Abstract
Mitochondrial complex II (succinate dehydrogenase [SDH]) is part of the tricarboxylic acid cycle and the respiratory electron transport chain. Its flavoprotein subunit is encoded by two nuclear genes, SDH1-1 and SDH1-2, in Arabidopsis (Arabidopsis thaliana). The SDH1-2 gene is significantly expressed only in roots, albeit at very low level, and its disruption has no effect on growth and development of homozygous mutant plants. In contrast, SDH1-1 transcripts are ubiquitously expressed, with highest expression in flowers. Disruption of the SDH1-1 gene results in alterations in gametophyte development. Indeed, heterozygous SDH1-1/sdh1-1 mutant plants showed normal vegetative growth, yet a reduced seed set. In the progeny of selfed SDH1-1/sdh1-1 plants, distorted segregation ratios were observed, and no homozygous mutant plants were obtained. Reciprocal test crosses with the wild type demonstrated that the mutated sdh1-1 allele is not transmitted through the male gametophyte and is only partially transmitted through the female gametophyte. Consistently, microscopic analysis showed that mutant microspores develop normally until the vacuolated microspore stage, but fail to undergo mitosis I, and then cell structures are degraded and cell content disappears. On the other hand, half the mutant embryo sacs showed arrested development, either at the two-nucleate stage or before polar nuclei fusion. Down-regulation of SDH1-1 by RNA interference results in pollen abortion and a reduced seed set, as in the insertional mutant. Altogether, our results show that SDH1-1, and therefore complex II, are essential for gametophyte development.
Succinate:ubiquinone oxidoreductase (succinate dehydrogenase [SDH]; EC 1.3.5.1), commonly referred to as mitochondrial complex II, has a central role in mitochondrial metabolism as a member of both the electron transport chain and the tricarboxylic acid (TCA) cycle. This membrane-associated complex catalyzes the oxidation of succinate to fumarate and the reduction of ubiquinone to ubiquinol, and has been characterized in bacteria and heterotrophic eukaryotes (Lemire and Oyedotun, 2002; Yankovskaya et al., 2003). In these organisms, complex II contains only four subunits: two peripheral membrane proteins, a flavoprotein (SDH1) and an iron-sulfur protein (SDH2), and two small integral membrane proteins (SDH3 and SDH4). The succinate-binding site is formed by the SDH1 protein, which is covalently linked to a FAD molecule acting as acceptor of a hydride ion at an early step of succinate oxidation. This flavoprotein subunit interacts with the SDH2 subunit, which transfers the electrons to the membrane through its three nonheme iron-sulfur centers. The two integral membrane proteins, SDH3 and SDH4, anchor the SDH1-SDH2 subcomplex to the matrix side of the inner mitochondrial membrane and contain a b-type heme and the ubiquinone-binding site (Yankovskaya et al., 2003). Surprisingly, it has been shown recently that plant complex II may contain additional subunits of unknown function along with the four classical subunits (Millar et al., 2004).
SDH subunits are all encoded in the nuclear genome in Arabidopsis (Arabidopsis thaliana), just like complex II from heterotrophic eukaryotes (Scheffler, 1998; Figueroa et al., 2001, 2002; Millar et al., 2004). Surprisingly, several of the complex II subunits are encoded by more than one gene in Arabidopsis. For instance, we have reported that three nuclear genes, named SDH2-1, SDH2-2 and SDH2-3, encode the iron-sulfur subunit, and two nuclear genes, designated SDH1-1 and SDH1-2, encode the flavoprotein (Figueroa et al., 2001, 2002). SDH2-1 and SDH2-2, which have similar structures and encode nearly identical proteins, have similar expression patterns (Figueroa et al., 2001; Elorza et al., 2004). These results are consistent with the fact that SDH2-1 and SDH2-2 probably arose via a recent gene duplication event and are redundant. In contrast, we have recently reported a highly tissue-specific expression for SDH2-3: this gene, which is structurally different and would have diverged for a long time from SDH2-1 and SDH2-2, is highly expressed in the embryo during seed development and its transcript accumulates in dry seeds (Elorza et al., 2006).
SDH1-1 (At5g66760) and SDH1-2 (At2g18450) have similar structures and encode highly similar proteins that would be functional as complex II flavoprotein since they are highly conserved when compared to their homologs in other organisms (Figueroa et al., 2002). Moreover, they contain the residues known to be involved in FAD binding and substrate binding, and in proton transfer during catalysis, and they are actively imported into isolated plant mitochondria (Figueroa et al., 2002). However, SDH1-1 and SDH1-2 appear to be expressed at very different levels: the SDH1-1 mRNA was easily detected by northern-blot hybridization in all tissues examined, whereas SDH1-2 transcripts were only detected in reverse transcription-PCR and 3′ RACE experiments (Figueroa et al., 2002).
The highest SDH1-1 mRNA levels were found in flowers (Figueroa et al., 2002). Analogous enhanced expression in flowers has been observed for other plant nuclear genes encoding mitochondrial proteins, including SDH2-1 and SDH2-2, and for mitochondrial transcripts (e.g. Huang et al., 1994; Smart et al., 1994; Heiser et al., 1996; Elorza et al., 2004). This is related to the fact that pollen development is one of the highest energy-requiring processes in plants. The most compelling evidence for an essential role of mitochondria during pollen development is the phenomenon of cytoplasmic male sterility (CMS). CMS is a maternal inherited trait in which mutations in the mitochondrial genome impair pollen development (Hanson and Bentolila, 2004). Furthermore, an increase in the number of mitochondria per cell has been reported in maize (Zea mays) and tobacco (Nicotiana tabacum) anthers during pollen development (Lee and Warmke, 1979; Huang et al., 1994).
To gain insight into the physiological role of complex II and to explore the function of the multiple genes encoding the same SDH subunit, our group has undertaken a reverse genetic analysis of the SDH genes. Here we report the analysis of T-DNA insertional mutants in SDH1-1 and SDH1-2, the flavoprotein genes. Our results reveal that SDH1-2 is dispensable and that the sdh1-1 null allele behaves as a general gametophytic mutation, demonstrating that complex II plays an essential role in gametophyte development.
RESULTS
Isolation of T-DNA Insertion Mutants of Arabidopsis SDH1 Genes
Several different mutant lines carrying T-DNA insertions in the SDH1-2 gene were identified as described in “Materials and Methods.” One mutant was isolated and further characterized: the T-DNA was confirmed to be in the eighth out of 15 exons, and interrupted codon 336. The precursor SDH1-2 polypeptide deduced from the gene sequence has 632 amino acids and disruption at codon 336 that is upstream of several residues involved in FAD and substrate binding, and in catalysis is expected to result in a null mutation. Homozygous sdh1-2 mutant plants were obtained and showed no apparent phenotypic defects during vegetative or reproductive growth when compared to wild-type plants, at least under the growth conditions used.
These results indicate that the loss of SDH1-2 has no impact on Arabidopsis growth and development, a fact consistent with our previous expression data (Figueroa et al., 2002). Furthermore, data on SDH1-2 expression in the existing large expression databases confirm that SDH1-2 is not expressed or is expressed at a very low level in most tissues and developmental stages (Supplemental Fig. S1; Schmid et al., 2005; see also https://www.genevestigator.ethz.ch, Zimmermann et al., 2004 for microarray data; and http://mpss.udel.edu/at, Meyers et al., 2004 for Massively Parallel Signature Sequencing experiments). Significant expression was only observed in roots, nevertheless its expression level is still less than 10% that of SDH1-1 in the same tissue (Supplemental Fig. S1).
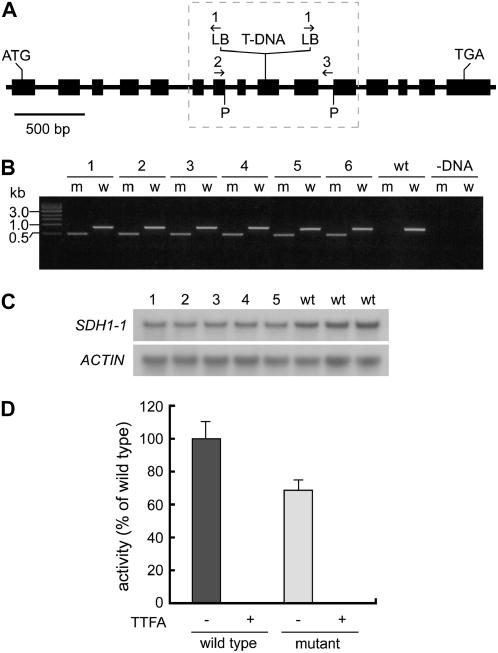
For SDH1-1, only one mutant line was identified: the T-DNA insertion was mapped to exon 9 and interrupts codon 317 (Fig. 1A). The SDH1-1 precursor polypeptide has 634 amino acids and disruption at codon 317 is expected to result in a null mutation. Genotyping of T2 plants obtained from the seed pool sent by the Arabidopsis Biological Resource Center (ABRC) led to the identification of several heterozygous SDH1-1/sdh1-1 mutant plants (Fig. 1B). However, attempts to identify homozygous sdh1-1/sdh1-1 mutant plants were unsuccessful. Moreover, no homozygous mutant seedlings were obtained in the progeny of selfed SDH1-1/sdh1-1 plants, suggesting that gametophyte and/or embryo development are altered and that SDH1-1 is an essential gene. Currently, there are no other mutant alleles available for SDH1-1, as confirmed by searching the Arabidopsis Insertion Data Base (http://atidb.org/cgi-perl/index).
Figure 1.
Identification of a mutant sdh1-1 allele and molecular characterization of heterozygous mutant plants. A, Genomic organization of the SDH1-1 gene. Exons are presented as black boxes. The T-DNA insertion site is indicated. Horizontal arrows show the position of primers. The T-DNA insert is not drawn to scale. LB, T-DNA left border; P, genomic PstI sites flanking the T-DNA insert. B, Identification of heterozygous mutant plants. Genotyping was performed by PCR amplification with primers 1 and 2 to identify the mutant allele (m), and primers 2 and 3 to identify the wild-type allele (w). Six (1–6) heterozygous mutant plants were found. wt lanes correspond to a control with DNA from a wild-type plant, and lane −DNA to a PCR control without template. C, Northern-blot analysis of SDH1-1 expression in five heterozygous mutant plants (lanes 1–5) and three wild-type plants (lanes wt). Each lane was loaded with 15 μg of total RNA isolated from flowers. The blot was hybridized with specific SDH1-1 probe (derived from the 3′ UTR) and then with an actin probe as loading control. The SDH1-1 transcript was present at a lower level in heterozygous mutant plants. D, Complex II activity is reduced in heterozygous mutant plants. SQR activity of wild-type and heterozygous mutant seedlings was determined in the presence or absence of thenoyltrifluoroacetone (TTFA), a complex II inhibitor. Three independent experiments were performed and the actual values were 17.8, 14.6, and 15.3 nmol of reduced DCIP min−1 mg−1 of protein for wild type seedlings, and 12.0, 10.5, and 10.1 nmol of reduced DCIP min−1 mg−1 of protein for mutant seedlings. Error bars correspond to sds.
Molecular Characterization of Heterozygous SDH1-1/sdh1-1 Mutant Plants
Northern-blot analysis was performed using RNA from three wild-type plants and six plants carrying the sdh1-1 mutated allele (Fig. 1C). As expected, the heterozygous mutant plants showed a reduced steady-state level of the SDH1-1 mRNA (roughly 50%). To assess whether this transcript decrease results in a lower complex II activity, succinate:quinone reductase (SQR) activity was measured in three independent experiments performed with mitochondrial fractions prepared from wild-type and SDH1-1/sdh1-1 18-d-old seedlings. Heterozygous mutant plants consistently showed a 32% reduction in SQR activity (Fig. 1D). When 1 mm thenoyltrifluoroacetone, a known complex II inhibitor, was included in the assays, the activity was completely abolished. Thus, these heterozygous mutant plants have a mild reduction in complex II activity, a result consistent with their normal phenotype during sporophytic growth (see below).
Since no other sdh1-1 mutant alleles are available to establish a correlation between a phenotype (see below) and different mutations in SDH1-1, it was important to characterize the mutated sdh1-1 locus and to analyze the number of T-DNA insertions in the mutant genome. PCR amplification of SDH1-1 gene/T-DNA junctions first suggested that the insertion is complex. Indeed, both insertion sides were amplified using the left-border primer (primer 1 in Fig. 1A) in combination with either an upstream (primer 2) or a downstream (primer 3) gene-specific primer. Sequencing of the PCR products demonstrated that the T-DNA left border was present at both ends of the insertion, suggesting the presence of at least two T-DNA molecules. More importantly, sequence analysis established that no major deletions or chromosomal rearrangements took place during the insertional event. Only a minor 23 bp deletion occurred in SDH1-1 at the insertion site.
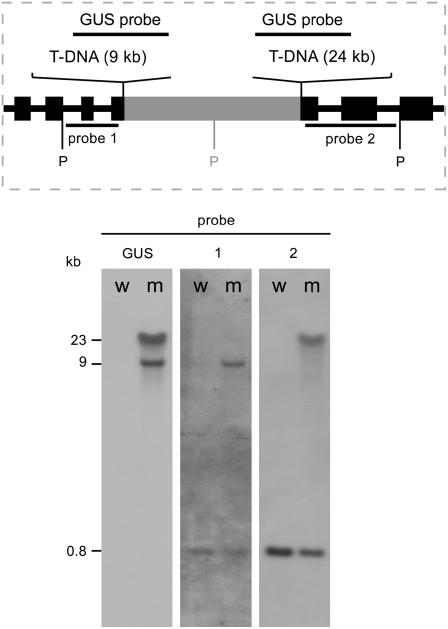
T-DNA copy number was determined through DNA gel-blot analysis, using a T-DNA noncutting enzyme (PstI). Hybridization of PstI-restricted DNA with a T-DNA-specific probe identified two DNA fragments in the heterozygous mutant plants (Fig. 2, β-glucuronidase [GUS] probe). Although this result may suggest two independent insertions, all our attempts to segregate them apart, for instance, by backcrossing to wild-type plants, were unsuccessful. It is known that insertional events may be very complex and generate tandem arrays of T-DNA molecules in the same locus (De Neve et al., 1997; De Buck et al., 1999). To determine whether this occurred in our mutant, we sequentially hybridized the blot in Figure 2 with probes directed to the SDH1-1 regions located upstream (probe 1) and downstream (probe 2) of the insertion (Fig. 2). Probe 1 (from a PstI site in SDH1-1 to the 5′ T-DNA insertion site) identified in the mutant DNA the 9 kb fragment also detected by the GUS probe, along with a 0.815 kb DNA fragment derived from the SDH1-1 wild-type allele. Probe 2 (from the 3′ T-DNA insertion site to a PstI site in SDH1-1) detected the other fragment (24 kb) identified by the GUS probe and the expected 0.815 kb DNA fragment of wild-type SDH1-1. These results are summarized in the scheme shown in Figure 2: both DNA fragments hybridizing to the T-DNA probe are derived from the SDH1-1 locus, clearly showing that no other loci were interrupted by T-DNA insertions in the SDH1-1/sdh1-1 heterozygous mutant plants. Therefore, Southern-blot analysis confirmed that these plants are heterozygous for the sdh1-1 mutation and contained only one complex T-DNA insertion. It may be concluded that any phenotypic alteration will be due to the presence of the sdh1-1 mutated allele.
Figure 2.
SDH1-1/sdh1-1 plants contain a unique complex T-DNA insertion. Southern-blot analysis was performed to characterize the insertion. Total DNA (6 μg) from wt (w) or mutant (m) plants was digested with PstI and hybridized sequentially with probes directed to the T-DNA (GUS probe), the SDH1-1 upstream region (probe 1), and the SDH1-1 downstream region (probe 2). The presence of a new PstI restriction site (P) was inferred from the hybridization results and is shown in gray, in a sequence of unknown origin.
Gametophytic Development Is Altered in SDH1-1/sdh1-1 Heterozygous Mutant Plants
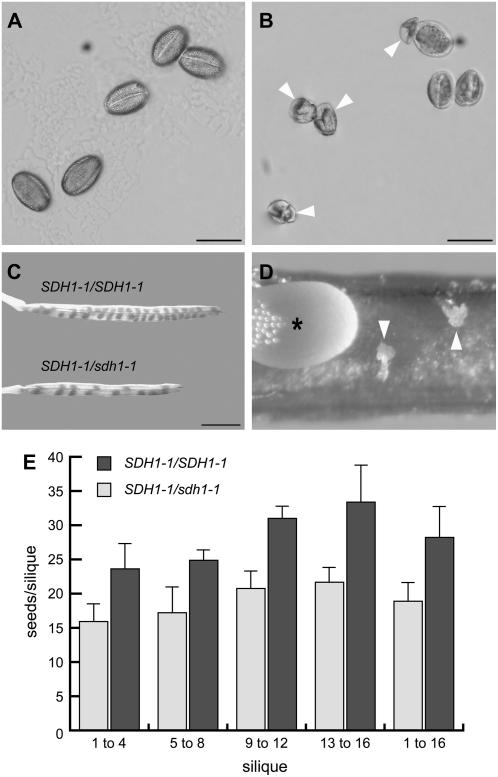
All the heterozygous SDH1-1/sdh1-1 mutant plants were phenotypically indistinguishable from the wild type during vegetative development, at least under the growth conditions used. To examine flower development we used some parameters of the phenotypic analysis platform described by Boyes et al. (2001) for Arabidopsis. For instance, we did not find differences between heterozygous mutant and wild-type plants in flowering time, flower number and size, number and size of stamens, anther size, and anther dehiscence. However, microscopic analysis of mature pollen grains showed that significant numbers of abnormal and collapsed pollen grains were detected in the SDH1-1/sdh1-1 mutant (Fig. 3B). Furthermore, SDH1-1/sdh1-1 siliques contained a reduced seed set and were shorter than those of wild-type plants (Fig. 3C). Closer examination of SDH1-1/sdh1-1 siliques revealed the presence of small, white, unfertilized ovules (Fig. 3D). No seed of an intermediate size indicative of abortion after fertilization was observed. Seed set per silique was quantitatively determined, using six T3 SDH1-1/sdh1-1 plants and two wild-type plants grown hydroponically in the same batch. The average seed set per silique was reduced by 33% in the SDH1-1/sdh1-1 mutant plants (last bars, Fig. 3E), suggesting that only 67% of the ovules were fertilized in these plants. Moreover, reduction of seed production in mutant plants was evenly distributed along the inflorescence axis, indicating that the overall seed set reduction in the mutant is not due to the presence of flowers not producing seeds along with flowers giving a normal seed number (Fig. 3E).
Figure 3.
Reproductive abnormalities in SDH1-1/sdh1-1 mutant plants. A, Mature pollen grains from a wild-type plant. Bar = 20 μm. B, Mature pollen grains from a heterozygous mutant plant. A substantial proportion of pollen grains are collapsed (arrowheads). Bar = 20 μm. C, Mature siliques of a SDH1-1/sdh1-1 plant are shorter and contain less seeds than mature siliques of the wild type. Bar = 2 mm. D, Closeup of a developing SDH1-1/sdh1-1 silique. Arrowheads denote aborted, white, and shrunken ovules. A wild-type seed is indicated by an asterisk. E, Seed set is reduced in SDH1-1/sdh1-1 siliques. Seed set per silique was scored along the main inflorescence axis for six T3 Kanr SDH1-1/sdh1-1 plants and two wild-type plants. Silique 1 (first, older flower) to silique 16 were grouped in four groups of four siliques for each plant. Total counts of 28.2 and 18.9 seeds/silique were obtained for wild type and SDH1-1/sdh1-1 plants, respectively (last two bars). Error bars are sds.
Altogether, these observations suggest that both pollen and embryo sac development are compromised in the heterozygous SDH1-1/sdh1-1 mutant plants, and that the sdh1-1 mutation is gametophytic (Park et al., 1998; Drews and Yadegari, 2002; Johnson-Brousseau and McCormick, 2004; Niewiadomski et al., 2005). Indeed, the presence of abnormal pollen (along with normal pollen) would not be sufficient to explain a reduced seed set, since pollen is supposed to be in great excess with respect to ovules.
Genetic Analysis of the Mutated sdh1-1 Allele
The T-DNA insertion in the SDH1-1/sdh1-1 mutant plants confers kanamycin resistance (Kanr). Segregation of Kanr was analyzed in the progeny of six selfed T2 heterozygous SDH1-1/sdh1-1 plants (Supplemental Table S1). From a total of 1,243 offspring, only 387 seedlings were kanamycin resistant. This segregation ratio of 0.5:1 for Kanr:Kans (kanamycin sensitive) is far below from the expected 3:1 segregation for a dominant resistant phenotype and from the 1:1 expected ratio for a fully penetrant mutation in either the male or female gametophyte, and is indicative of a gametophytic defect affecting both the male and female parents (Howden et al., 1998). To test this, male and female transmission efficiencies (TEs) of the mutated sdh1-1 allele were determined by performing reciprocal test crosses between the heterozygous SDH1-1/sdh1-1 mutant and wild-type plants (Park et al., 1998). The F1 progenies were genotyped by PCR and analyzed for Kanr or sensitivity (Supplemental Table S2). As expected, all Kanr seedlings carried the sdh1-1 mutated allele along with the SDH1-1 wild-type allele, and all Kans plants only bore the wild-type allele. When the heterozygous SDH1-1/sdh1-1 mutant was used as the male parent, no seedlings bearing the mutated sdh1-1 allele were found (male TE of 0%). When the heterozygous mutant was used as the female parent, a female TE of 60% was measured (33 instead of 55 seedlings bearing the mutated allele). Thus, the observed segregation ratio of 0.5:1 in selfed heterozygous plants was caused by defects in both male and female gametophytes, resulting in reduced TE of the mutated allele. Altogether, these results showed that all the pollen grains and about half the ovules bearing the mutated sdh1-1 allele were unable to transmit this allele.
Therefore, the sdh1-1 mutation is partially penetrant in the female gametophyte and fully penetrant in the male gametophyte, strongly suggesting that complex II activity is essential for pollen development and important for embryo sac development.
Pollen Development Is Perturbed in SDH1-1/sdh1-1 Mutant Plants
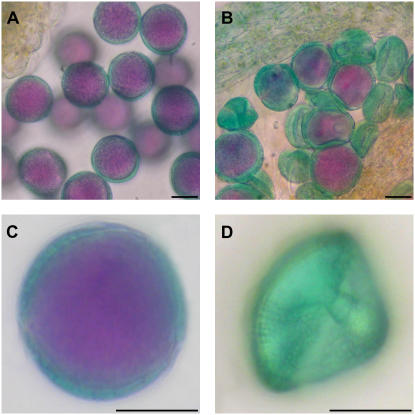
Our genetic analyses demonstrated that the male germ line was unable to transmit the sdh1-1 mutated allele and prompted us to analyze pollen development in detail. First, pollen of wild-type and SDH1-1/sdh1-1 plants was analyzed for viability by the staining method of Alexander (1969). In this assay, mature viable pollen grains show an intensive purple stain in the cytoplasm, surrounded by a thin green stain of the external exine layer (Fig. 4, A and C). In the SDH1-1/sdh1-1mutant, mature viable pollen grains were found (Fig. 4B). However, aborted pollen grains showing only the green staining of the external exine layer were abundant (Fig. 4, B and D). To determine the frequency of dead pollen in the SDH1-1/sdh1-1 mutant, pollen from 12 heterozygous T3 mutant plants was stained with Alexander's solution. Nonviable pollen grains occurred with a frequency of 41.6% to 49.4% (Supplemental Table S3). From a total of 3,133 pollen grains, 1,448 (46.2%) were not viable according to the Alexander staining test. Thus, these figures show low variability of the pollen phenotype among different plants and are consistent with the genetic data in the sense that near half of the pollen grains, those carrying the mutated sdh1-1 allele are not viable.
Figure 4.
Loss of SDH1 function affects pollen viability. Alexander's staining was performed with wild type (A and C) and heterozygous SDH1-1/sdh1-1 mutant (B and D) anthers. In these bright-field images, wild-type, viable pollen grains show intensive purple staining in the cytoplasm, whereas dead, unviable pollen grains are devoid of cytoplasm and exhibit only the green staining of the exine outer layer. Bars = 10 μm.
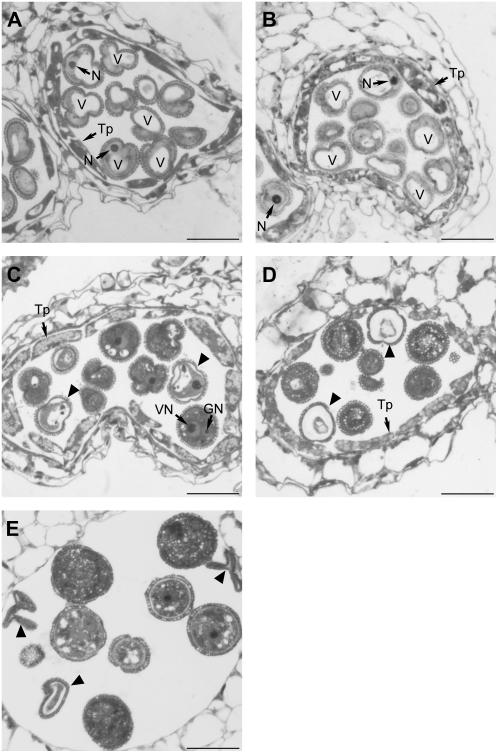
Second, we compared anther and pollen development in SDH1-1/sdh1-1 and wild-type plants (Fig. 5). Anther development has been divided into 14 stages based on morphological landmarks and cellular events visible under the light microscope (Sanders et al., 1999). Anthers of the SDH1-1/sdh1-1 mutant were indistinguishable from those of wild type at stage 9 (Fig. 5, A and B). At this stage, meiosis has already been completed, the exine wall is deposited on microspore surface, microspores contain a large vacuole, and the microspore nucleus is located at the cell periphery. Anthers of the mutant deviated from wild type at stage 11, when pollen mitotic divisions and vacuole resorption occur (Fig. 5, C and D). Besides normal pollen grains, which have both vegetative and generative nuclei, a dense cytoplasm, and small vacuoles, abnormal pollen grains were clearly visible. In these altered pollen grains, mitosis did not occur and the vacuole persisted. Moreover, the cytoplasm detached from the external exine layer and was progressively degraded (compare early stage 11 in Fig. 5C with late stage 11 in Fig. 5D). At stage 12, just prior to dehiscence, the abnormal pollen grains showed varying degrees of damage, including some in which the cellular material disappeared and the exine layer collapsed (Fig. 5E). It has to be pointed out that mutant SDH1-1/sdh1-1 anthers were not detectably different from those of wild-type plants, except for these described pollen abnormalities. For instance, other anther tissues like the epidermis, endothecium, and vascular bundle were not affected. Furthermore, mutant and wild-type anthers underwent the same events at similar stages, including disappearance of the tapetum and normal dehiscence.
Figure 5.
Pollen development in SDH1-1/sdh1-1 mutant plants. Anther sections were stained with Toluidine blue and photographed by bright-field microscopy. Abnormal pollen grains are indicated by arrowheads. Magnification is the same in all micrographs (Bars = 20 μm). GN, Generative cell nucleus; N, nucleus; Tp, tapetum; V, vacuole; VN, vegetative cell nucleus. A, Anther from the wild type at the vacuolated microspore stage. B, Anther from the SDH1-1/sdh1-1 mutant, at the vacuolated microspore stage. All microspores are indistinguishable from those of wild-type anthers. C, Anther from the SDH1-1/sdh1-1 mutant after pollen mitosis I, showing normal pollen grains with two nuclei, and abnormal pollen grains (arrowheads) not entering mitosis. D, Anther from the SDH1-1/sdh1-1 mutant after pollen mitosis II, showing normal pollen grains with a dense cytoplasm, and abnormal pollen grains (arrowheads) containing cell debris. E, Anther just before dehiscence, containing mature pollen grains and collapsed pollen grains devoid of cytoplasm (arrowheads).
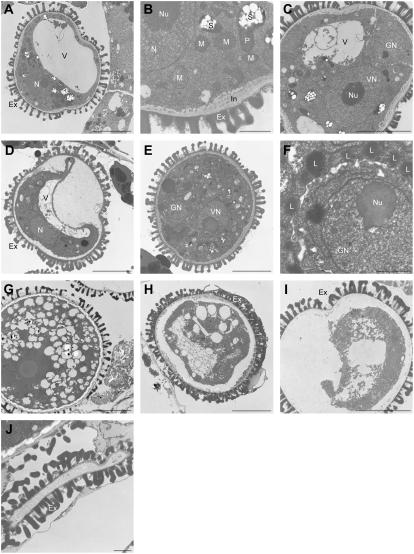
Transmission electron microscopy was conducted on cross sections of wild-type and mutant SDH1-1/sdh1-1 anthers (Fig. 6). By the vacuolated microspore stage, mutant microspores were not detectably different from those of wild-type plants (Fig. 6, A and B; Owen and Makaroff, 1995). Numerous mitochondria and starch-containing plastids are present throughout the cytoplasm, the exine is essentially complete and just below it vesicles produced by the microspores add material to the intine. While the nucleus is located near the side of the microspore, the asymmetric first mitotic division occurs, producing bicellular pollen grains (Fig. 6C). The generative cell is located at one end of the pollen grain and is surrounded by a wall that is continuous with the intine. At this stage, however, the SDH1-1 defect was evident in pollen sacs of SDH1-1/sdh1-1 anthers, since a mixture of wild-type-like (Fig. 6C) and altered microspores (Fig. 6D) were found within the locule. Abnormal pollen grains never underwent microspore mitosis, the large vacuole persisted, and the plasma membrane was withdrawn from the cell wall.
Figure 6.
Transmission electron microscopy analysis of anthers in the SDH1-1/sdh1-1 mutant. Ex, Exine layer; GN, generative nucleus; L, lipid body; M, mitochondrion; N, nucleus; Nu, nucleolus; S, starch granules within plastids; V, vacuole; VN, vegetative nucleus. Bar = 5 μm in A, C to E, and G to I; bar = 1 μm in B, F, and J. A, Microspores at the vacuolated stage (anther stage 9 according to Sanders et al., 1999). All microspores had wild-type morphology, with numerous mitochondria, starch granules within plastids, and a normal exine outer layer. B, Closeup of the microspore shown in A. C, Bicellular pollen grain from SDH1-1/sdh1-1 anthers. The asymmetric first mitotic division has occurred (early anther stage 11). D, Same anther as in C. In addition to pollen with wild-type morphology (C), abnormal pollen grains were present. They did not undergo mitosis and cell content detached from the outer layers. E and F, Later developmental stage of wild-type pollen grains from SDH1-1/sdh1-1 anthers. G, Mature wild-type pollen grain from SDH1-1/sdh1-1 anthers. H and I, Later developmental stages of aberrant pollen grains from SDH1-1/sdh1-1 anthers. Cellular structures and content progressively disappeared and internal structures were no longer distinguishable. J, Collapsed pollen grain in the same anther as in G. The exine layer, of sporophytic origin, remained wild-type like.
In SDH1-1/sdh1-1 anthers, wild-type-like pollen grains followed a normal developmental fate: generative cell occupies a more central location in the cytoplasm of the vegetative cell, the large vacuole is resorbed (Fig. 6E), and lipid bodies accumulate in the vegetative cells at the surface of the generative cell (Fig. 6F). Later, the cytoplasm of the vegetative cell becomes highly vacuolated (Fig. 6G). Neither of these landmarks of normal pollen development was observed in altered pollen grains. In these grains, autolysis led to the disintegration of cellular structures and progressive disappearance of cellular content (Fig. 6, H and I). Finally, pollen grains had collapsed completely (Fig. 6J).
Embryo Sac Development Is Defective in SDH1-1/sdh1-1 Mutant Plants
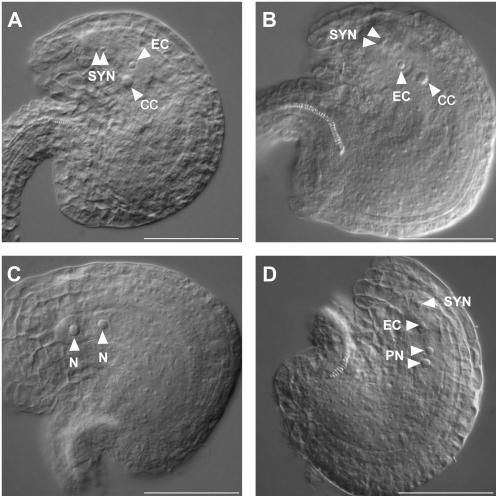
Reduced seed set, accompanied by the presence of white, small senescing ovules, and reduced transmission of the sdh1-1 mutation through the female gametophyte, suggested defects in embryo sac maturation and led us to analyze the phenotype of female gametophytes. To do this, we allowed the female gametophytes within wild-type and heterozygous pistils to progress to the terminal developmental stage, in the absence of fertilization, and embryo sacs were examined under Nomarski optics. In ovaries of wild-type plants, all embryo sacs reached the terminal developmental stage, containing one secondary central cell nucleus, one egg cell nucleus, and two synergid cell nuclei (Fig. 7A). In mutant plants, only 72.75% (n = 291) of embryo sacs displayed the wild-type phenotype (Fig. 7B). In addition, two abnormal phenotypes were observed. In 14.75% (n = 59) of the analyzed ovules, embryo sacs were arrested at the two-nucleate stage (Fig. 7C), and in 12.5% of the embryo sacs (n = 50) the overall morphology of the egg cell, central cell, and synergid cells was normal, yet polar nuclei failed to fuse (Fig. 7D). Therefore, 27% of the embryo sacs in SDH1-1/sdh1-1 plants were phenotypically mutant, a value in good agreement with the seed set reduction (33%).
Figure 7.
Embryo sac development defects in SDH1-1/sdh1-1 mutant plants. Flowers of wild-type and SDH1-1/sdh1-1 plants were emasculated, and whole-mount preparations of ovules were analyzed by DIC microscopy 48 h after emasculation. Embryo sac nuclei are indicated by arrows. A, Ovule from a wild-type Ws plant. The embryo sac is at the terminal developmental stage and contains one secondary nucleus in the central cell, one egg cell nucleus, and two synergid cell nuclei at the micropylar end. B, Ovule harboring a wild-type-like embryo sac in a SDH1-1/sdh1-1 mutant plant. C, Ovule from the same pistil of the ovule shown in B. The embryo sac displays a mutant phenotype, being arrested at the two-nucleate stage. D, Ovule from the same pistil of the ovule shown in B and C, but showing a second mutant phenotype. The two polar nuclei lie side by side but failed to fuse. CC, Central cell nucleus; EC, egg cell nucleus; N, nuclei from an embryo sac arrested at the two-nucleate stage; PN, polar nuclei; SYN, synergid cell nuclei. Bars = 50 μm.
Down-Regulation of SDH1-1 by RNA Interference Results in a Phenotype Similar to That of the Insertional Mutant
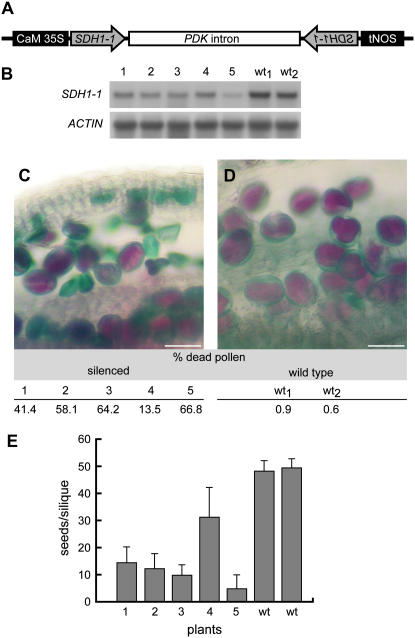
Since no other mutant alleles were available for SDH1-1, an RNA interference approach was used. Figure 8A illustrates the construct: the 35S RNA promoter of the Cauliflower mosaic virus drives the transcription of partial SDH1-1 sequences cloned in sense and antisense orientations and separated by a plant intron. After formation of hairpin RNA structures (ihpRNA) and splicing, the resulting double-stranded RNA transcripts can cause posttranscriptional silencing of endogenous gene activity (Smith et al., 2000). After floral-dipping transformation, only two independent transgenic T1 plants were obtained. Since mature gametophytes or recently fertilized embryos are the likely targets of this transformation procedure (Clough and Bent, 1998), this low efficiency of transformation is consistent with our results showing that gametophyte development is affected in the SDH1-1/sdh1-1 mutant. Moreover, northern-blot analysis of these primary transformants showed that SDH1-1 mRNA levels are only partially reduced (roughly 50%, data not shown), suggesting that a drastic reduction in SDH1-1 expression may lead to unviable transformed embryos.
Figure 8.
Phenotype of SDH1-1 RNAi transgenic plants. A, Schematic diagram of the construct used to down-regulate SDH1-1. The arrows indicate the SDH1-1 sequence (first exon) cloned in both orientations in the RNAi pHELLSGATE4 vector. Sense and antisense SDH1-1 transcribed sequences would be separated by the plant pyruvate orthophosphate dikinase (PDK) intron, generating an ihpRNA. B, Northern-blot analysis of SDH1-1 expression in five RNAi T2 plants and two wild-type controls. Each lane was loaded with 10 μg of total RNA isolated from flowers. The blot was hybridized with a SDH1-1 specific probe and then with an actin probe as loading control. RNAi plants had decreased SDH1-1 mRNA levels. C and D, Pollen abortion in SDH1-1 RNAi plants. Alexander staining was performed on two wild-type and five RNAi T2 anthers. Sections C and D show the staining of one RNAi plant and one wild-type plant, respectively. Quantitative determination of pollen abortion was done by counting viable, purple pollen grains, and aborted, green pollen grains, and the results obtained are shown under the images. Bars = 20 μm. E, Seed set is reduced in SDH1-1 RNAi plants. Seed set per silique was scored for the first eight siliques from five T2 RNAi plants (1–5) and two wild-type control plants (wt). Error bars represent sds.
T2 seeds from one of these two lines were germinated on kanamycin and five resistant seedlings were transferred to hydroponic medium. Northern-blot analysis was performed using RNA from these five transgenic plants and two wild-type plants. The results are shown in Figure 8B, with actin as a constitutive control. Compared to wild-type plants, all transgenic plants showed a decrease in the SDH1-1 mRNA levels. These RNAi plants were phenotypically indistinguishable from the wild type during sporophytic development. However, analysis of pollen viability by the staining method of Alexander showed varying percentages, ranging from 13.5% to 66.8% of dead, unviable pollen grains (Fig. 8, C and D). Moreover, seed set was significantly reduced in the transgenic plants bearing the RNAi construct (Fig. 8E). Interestingly, a correlation was found between the level of SDH1-1 mRNA expression and the degree of seed set reduction and pollen abortion (for instance, plant no. 5 showed the strongest reduction in mRNA, pollen viability, and seed set).
In conclusion, plants in which SDH1-1 gene expression is decreased by dsRNA showed the same phenotype as the heterozygous insertional mutant plants, i.e. reduced seed set and pollen abortion.
DISCUSSION
SDH1 genes are interesting targets for reverse genetics analysis, to gain insights into the role of complex II in plants. Thus, we have characterized knockout alleles of SDH1-1 and SDH1-2 genes. The SDH1-2 gene appears to be dispensable, at least under the growth conditions used, presumably because the SDH1-1 gene encodes the predominant flavoprotein. Our results do not rule out a SDH1-2 nonessential role in a restricted group of cells or in a particular developmental stage, resulting in subtle beneficial characteristics. Such a hypothesis may provide an explanation for SDH1-2 integrity and conservation when compared to SDH1-1. Alternatively, SDH1 duplication may be very recent, evolutionarily speaking, and either SDH1-2 inactivation or SDH1-2 gain of a new function are under way.
Molecular and genetic characterization of heterozygous SDH1-1/sdh1-1 mutant plants permit us to conclude that sdh1-1 is a gametophytic mutation affecting both male and female gametophyte development. SDH1-1 is essential for pollen development and the first defect was evident in sdh1-1 microspores at pollen mitosis I, the asymmetric mitosis that results in bicellular pollen grains. Mutant microspores never underwent mitosis, the large vacuole was not resorbed, and cellular content progressively disappeared. These results formally established that microspore mitochondria (and complex II) have an essential role in pollen development.
During male gametophyte development, defined as development of the haploid microspores, pollen grains show a very high metabolic activity. For instance, respiration rates are 10 times higher in pollen grains than in green tissues (Tadege and Kuhlemeier, 1997). The uninucleate microspore develops to trinucleate pollen through two mitotic divisions, and simultaneously starch, proteins, and other nutrients enrich the pollen of the vegetative cell (McCormick, 2004). These processes put demands on energetic and metabolic resources for the synthesis of proteins and other biomolecules. In Arabidopsis, all the genes for complex II components, with the exception of SDH1-2 and SDH2-3, show enhanced expression in floral tissues (Figueroa et al., 2001, 2002). Furthermore, digital northern analysis of developing pollen grains (Honys and Twell, 2004) shows that these genes are expressed in the male gametophyte (Supplemental Fig. S2). Considering that SDH is a component of the respiratory chain and the TCA cycle, loss of SDH1 in microspores would severely compromise both ATP production and provision of carbon skeletons for biosynthesis, leading to their abortion.
In addition to its essential role during pollen development, SDH1-1 is important for normal embryo sac development. Although our data indicate that the SDH1-1 protein is essential for efficient megagametophyte development, a fraction of the gametophytes still remains functional. Therefore, these mutant gametophytes cannot be completely energy deficient. The more plausible explanations for this observation are (1) a stochastic inheritance and persistence of wild-type mitochondria from the diploid megaspore mother cell, which would result in variable depletion rates of SDH1-1 through cell division and protein turnover during embryo sac development, and/or (2) variable provision of nutrients (ATP, carbon skeletons) to the female gametophyte from the surrounding sporophytic cells. In this context, it is tempting to speculate that male mutant gametophytes have a more severe phenotype because more nutrients are carried from sporophytic tissues to the female than to the male gametophyte. These arguments may also explain why more than one phenotype was found in the mutant embryo sacs (see below).
During megagametogenesis, a functional haploid megaspore gives rise to the mature female gametophyte (Yadegari and Drews, 2004). This process comprises three successive rounds of nuclear division, followed by cellularization. During cellularization, the two polar nuclei migrate to the center of the female gametophyte and fused together in the central cell. The result is a seven-cell embryo sac that contains three antipodal, two synergid, one egg, and one central cell. Two abnormal phenotypes were observed in sdh1-1 embryo sacs, developmental arrest at the two-nucleate stage, and failure of the two polar nuclei to fuse after their proper migration.
Large collections of gametophytic mutants defective in female gametophyte development have been reported, and in some cases characterized with respect to the genes responsible for the observed defects (e.g. Christensen et al., 2002; Yadegari and Drews, 2004; Niewiadomski et al., 2005; Pagnussat et al., 2005; Portereiko et al., 2006). A wide variety of phenotypes were observed, including mutants arrested at the two-nucleate stage and mutants cellularized but with defects in fusion of the polar nuclei. Interestingly, among the mutants affected in fusion of polar nuclei, several of the mutated genes encode mitochondrial proteins or predicted mitochondrial proteins (Christensen et al., 2002; Portereiko et al., 2006). The best characterized are GFA2, a mitochondrial matrix chaperone of the DnaJ protein family (Christensen et al., 2002), RPL21M, a mitochondrial 50S ribosomal subunit L21 (Portereiko et al., 2006), and now SDH1-1 (this work). Altogether these results strongly suggest a role for mitochondria in nuclear fusion (karyogamy). At this time, it is unclear why these mutants have a nuclear fusion defect. However, the fact that mutations in a ribosomal protein, a chaperone, and a complex II subunit lead to a similar phenotype favors the hypothesis that the karyogamy defect is a consequence of impairment in basic mitochondrial functions, like electron transport, ATP production, and/or provision of carbon skeletons.
Few defects in the respiratory chain and TCA cycle have been reported. For instance, a knockout mutation in a subunit of the NAD+-dependent isocitrate dehydrogenase has been described in Arabidopsis (Lin et al., 2004). The homozygous mutant plants showed no detectable differences in vegetative growth and reproductive development compared to wild-type plants. However, Arabidopsis contains multiple genes encoding catalytic and noncatalytic subunits of this enzyme, and in mitochondrial extracts from homozygous mutant plants there was still half as much enzyme activity as in wild-type plants. Antisense repression of the potato (Solanum tuberosum) mitochondrial citrate synthase gene resulted in a delaying flowering phenotype and a specific disintegration of the ovary tissues, indicating an important role of the TCA cycle during the early stages of flower development and the formation of the sporophytic female tissues (Landschütze et al., 1995). Additional efforts to analyze the function of the TCA cycle have been performed in a mutant (Aco1) that exhibits a deficiency in expression of one of two isoforms of aconitase present in tomato (Lycopersicon esculentum; Carrari et al., 2003), and in antisense transgenic plants exhibiting reduced activity of mitochondrial malate dehydrogenase (Nunes-Nesi et al., 2005). However, the possible presence of multiple genes has not been analyzed, these plants still maintain mitochondrial aconitase and malate dehydrogenase activities, and their reproductive development is not affected.
The best-characterized respiratory chain mutant is a Nicotiana sylvestris mitochondrial mutant, CMS II. In this mutant, the mitochondrial nad7 gene encoding the NAD7 subunit of complex I is deleted, and mitochondria are impaired in complex I structure and function (Pla et al., 1995; Gutierres et al., 1997). The absence of a competent complex I leads to impaired photosynthesis and slower growth, however plants attain biomass similar to that of wild-type plants and undergo reproductive development, although they are partially male sterile (Gutierres et al., 1997; Dutilleul et al., 2003a). Therefore, mutant plants were acclimated to this defect and this acclimation includes enhanced activity of nonphosphorylating NAD(P)H dehydrogenases, which bypass complex I (Sabar et al., 2000; Dutilleul et al., 2003a, 2003b). An Arabidopsis mutant lacking the 18 kD Fe-S subunit of complex I has also been described and shown to be affected in cold acclimation (Lee et al., 2002). Homozygous mutant plants grow more slowly than wild-type plants and a delay was observed in flowering. Nevertheless, these plants eventually reach heights similar to those of wild-type plants and were fully fertile. Altogether, data with these mutants indicate that a defect in complex I in plants is not lethal. In contrast, loss of complex II leads to a more severe phenotype, at least in gametophytic development, and this is probably due to the role of SDH in the TCA cycle.
Deficiencies of any of the four subunits of SDH are associated with a broad spectrum of human diseases, ranging from myo- and encephalopathies to aging and tumor formation (Rustin and Rötig, 2002). To our knowledge, the first reported nuclear gene mutation causing a respiratory chain defect in humans was a mutation in the gene encoding the complex II flavoprotein subunit (Bourgeron et al., 1995). The mutation was identified in two siblings presenting Leigh syndrome (necrosis of specific brain territories, causing encephalopathy) and SDH deficiency, and the patients were homozygous for an amino acid substitution in a conserved domain of the protein. It has to be emphasized that the available data suggest that complex II is essential in mammals and could not be completely lost. For instance, Piruat et al. (2004) reported the generation of a SDH4 knockout mouse, the first mammalian model lacking a protein of the mitochondrial electron transport chain, and showed that homozygous sdh4/sdh4 animals die at early embryonic stages.
Although development and function of gametophytes are crucial for plant reproduction, relatively little is known about the genes required for, and the pathways involved in gametophytic development in flowering plants. An important question is whether plants exploit metabolism as a way to control development. We have found here that an apparent metabolic mutant shows developmental effects, raising the possibility that some developmental regulators exert their effects through the modulation of basic metabolic processes. Metabolic regulation thus should be considered as one possible effector pathway in models of genetic regulation of development.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Wassilewskija (Ws) seeds were cold treated for 48 h at 4°C in darkness and then germinated and grown hydroponically at 20°C to 24°C, under a 16-h light/8-h dark cycle (Gibeaut et al., 1997). To grow seedlings in vitro and for Kanr assays, seeds were surface sterilized for 10 min with a solution of 10% (v/v) commercial bleach and 0.05% (v/v) Tween 20. Then seeds were rinsed thoroughly with sterile distilled water and sown on one-half-concentrated Murashige and Skoog medium solidified with 0.8% (w/v) agar and supplemented or not with 50 mg L−1 kanamycin. Finally seeds were cold treated, germinated, and grown as described above.
Isolation of Insertional SDH1 Mutants
The Arabidopsis Knockout Facility (AKF) alpha and BASTA populations (Sussman et al., 2000) were screened by PCR, using a primer specific for the T-DNA left border (JL 202, 5′ CATTTTATAATAACGCTGCGGACATCTAC 3′) in combination with either a forward or a reverse gene-specific primer. For SDH1-1, the forward primer was 5′ GGAGTGAGAATAGGGGTAATTAAGGGAAT 3′ and the reverse primer was 5′ CTGGAGTAAAACGTAAACCCAGTTTCAGA 3′. For SDH1-2, forward and reverse primers were 5′ TATTTCACTGCTGCAACATTATGGGCTTT 3′ and 5′ GATGGACGGTCTCGATTAAACGGTTAGAT 3′, respectively. Identification of DNA positive pools was performed by DNA gel-blot analysis of the PCR products, using SDH1-1 or SDH1-2 gene fragments as probes, and confirmed by sequencing of the amplified DNA (see Sussman et al., 2000 and http://www.biotech.wisc.edu/NewServicesAndResearch/Arabidopsis/OverviewIndex.html for methods). Only one insertion in each SDH1-1 and SDH1-2 were found (both in the AKF alpha mutant collection).
The Salk Institute collection (Alonso et al., 2003), the Torrey Mesa Research Institute collection (Sessions et al., 2002), and the Wisconsin DsLox T-DNA lines (http://www.hort.wisc.edu/krysan/DS-Lox) were searched in silico at two sites: the Arabidopsis Insertion Database (http://atidb.org/cgi-perl/index) and the T-DNAExpress Web sites (http://signal.salk.edu/cgi-bin/tdnaexpress). Although several mutant alleles were identified for SDH1-2 (e.g. SALK_015173 and SALK_087371), no T-DNA additional insertions were found in the SDH1-1 gene. A line in the Wisconsin DsLox T-DNA population (DsLox501H11) indexed as a SDH1-1 mutant was further characterized and found to carry the T-DNA insertion downstream of the SDH1-1 3′ untranslated region (UTR; data not shown).
Arabidopsis Ws seeds from the positive AKF pools containing the sdh1-1 and sdh1-2 mutant alleles were obtained from the ABRC (Ohio State University, Columbus, OH). These T2 seeds were stratified, germinated, and grown as described above. To identify SDH1 mutant plants and eventually isolate plants homozygous for a mutation, seedlings were genotyped by a PCR-based approach, using total DNA extracted from one cotyledon or one small leaf. Briefly, tissue was homogenized in 100 μL of TNE/SDS buffer (0.2 m Tris-HCl pH 8.0, 0.25 m NaCl, 0.02 m EDTA, 0.5% [w/v] SDS), using a disposable pestle adapted to a portable drill. After a 5 min centrifugation at 14,000g, the DNA in the supernatant was precipitated with 1 volume of isopropanol, washed with 70% (v/v) ethanol, and resuspended in sterile water. The genotype of plants was determined by PCR using primers flanking the insertion point for the wild-type allele and a gene-specific and left border-specific (JL202) primer pair for the insertional mutant alleles. For SDH1-1, the wild-type allele was amplified with 5′ CAACCTCAGCACATACATGCACAG 3′ (primer 2 in Fig. 1A) and 5′ CCATCTCATGCTTAACTCCACACA 3′ (primer 3 in Fig. 1A), and the mutant allele with JL202 (primer 1 in Fig. 1A) and primer 2. For SDH1-2, the wild-type allele was amplified with 5′ TATTTCACTGCTGCAACATTATGGGCTTT 3′ (primer sdha3) and 5′ GATGGACGGTCTCGATTAAACGGTTAGAT 3′ (primer sdha4), and the mutant allele with JL202 and sdha4.
Nucleic Acids Preparation and Analysis
Total DNA was prepared from green leaves according to Ausubel et al. (1994). Total RNA was isolated from tissues harvested into liquid nitrogen using TRIzol reagent and the manufacturer's protocol (Invitrogen). Southern and northern analyses were performed by standard procedures with 32P-labeled probes (Ausubel et al., 1994). To obtain specific probes, DNA fragments were PCR amplified using the following primer pairs: for a T-DNA GUS probe (Fig. 2), 5′ TCTACACCACGCCGAACACCTGG 3′ and 5′ TTCGGTGATGATAATCGGCTGATGC 3′; for probe 1 in Figure 2, 5′ CTGCAGGTTTTCCACCTGTTTC 3′ and 5′ ACGTTCACCTTCACTGTTTCTA 3′; for probe 2 in Figure 2, 5′ AACGATATGCTCCTACAGCCAA 3′ and 5′ CCATCTCATGCTTAACTCCACACA 3′; for a SDH1-1 probe derived from the 3′ UTR, 5′ TGAGATTGGATTATAGGCCTGTT 3′ and 5′ CTGGAGTAAAACGTAAACCCAGTTTCAGA 3′; and for an actin probe, 5′ GCTATGTATGTCGCCATTCAAGC 3′ and 5′ CATCATATTCTGCCTTTGC(A/G)ATCC 3′.
Preparation of Mitochondria and SQR Activity Assay
A crude mitochondrial fraction was prepared from Arabidopsis in vitro-grown 18-d-old seedlings. All steps were performed at 2°C to 4°C. The tissue (1 g fresh weight) was chilled in 10 mL of ice-cold grinding buffer (0.03 m MOPS/KOH pH 7.5, 0.3 m Mannitol, 0.001 m EDTA, 0.004 m l-Cysteine, 0.1% [w/v] fatty acid-free bovine serum albumin [fraction V], 0.6% [w/v] Polyvinylpyrrolidone-40) and homogenized with a Dounce potter. The homogenate was filtered through Miracloth and the filtrate was centrifuged at 1,000g for 20 min. The supernatant was then centrifuged at 12,500g for 20 min and the pellet was suspended in washing buffer (0.03 m MOPS/KOH pH 7.5, 0.3 m Mannitol, 0.001 m EDTA). Both centrifugation steps were repeated. Finally, the pellet (crude mitochondrial fraction) was resuspended in 200 μL of washing buffer. Protein concentration was determined by the Bradford method (Bradford, 1976) and SQR activity assayed immediately.
SQR activity was determined as described by Miyadera et al. (2003). Mitochondrial fractions (25 μg protein) were assayed for activity spectrophometrically at 600 nm, in 1 mL of a reaction medium containing 0.05 m potassium phosphate pH 7.5, 90 μm ubiquinone 2 (UQ2, Sigma-Aldrich), 55 μm dichlorophenolindophenol (DCIP; Merck; extinction coefficient 21 mm−1 cm−1), and 0.01 m sodium succinate (Merck). The reaction was performed at 30°C for 30 min, conditions in which DCIP reduction was proportional to incubation time and protein concentration.
Microscopic Analysis
Alexander staining of pollen was performed by incubating anthers from recently opened flowers in Alexander's solution for 15 min (Alexander, 1969; Johnson-Brousseau and McCormick, 2004). For anther structure analysis, flowers were fixed overnight in 3% (v/v) glutaraldehyde, 0.1 m sodium cacodylate, pH 7.2, dehydrated in acetone series to 100%, and embedded in Embed812 resin according to the recommendations of the manufacturer (EMS). Anther transverse sections (2 μm) were stained with 1% toluidine blue. Alexander stained pollen and anther cross sections were viewed using a light microscope (Optiphot-2, Nikon), and bright-field photographs were taken with a Nikon Coolpix 4500 CCD digital camera.
For transmission electron microscopy, samples were fixed in 3% (v/v) glutaraldehyde, 0.1 m sodium cacodylate, pH 7.2, and postfixed in 1% osmium tetroxide for 1 h, dehydrated in a graduate acetone series, embedded in Epon resin, and polymerized at 60°C for 24 h. Ultrathin sections (60 nm) were cut with a diamond knife (Micro Star) on a Sorvall MT-5000 microtome and mounted on 300 mesh copper grids. Sections were stained with 4% (w/v) uranyl acetate and lead citrate in methanol (Reynolds, 1963) and viewed with a Philips Tecnai 12 Bio Twin transmission electron microscope operating at 80 kV. Micrographs were taken using Kodak Electron Image SO163 film (Kodak).
To analyze the terminal phenotype of ovules, flowers were emasculated and ovules observed 48 h after emasculation. For this, wild-type and mutant pistils were dissected longitudinally with hypodermic needles, fixed overnight at room temperature in 50% (v/v) ethanol, 5% (v/v) glacial acetic acid, 10% (v/v) formaldehyde, and then cleared in chloral hydrate:glycerol:water (8g:2 mL:1 mL). Single ovules were dissected and viewed with a light microscope (Optiphot-2, Nikon) equipped for differential interference contrast (DIC). Images were captured using a Nikon Coolpix 4500 CCD digital camera.
Generation of ihpRNA Lines
A 225-bp fragment containing the 5′ UTR and the first exon of SDH1-1 was PCR amplified using the forward primer 5′ AAGCAGGCTACTAGTGTTCGCGTCTTCCT 3′ and the reverse primer 5′ CAAGAAAGCTGGGTAGAACCAGTGGAGA 3′ (partial attB sites in italics, SpeI restriction site underlined). The resulting product, which contained at both ends partial attB recombination sites of the Gateway system (Invitrogen), was reamplified using primers attB1 (5′ GGGGACAAGTTTGTACAAAAAAGCAGGCT 3′) and attB2 (5′ GGGGACCACTTTGTACAAGAAAGCTGGGT 3′). The PCR product (attB1:SDH1-1:attB2) was cloned into a pGEM-T easy vector (Promega) and sequenced. An in vitro recombination reaction was performed according to the manufacturer's protocol, using BP Clonase (Invitrogen), the vector pHELLSGATE4 (Helliwell et al., 2002), and gel-purified attB1:SDH1-1:attB2 DNA fragment. Spectinomycin-resistant Escherichia coli XL-1blue colonies bearing the recombinant plasmid were selected. The resulting plasmid, containing SDH1-1 sequences in both sense and antisense orientations separated by a pyruvate orthophosphate dikinase intron and under the control of the cauliflower mosaic virus 35S promoter, was transferred to Agrobacterium tumefaciens C58. A. tumefaciens-mediated transformation of Arabidopsis Columbia 0 plants was accomplished using the floral dip protocol (Clough and Bent, 1998). Seeds of the T1 generation were selected for resistance to kanamycin.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SDH1-1 and SDH1-2 expression in different tissues and developmental stages.
Supplemental Figure S2. Expression of the complex II genes during pollen development.
Supplemental Table S1. Segregation analysis of sdh1-1.
Supplemental Table S2. TE of sdh1-1 in reciprocal crosses.
Supplemental Table S3. Frequency of aborted pollen in SDH1-1/sdh1-1 plants.
Supplementary Material
Acknowledgments
The authors are greatly thankful to Alejandro Araya, Simon Litvak, and Laura Tarragó-Litvak for their constant encouragement. We also would like to thank Hannetz Roschzttardtz and Rodrigo Tapia for critical reading of the manuscript and constant discussion of results. Also, we thank to Bernard Hausser for technical advice on DIC microscopy and Sheila Johnson-Brousseau for experimental guidelines. We thank the AKF (University of Wisconsin) for mutant screens and for making T-DNA insertion lines publicly available through the ABRC.
This work was supported by the Fondecyt grant number 1060485 and Beca Apoyo Tesis Doctoral 2003 to G.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (http://www.plantphysiol.org) is: Xavier Jordana (xjordana@bio.puc.cl).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alexander MP (1969) Differential staining of aborted and non-aborted pollen. Stain Technol 44 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current Protocols in Molecular Biology. Wiley Interscience, New York
- Bourgeron T, Rustin P, Chretien D, Birch-Machin M, Bourgeois M, Viegas-Péquignot E, Munnich A, Rötig A (1995) Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat Genet 11 144–149 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Ehlers Loureiro M, Fernie AR (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, Gorsich SW, Brown RH, Jones LG, Brown J, Shaw JM, Drews GN (2002) Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14 2215–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- De Buck S, Jacobs A, Van Montagu M, Depicker A (1999) The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J 20 295–304 [DOI] [PubMed] [Google Scholar]
- De Neve M, De Buck S, Jacobs A, Van Montagu M, Depicker A (1997) T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from co-integration of separate T-DNAs. Plant J 11 15–29 [DOI] [PubMed] [Google Scholar]
- Drews GN, Yadegari R (2002) Development and function of the angiosperm female gametophyte. Annu Rev Genet 36 99–124 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G (2003. a) Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chétrit P, Foyer CH, De Paepe R (2003. b) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elorza A, León G, Gómez I, Mouras A, Holuigue L, Araya A, Jordana X (2004) Nuclear SDH2-1 and SDH2-2 genes, encoding the iron-sulfur subunit of mitochondrial complex II in Arabidopsis, have distinct cell-specific expression patterns and promoter activities. Plant Physiol 136 4072–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elorza A, Roschzttardtz H, Gómez I, Mouras A, Holuigue L, Araya A, Jordana X (2006) A nuclear gene for the iron-sulfur subunit of mitochondrial complex II is specifically expressed during Arabidopsis seed development and germination. Plant Cell Physiol 47 14–21 [DOI] [PubMed] [Google Scholar]
- Figueroa P, León G, Elorza A, Holuigue L, Araya A, Jordana X (2002) The four subunits of mitochondrial respiratory complex II are encoded by multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol Biol 50 725–734 [DOI] [PubMed] [Google Scholar]
- Figueroa P, León G, Elorza A, Holuigue L, Jordana X (2001) Three different genes encode the iron-sulfur subunit of succinate dehydrogenase in Arabidopsis thaliana. Plant Mol Biol 46 241–250 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chétrit P, Diolez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, De Paepe R (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MR, Bentolila S (2004) Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell (Suppl) 16 S154–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser V, Brennicke A, Grohmann L (1996) The plant mitochondrial 22 kDa (PSST) subunit of respiratory chain complex I is encoded by a nuclear gene with enhanced transcript levels in flowers. Plant Mol Biol 31 1195–1204 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM (2002) High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol 29 1217–1225 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85 [DOI] [PMC free article] [PubMed]
- Howden R, Park SK, Moore JM, Orme J, Grossniklaus U, Twell D (1998) Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Friedhelm S, Matzinger DF, Levings CS III (1994) Flower-enhanced expression of a nuclear-encoded mitochondrial respiratory protein is associated with changes in mitochondrion number. Plant Cell 6 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39 761–775 [DOI] [PubMed] [Google Scholar]
- Landschütze V, Willmitzer L, Müller-Rober B (1995) Inhibition of flower formation by antisense repression of mitochondrial citrate synthase in transgenic potato plants leads to a specific disintegration of the ovary tissues of flowers. EMBO J 14 660–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Lee H, Xiong L, Zhu J-K (2002) A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SLJ, Warmke HE (1979) Organelle size and number in fertile and T-cytoplasmic male-sterile corn. Am J Bot 60 141–148 [Google Scholar]
- Lemire BD, Oyedotun KS (2002) The Saccharomyces cerevisiae mitochondrial succinate: ubiquinone oxidoreductase. Biochim Biophys Acta 1553 102–116 [DOI] [PubMed] [Google Scholar]
- Lin M, Behal RH, Oliver DJ (2004) Characterization of a mutation in the IDH-II subunit of the NAD+-dependent isocitrate dehydrogenase from Arabidopsis thaliana. Plant Sci 166 983–988 [Google Scholar]
- McCormick S (2004) Control of male gametophyte development. Plant Cell (Suppl) 16 S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD (2004) Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol 22 1006–1011 [DOI] [PubMed] [Google Scholar]
- Millar HA, Eubel H, Jänsch L, Kruft V, Heazlewood JL, Braun HP (2004) Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol Biol 56 77–90 [DOI] [PubMed] [Google Scholar]
- Miyadera H, Shiomi K, Ui H, Yamaguchi Y, Masuma R, Tomoda H, Miyoshi H, Osanai A, Kita K, Omura S (2003) Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proc Natl Acad Sci USA 100 473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge U-I, Schneider A (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AMO, Ehlers Loureiro M, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen HA, Makaroff CA (1995) Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185 7–21 [Google Scholar]
- Pagnussat GC, Yu H-J, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie L-F, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132 603–614 [DOI] [PubMed] [Google Scholar]
- Park SK, Howden R, Twell D (1998) The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125 3789–3799 [DOI] [PubMed] [Google Scholar]
- Piruat JI, Pintado CO, Ortega-Sáenz P, Roche M, López-Barneo J (2004) The mitochondrial SDHD gene is required for early embryogenesis, and its partial deficiency results in persistent carotid body glomus cell activation with full responsiveness to hypoxia. Mol Cell Biol 24 10933–10940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla M, Mathieu C, De Paepe R, Chétrit P, Vedel F (1995) Deletion of the last two exons of the mitochondrial nad7 gene results in lack of the NAD7 polypeptide in a Nicotiana sylvestris CMS mutant. Mol Gen Genet 248 79–88 [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Sandaklie-Nikolova L, Lloyd A, Dever CA, Otsuga D, Drews GN (2006) NUCLEAR FUSION DEFECTIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol 141 957–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds ES (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin P, Rötig A (2002) Inborn errors of complex II—unusual human mitochondrial diseases. Biochim Biophys Acta 1553 117–122 [DOI] [PubMed] [Google Scholar]
- Sabar M, De Paepe R, de Kouchkovsky Y (2000) Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol 124 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu Y-C, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11 297–322 [Google Scholar]
- Scheffler IE (1998) Molecular genetics of succinate:quinone oxidoreductase in eukaryotes. Progr Nucleic Acid Res Mol Biol 60 267–315 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart CJ, Monéger F, Leaver CJ (1994) Cell-specific regulation of gene expression in mitochondria during anther development in sunflower. Plant Cell 6 811–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang M-B, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407 319–320 [DOI] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Kuhlemeier C (1997) Aerobic fermentation during tobacco pollen development. Plant Mol Biol 35 343–354 [DOI] [PubMed] [Google Scholar]
- Yadegari R, Drews GN (2004) Female gametophyte development. Plant Cell (Suppl) 16 S133–S141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, Byrne B, Cecchini G, Iwata S (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299 700–704 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) Genevestigator: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.