, were also deleted. Thus, only the spots whose signal intensity was at least 4-fold that of its background were further analyzed, whereas those less than 4-fold that of the background were removed. In addition, we ruled out spots whose regulation pattern was contradictory in two dye-exchange replicates. In this article, only data with |Log2 ratio| ≥ 1 in two replicates were selected as candidate cold-related genes. The reliability of the candidate genes was tested by semiquantitative RT-PCR or real-time PCR.
To further annotate the genes differentially expressed during cold stress, similarity analysis for each sequence involved use of databases from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and The Institute for Genomic Research rice genome project (http://www.tigr.org) with both BLASTn and BLASTp. Functional classification involved gene ontology searches (http://www.geneontology.org).
Semiquantitative RT-PCR and Quantitative Real-Time PCR
Total RNA was extracted from Arabidopsis or rice seedlings with use of Trizol reagent (Invitrogen) and treated with RNase free DNase (Progma).
To confirm the reliability of microarray hybridization, semiquantitative RT-PCR involved use of the One Step RNA PCR kit (AMV; TaKaRa) with gene-specific primers (Supplemental Table S2). Total RNA was isolated from materials collected for microarray hybridization. One microgram of total RNA was used as template in one reaction. The same amplification reaction was conducted with a rice Tubulin gene used as template RNA loading control. RT-PCR reactions were repeated five times.
For real-time PCR, 2 μg total RNA was used for RT with SuperScripts II reverse transcriptase (Invitrogen). The cDNA samples were diluted to 2 and 8 ng/μL. Triplicate quantitative assays were performed on 1 μL of each cDNA dilution with the SYBR GreenMaster mix and an ABI 7900 sequence detection system according to the manufacturer's protocol (Applied Biosystems). The relative quantification method (Delta-Delta CT) was used to evaluate quantitative variation between the replicates examined. The amplification of Actin was used as an internal control to normalize all data. Gene-specific primers for OsMYB3R-2 were 5′-CAG GGT TTC TAT CTC GTT CC-3′ and 5′-ATT TCC AAG CCC TTA CCA C-3′; for OSISAP1, 5′-GAT CAG GAG CCG ACG GAG CT-3′ and 5′-GAC AAA GAA GAC GGC GAC GAG-3′; for DREB2A, 5′-AAG GTA AAG GAG GAC CAG AG-3′ and 5′-ACA CAA CCA GGA GTC TCA AC-3′; for COR15a, 5′-CTC AGT TCG TCG TCG TTT C-3′ and 5′-CAT CTG CTA ATG CCT CTT T-3′; for RCI2A, 5′-ATC GCC ATC CTC TTG CCT CC-3′ and 5′-TAG GAG AAC ACG ACG GAA C-3′; for CBF1, 5′-CTT CGC TGA CTC GGC TTG G-3′ and 5′-ACG CAC CTT CAC TCT GTT CC-3′; for CBF2, 5′-AAC CAG CGG GAA GGA AGA AGT-3′ and 5′-TTT CCT TGG CAC AGG TTG ATT-3′; for CBF3, 5′-GAT CAG CCT GTC TCA ATT TC-3′ and 5′-CTT CTG CCA TAT TAG CCA AC-3′; for COR 47, 5′-TAT CAT GCC AAG ACC ACT GAA-3′ and 5′-CAA CGA AAG CCA CAA TAA CAA-3′; for RD29A, 5′-ATC ACT TGG CTC CAC TGT TGT TC-3′ and 5′-ACA AAA CAC ACA TAA ACA TCC AAA GT-3′; for KIN1, 5′-ACC AAC AAG AAT GCC TTC CA-3′ and 5′-CCG CAT CCG ATA CAC TCT TT-3′; for Actin in Arabidopsis, 5′-GGT AAC ATT GTG CTC AGT GGT GG-3′ and 5′-AAC GAC CTT AAT CTT CAT GCT GC-3′; and for Actin in rice, 5′-GAA CTG GTA TGG TCA AGG CTG-3′ and 5′-ACA CGG AGC TCG TTG TAG AAG-3′.
Localization of OsMYB3R-2-GFP Fusion Proteins
The localization assay was performed as described by Wang et al. (2004). The whole coding sequence of OsMYB3R-2 was amplified with two primers (5′-GCT CTA GAA TGG CGA TGG TGG AGC AGG AGG-3′, XbaI site underlined) and (5′-CGG GGT ACC GGT TAC ATC CAA ATT GGT TG-3′, KpnI site underlined). The PCR product was subcloned into the pBI221 vector to generate pBI221-OsMYB3R-2-GFP containing an OsMYB3R-2-GFP fusion construct under the control of CaMV 35S. The construct was confirmed by sequencing and used for transient transformation of onion (Allium cepa) epidermis via a gene gun (Bio-Rad). Transformed onion cells were observed under a confocal microscope (Nikon).
Transformation of OsMYB3R-2 in Arabidopsis
The digestion product OsMYB3R-2 from pT-OsMYB3R-2 was directionally cloned into the KpnI-BamHI sites of an SN1301 vector to create SN1301-OsMYB3R-2, which carried a GUS marker. OsMYB3R-2 was driven by a CaMV 35S promoter in the construct. The construct was electroporated into the Agrobacterium tumefaciens C58. Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998).
DNA Gel-Blot Analysis
DNA gel-blot analysis was performed as described by Wang et al. (2004). Genomic DNA isolated from 3-week-old Arabidopsis seedlings was digested with EcoRI or HindIII, fractioned electrophoretically on 0.8% (w/v) agarose gel, and blotted onto a nylon membrane (Amersham Pharmacia Biotech). The membrane was prehybridized at 65°C for 2 h and hybridized in the same solution containing α-32P-ATP- and CTP-labeled GUS for 20 h at 65°C. After hybridization, the membrane was washed once with 2× SSC plus 0.1% SDS at 65°C for 20 min, then twice with 1× SSC plus 0.1% SDS at 37°C for 30 min. The membrane was exposed to x-ray film (Eastman-Kodak) at −70°C for 3 to 7 d.
RNA Gel-Blot Analysis
Total RNA was extracted from 3-week-old seedlings of Arabidopsis with use of Trizol reagent (Invitrogen). Total RNA of 20 μg was electrophoresed on 1.2% agarose gel containing 0.4 m formaldehyde and transferred to Hybond-N+ membrane (Amersham Pharmacia Biotech). Probes labeled with α-32P were prepared from OsMYB3R-2 cDNA. Hybridization was the same as for DNA gel blot. The ethidium bromide-stained ribosomal RNA was used as loading control.
Freezing, Drought, and Salt Stress Treatment in Transgenic Arabidopsis
Arabidopsis was grown in 10-cm pots filled with a 1:1 mixture of perlite and vermiculite under a long-day photoperiod (16-h light/8-h dark) at 22°C. Freezing stress involved transferring the 4-week-old or 3-week-old plants into a chamber, decreasing the temperature to −2°C, −4°C, −6°C, −8°C, and −10°C for 10 h respectively, then returning the temperature to 22°C. Plants were analyzed after recovery for 6 d in normal growth conditions.
For the drought stress evaluation, seedlings were grown in pots (10-cm diameter) filled with vermiculite for 2 weeks with constant watering before water was withheld. After 2 weeks without water, all the pots were rewatered simultaneously, and the plant regrowth was scored 7 d later. Plants were considered dead if all the leaves were brown and there was no regrowth 7 d after rewatering.
For water-loss analysis, 10 fully expanded leaves from wild-type and 35S∷OsMYB3R-2 plants that had developed approximately 14 leaves were detached and weighed at different times to determine the rate of water loss. Each experiment was carried out at least three times.
For the salt tolerance assay, transgenic and wild-type seeds were planted on MS agar plates for germination. Two days after germination, seedlings from each line were carefully transferred to a new MS agar plate supplemented with different concentrations of NaCl. After 7-d growth in treatment media, plants with absent green or dead cotyledons were scored. The root length of the seedlings was measured. We repeated freezing, drought, and salt tolerance experiments three times.
Germination Assay
The sensitivity of seed germination to ABA and NaCl was assayed on MS agar plates saturated with ABA and NaCl solution (Xiong et al., 2001). Seeds from wild-type and transgenic plants were placed on MS agar plates saturated with distilled water or different concentrations of ABA or NaCl and incubated at 0°C for 48 h before being placed at room temperature under cool-white light for germination. Seeds were considered germinated when radicles completely penetrated the seed coat. Germination was scored daily up to 10 d after seeds were placed at room temperature.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number BAD81765.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. List of up- or down-regulated genes in Yuedongdao rice under cold stress.
Supplemental Table S2. Primers used in RT-PCR to verify the gene expression in microarray.
Supplementary Material
Acknowledgments
We are grateful to Joshua Gendron and Dr. Zhiyong Wang (Stanford University) for their critical reading, Prof. Dazhou Chen (Rice Research Institute, Jiangxi Academy of Agricultural Sciences) for supplying the rice seeds of Yuedongdao, and Mr. Cheng Yuan (Institute of Botany, Chinese Academy of Sciences) for assisting with microscope techniques.
This work was supported by the Major State Basic Research Program of the People's Republic of China (grant no. 2005CB120806), the National Natural Science Foundation of China (grant nos. 30525026 and 30470866), the State Project of Transgenic Plants (grant no. JA03–A–09), and by the State High-Tech Project (grant no. 2006AA10Z169).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kang Chong (chongk@ibcas.ac.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF (1996) Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA 93 13404–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avonce N, Leyman B, Mascorro-Gallardo JO, Van Dijck P, Thevelein JM, Iturriaga G (2004) The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol 136 3649–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24 701–713 [DOI] [PubMed] [Google Scholar]
- Braun EL, Grotewold E (1999) Newly discovered plant c-myb-like genes rewrite the evolution of the plant myb gene family. Plant Physiol 121 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, Xin Z (2001) Temperature sensing and cold acclimation. Curr Opin Plant Biol 4 241–246 [DOI] [PubMed] [Google Scholar]
- Cantrell RP, Reeves TG (2002) The rice genome: the cereal of the world's poor takes center stage. Science 296 53. [DOI] [PubMed] [Google Scholar]
- Carr MD, Mott RF (1991) The transcriptional control proteins c-Myb and v-Myb contain a basic region DNA binding motif. FEBS Lett 282 293–294 [DOI] [PubMed] [Google Scholar]
- Cho SK, Kim JE, Park JA, Eom TJ, Kim WT (2006) Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett 580 3136–3144 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33 751–763 [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Artus NN, Thomashow MF (1992) cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol Biol 18 13–21 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16 433–442 [DOI] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA 99 11507–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51 463–499 [DOI] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba K (2002) Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu Rev Plant Biol 53 225–245 [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47 377–403 [DOI] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A (2001) G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13 1891–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106 [DOI] [PubMed] [Google Scholar]
- Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41 577–585 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17 287–291 [DOI] [PubMed] [Google Scholar]
- Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35 25–34 [PubMed] [Google Scholar]
- Khush GS (1999) Green revolution: preparing for the 21st century. Genome 42 646–655 [PubMed] [Google Scholar]
- Ko JH, Yang SH, Han KH (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47 343–355 [DOI] [PubMed] [Google Scholar]
- Kranz H, Scholz K, Weisshaar B (2000) c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J 21 231–235 [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Franck M (1990) Cloning and characterization of a cold- and ABA-inducible Arabidopsis gene. Plant Mol Biol 15 137–144 [DOI] [PubMed] [Google Scholar]
- Liu F, Sun C, Tan L, Fu Y, Li D, Wang X (2003. a) Identification and mapping of quantitative trait loci controlling cold-tolerance of Chinese common wild rice (O. rufipogon Griff.) at booting to flowering stages. Chin Sci Bull 48 2068–2071 [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xu ZH, Luo D, Xue HW (2003. b) Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J 36 189–202 [DOI] [PubMed] [Google Scholar]
- Medina J, Rodriguez-Franco M, Penalosa A, Carrascosa MJ, Neuhaus G, Salinas J (2005) Arabidopsis mutants deregulated in RCI2A expression reveal new signaling pathways in abiotic stress responses. Plant J 42 586–597 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101 6309–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3 217–223 [PubMed] [Google Scholar]
- Sridha S, Wu K (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J 46 124–133 [DOI] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF (1998) Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA 95 14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447–456 [DOI] [PubMed] [Google Scholar]
- Thomashow M (2001) So what's new in the filed of plant cold acclimation? Lots. Plant Physiol 125 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1998) Role of cold-responsive genes in plant freezing tolerance. Plant Physiol 118 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50 571–599 [DOI] [PubMed] [Google Scholar]
- Tyagi A, Mohanty A, Bajaj S, Chaudhury A, Maheshwari S (1999) Transgenic rice: a valuable monocot system for crop improvement and gene research. Crit Rev Biotechnol 19 41–79 [Google Scholar]
- Tyagi AK, Mohanty A (2000) Rice transformation for crop improvement and functional genomics. Plant Sci 158 1–18 [DOI] [PubMed] [Google Scholar]
- Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37 115–127 [DOI] [PubMed] [Google Scholar]
- Wang X, Xu W, Xu Y, Chong K, Xu Z, Xia G (2004) Wheat RAN1, a nuclear small G protein, is involved in regulation of cell division in yeast. Plant Sci 167 1183–1190 [Google Scholar]
- Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK (2001) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236 331–340 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57 781–803 [DOI] [PubMed] [Google Scholar]
- Zhu J, Shi H, Lee BH, Damsz B, Cheng S, Stirm V, Zhu JK, Hasegawa PM, Bressan RA (2004) An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc Natl Acad Sci USA 101 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Verslues PE, Zheng X, Lee BH, Zhan X, Manabe Y, Sokolchik I, Zhu Y, Dong CH, Zhu JK, et al (2005) HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc Natl Acad Sci USA 102 9966–9971 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.