Abstract
Background
Several members of the R2R3-MYB family of transcription factors act as regulators of lignin and phenylpropanoid metabolism during wood formation in angiosperm and gymnosperm plants. The angiosperm Arabidopsis has over one hundred R2R3-MYBs genes; however, only a few members of this family have been discovered in gymnosperms.
Results
We isolated and characterised full-length cDNAs encoding R2R3-MYB genes from the gymnosperms white spruce, Picea glauca (13 sequences), and loblolly pine, Pinus taeda L. (five sequences). Sequence similarities and phylogenetic analyses placed the spruce and pine sequences in diverse subgroups of the large R2R3-MYB family, although several of the sequences clustered closely together. We searched the highly variable C-terminal region of diverse plant MYBs for conserved amino acid sequences and identified 20 motifs in the spruce MYBs, nine of which have not previously been reported and three of which are specific to conifers. The number and length of the introns in spruce MYB genes varied significantly, but their positions were well conserved relative to angiosperm MYB genes. Quantitative RTPCR of MYB genes transcript abundance in root and stem tissues revealed diverse expression patterns; three MYB genes were preferentially expressed in secondary xylem, whereas others were preferentially expressed in phloem or were ubiquitous. The MYB genes expressed in xylem, and three others, were up-regulated in the compression wood of leaning trees within 76 hours of induction.
Conclusion
Our survey of 18 conifer R2R3-MYB genes clearly showed a gene family structure similar to that of Arabidopsis. Three of the sequences are likely to play a role in lignin metabolism and/or wood formation in gymnosperm trees, including a close homolog of the loblolly pine PtMYB4, shown to regulate lignin biosynthesis in transgenic tobacco.
Background
Insights into the regulation of lignin biosynthesis during vascular development of plants are being derived from angiosperm model plants like Arabidopsis thaliana (reviewed by [1]) and from investigations unravelling the molecular basis of wood formation in trees like Populus (reviewed by [2]). Members of the R2R3-MYB transcription factor family have been implicated as regulators of phenylpropanoid and lignin metabolism [1] as well as pattern formation and differentiation of primary and secondary vascular tissues, (reviewed by [3]). The MYB proteins comprise one of the largest families of plant transcription factors, which is represented by over one hundred members in the model plant Arabidopsis [4]. The biological roles of MYBs have been deduced primarily from flowering plants (angiosperms) including snapdragon [5], maize [6], Arabidopsis [7,8] and eucalyptus [9]. By contrast, the biological roles of only a few R2R3-MYBs has been examined in non-flowering plants (gymnosperms) and relatively little is known about of their gene family structure [10-12]. The loblolly pine genes PtMYB1 and PtMYB4 were shown to be transcriptional activators which have the ability to regulate lignin synthesis enzymes [10,11]. They are expressed in xylem tissues, bind AC elements and activate transcription in transient assays in yeast or plant cells [10,11,13]. Overexpression of pine MYBs resulted in ectopic lignification in tobacco [10] and in Arabidopsis [8] The reports are strong evidence supporting a role for MYBs in the lignifying process in gymnosperm trees. Lignins play an important role in trees because they confer rigidity and impermeability to wood by accumulating in thickened secondary vascular tissues [1,14], therefore its regulation is of interest for understanding the genetic basis of wood properties. However, the number of MYB transcription factors that may participate in regulating lignification in gymnosperms, and their potential roles remain an open question.
Gymnosperms, especially conifers of the Pinaceae familyare ecologically and economically important due to their abundance in forests in many parts of the world (North America, Europe, Asia) and because of their use in diverse wood products (pulp and paper, solid wood and engineered lumber). Despite recent large-scale gene discovery initiatives for conifer trees like pine and spruce (e.g. [15,16]), only a few regulatory gene families have been characterised systematically in any conifer species. In one such study, it was recently shown that the structure of the knox-I gene family appears to be monophyletic in the Pinacea, whereas angiosperms have several distinct clades (four in dicots and three in monocots) [17]. The R2R3-MYB family is very large with over 120 members in angiosperms [4] and has been divided into several subgroups [18,19]. One may predict that several MYBs are likely to regulate lignin metabolism and other aspects of wood formation in conifer trees; however no data have been available from which to infer the size or the structure of the family in gymnosperms. Therefore, a broader survey of MYB genes expressed in the vascular and other tissues of gymnosperms seems essential for developing a better understanding of their roles in gymnosperm lignin biosynthesis and wood formation.
MYB proteins have two structural regions, an N-terminal DNA-binding domain (DBD or MYB domain) and a C-terminal modulator region that is responsible for the regulatory activity of the protein. The MYB domain is well conserved among plants, yeast and animals [20]. Its consensus sequence contains around 50 amino acid residues with regularly spaced tryptophans giving rise to a helix-turn-helix structure [21]. There are usually one to three imperfect repeats of the MYB domain. Proteins with two repeats (R2R3-MYBs) are specific to plants and yeast [22] and are the most abundant type in plants [4]. Plant R2R3-MYBs take part in many biological processes including seed development and germination [23], the stress response [24] and epidermal cell fate in addition to their involvement in phenylpropanoid and lignin biosynthesis [5,8-11] and vascular organisation [3] (for a review, see Ref. [25]).
The genetic selection and breeding activities of a few commercial conifer species are being expanded to include genetic mapping and marker development. Candidate gene approaches are being adopted to identify robust genetic markers derived from genes that have a physiological role in the traits that are targeted by breeders [26]. Our goal is to characterise several members of a gene family proposed to play a role in controlling lignin synthesis and wood properties in conifer trees, in order to support candidate gene approaches for marker discovery. In this report, we characterize 13 different R2R3-MYB gene sequences from the white spruce, Picea glauca, (designated PgMYB) and five from loblolly pine, Pinus taeda L (designated PtMYB). The full-length coding sequences we obtained enabled us to explore their phylogenetic relationships to other plant MYB genes and to search for novel amino acid motifs within this large protein family. We also compared the gene structures, i.e. number, size, position and splice sequences of introns, to gain further insights into their evolution. The steady-state levels of MYB and cell wall-related gene mRNAs were examined by Q-RTPCR in various spruce tissues and organs with an emphasis on wood-forming tissues and compression wood formation. We identify three MYBs that are preferentially expressed in secondary xylem and are also upregulated during the formation of compression wood.
Results
Isolation and sequence analysis of 18 R2R3-MYB genes from spruce and pine
We isolated and sequenced 18 full-length cDNAs encoding R2R3-MYB genes from conifer trees: 13 from spruce (PgMYB1-13) and five from pine (PtMYB2, 3, 7, 8 and 14). Each of the full length cDNA sequences were obtained starting from partial or full length clones identified by EST database mining (all of the pine sequences and most of the spruce sequences), or starting from the pine sequence and using RT-PCR amplification with conserved primers to amplify a spruce fragment (Table 1). For partial clones, we used RACE cloning to identify flanking sequences and, full length PCR amplification to generate a single full length cDNA. Their predicted amino acid sequences were aligned together with the three available full-length MYB sequences from gymnosperms [10-12]. The DNA-binding domains of these 21 gymnosperm sequences showed a high level of amino acid conservation particularly in the R3 helix-turn-helix repeat, consistent with its involvement in DNA binding (Fig. 1). Most of the variations among the spruce PgMYB sequences were located in the turn of each R repeat.
Table 1.
Predicted lengths and C-terminal motifs of spruce MYB proteins
| Sg1 | Full length cDNA2 | DNA Binding Domain3 | C-terminal Domain3 | Motifs | Consensus sequences | Angio-Gymno4 | MEME E-value | Start motif5 | Ref. | |
| 4 | Pg MYB5 | -b- | 115 | 142 | F | LlsrGiDP(at)tHrp(li)n | 13/13-5/5 | 6.00e-14 | 1 | a) |
| G | e(re)cpdLNLel(cr)ispp | 13/13-4/5 | 3.31e-16 | 67 | a), b) | |||||
| Pg MYB10 | -a- | 115 | 95 | F | LlsrGiDP(at)tHrp(li)n | 13/13-5/5 | 6.52e-14 | 1 | a) | |
| G | e(re)cpdLNLel(cr)ispp | 13/13-4/5 | 4.32e-15 | 67 | a), b) | |||||
| Pg MYB13 | -b- | 116 | 80 | F | LlsrGiDP(at)tHrp(li)n | 13/13-5/5 | 1.18e-14 | 1 | a) | |
| G | e(re)cpdLNLel(cr)ispp | 13/13-4/5 | 5.43e-14 | 65 | a), b) | |||||
| 22 | Pg MYB6 | -a- | 115 | 235 | H | (cs)s(sv)DPpT(ls)LsLslPg | 7/7-14/14 | 2.02e-14 | 99 | d) |
| I | YlkaedaismmsaAv | 0/7-13/14 | 1.87e-13 | 141 | d) | |||||
| J | vmremvakEVrsYmn | 7/7-14/14 | 1.07e-17 | 188 | a), b), c) | |||||
| Pg MYB7 | -c- | 116 | 257 | K | egdyEVesrgLKRln | 0/7-13/14 | 1.34e-12 | 43 | d) | |
| H | (cs)s(sv)DPpT(ls)LsLslPg | 7/7-14/14 | 4.28e-13 | 113 | d) | |||||
| I | YlkaedaismmsaAv | 0/7-13/14 | 2.30e-10 | 161 | d) | |||||
| J | vmremvakEVrsYmn | 7/7-14/14 | 6.15e-16 | 206 | a), b), c) | |||||
| Pg MYB9 | -a- | 119 | 297 | K | egdyEVesrgLKRln | 0/7-13/14 | 6.03e-16 | 60 | d) | |
| P | hRQSAFksYesqktp | 0/7-11/14 | 1.19e-13 | 116 | d) | |||||
| H | (cs)s(sv)DPpT(ls)LsLslPg | 7/7-14/14 | 2.91e-13 | 144 | d) | |||||
| I | YlkaedaismmsaAv | 0/7-13/14 | 9.50e-16 | 205 | d) | |||||
| J | vmremvakEVrsYmn | 7/7-14/14 | 1.09e-16 | 256 | a), b), c) | |||||
| 8 | Pg MYB1 | -b- | 115 | 217 | A | lr(kq)mGiDP(lv)THkpl | 5/5-2/2 | 1.79e-18 | 1 | a) |
| 21 | Pg MYB3 | -c- | 130 | 177 | C | (fg)Re(rq)S(rs)(is)(rg)(kr)R | 4/5-2/2 | 4.69e-14 | 1 | d) |
| D | e(en)s(l)(vs)(pt)ffDfl(g)vG(cn) | 5/5-2/2 | 1.26e-13 | 35 | a), b) | |||||
| E | (cy)xi(sg)h(in)nh(v)q(sf)(jr)Kef | 3/5-2/2 | 4.76e-14 | 123 | d) | |||||
| 13 | Pg MYB8 | -c- | 115 | 411 | L | LrrGIDP(n)THkpl | 4/4-2/2 | 2.54e-17 | 1 | a) |
| M | VC(dv)(yk)(np)SIm(al)nPsm(yn) | 2/4-2/2 | 1.94e-18 | 199 | d) | |||||
| N | e(ye)(ae)vKWSEml | 2/4-2/2 | 6.45e-14 | 317 | d) | |||||
| O | (pk)D(fl)(hq)R(im)Aa(vs)(lf)(dg)q | 2/4-2/2 | 4.89e-15 | 399 | a) | |||||
| 9 | Pg MYB11 | -a- | 115 | 384 | Q | L(lv)kMGIDPvTHkp(k) | 6/6-1/1 | 4.08e-16 | 1 | a), b), c) |
| R | h(m)AQWEsARleAear | 6/6-1/1 | 3.10e-13 | 35 | a), b), c) | |||||
| S | (yc)eDnknYw(nd)silnlV | 4/6-1/1 | 6.79e-12 | 360 | c) | |||||
| 2 | Pg MYB12 | -a- | 115 | 254 | T | MdfW(fl)(dn)v(fl)(t) | 5/5-1/1 | 2.39e-09 | 237 | a) |
| nd | Pg MYB2 | -c- | 115 | 333 | B | (c)SylPPL(y)d(v) | 2/2-2/2 | 3.29e-13 | 249 | d) |
| Pg MYB4 | -c- | 120 | 214 | none | none | 0/3-0/2 | none | none | none |
Conserved amino acid regions were identified in angiosperm and gymnosperm C-terminal sequences by the use of MEME software (setting described in Methods). Motifs were detected among the sequences belonging to each phylogenetic clade comprised of at least one spruce MYB (Additional File 2). Sequences from Additional File 3 were used to identify more conifer members of the PgMYB6, 7, 9 clade. Within the consensus sequences, upper-case letters indicate amino acids found in all members of a subgroup, lower-case letters indicate amino acids conserved in more than 50% of the members, pairs of lower-case amino acid in brackets show the two most abundant amino acids present for 50% each and above, x indicates that no amino acid is conserved among the sequences.
1Sg, MYB subgroups identified by Kranz et al. [18].
2Source of full length cDNA sequence: -a-, full length cDNA clone identified from EST of Picea glauca database; b- partial cDNA clone identified from EST database of P. glauca, extended by RACE amplifications and finally amplified as a single clone by PCR with gene specific primers, and -c-, from non degenerates primers based on Pinus taeda MYB sequences and used on spruce cDNA followed by RACE amplifications.
3Lengths are expressed in amino acid (aa) residues.
4The number of MYB sequences, separately from angiosperm and gymnosperms, sharing the motif among all those used in each case.
5The position of the motif relative to the beginning of the C-terminal domain (5' end).
Ref: references for previously reported motifs, a) Kranz et al. [18], b) Stracke et al. [31] and c) Jiang et al. [30] and d) new motifs.
Figure 1.
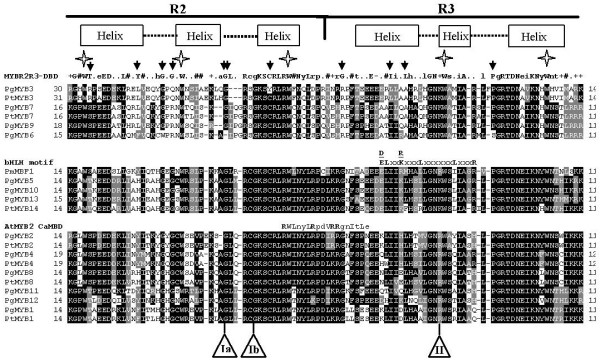
Alignment of predicted MYB domain protein sequences from spruce and pine. Amino acid sequence alignments of the 21 conifer MYB R2R3 domains were obtained with Clustal W (see Methods) and then separated into three groups based on their homologies to the consensus R2R3-MYB DNA-binding domain (MYBR2R3-DBD, top panel), the bHLH protein-binding motif (bHLH motif, middle panel) or the Arabidopsis calmodulin-interaction motif (AtMYB2 CaMBD, bottom panel), as indicated. Black shading indicates identical amino acid residues and grey shading the similar residues that agree with the fraction sequence of 0,4 (BoxShade 3.21) and dashes indicate gaps. The numbers on the left and right indicate the amino acid position relative to the translation start codon. The boxes and dotted line above the sequences show the predicted helix and turn structures in the R2 and R3 regions of the MYB domain. Stars show positions of conserved tryptophan residues and black arrows indicate unusual amino acid residues compared to the consensus amino acid sequence of the MYB DNA-binding domains of several plant R2R3-MYB proteins described by Avila et al. [27]. The bHLH protein-binding motif ([DE]L × 2 [RK] × 3L × 6L × 3R) identified by Zimmerman et al. [28] and the calmodulin-interaction motif [29] are shown above the middle and bottom panels, respectively (major amino acids in upper-case, bold). Ia or Ib and II indicate the positions of the first and second introns, respectively (Ib is specific to PgMYB3). Accession numbers of the newly identified spruce and pine MYBs are listed in Methods. Pg, Picea glauca; Pt, Pinus taeda; Pm, Picea mariana; At, Arabidopsis thaliana.
The conifer sequences were consistent with the consensus DBD sequence identified by Avila et al. [27], which was largely based on angiosperm sequences. Only a few amino acid residues differed from this consensus; these were mainly in PgMYB 3, 6, 7 and 9 and PtMYB3 (black arrows in Fig. 1). We found a motif similar to that involved in the interaction with basic helix-loop-helix (bHLH) proteins in Arabidopsis ([DE]L × 2 [RK] × 3L × 6L × 3R; [28]) in the R3 repeat of three spruce MYBs (PgMYB5, 10 and 13) as well as in PmMBF1 ([12]; Fig. 1). PtMYB14 had a similar motif but with two differences: an R instead of an L, and a gap before the last R residue. In addition, several conifer MYBs, including those with the bHLH motif (except PmMBF1), encoded an R × 5R × 3RR motif similar to the calmodulin-interaction site previously described in the DBD of Arabidopsis MYB2 [29]. The highest level of conservation with the calmodulin-binding motif was observed in PgMYB2 and PtMYB2, but most of the conifer MYB genes shown in Figure 1 had a similar motif.
Phylogenetic relationships and gene family structure of conifer R2R3-MYBs
We used the Mega 2.0 method to construct a phylogenetic tree using full length cDNA sequences (Fig. 2). The result of our analysis is congruent with the three major groups of R2R3-MYBs (A, B and C) defined by Romero et al. [19] on the basis of their binding affinities to MYB recognition elements. On this phylogenetic tree, the predicted spruce MYB proteins sequences fell into several subgroups with bootstrap values ranging from 96–100%, indicating the high grouping robustness. All of the spruce and pine MYB sequences fell into group A (PgMYB3, 6, 7 and 9, and putative pine orthologues) or C (all the other conifer sequences in Fig. 2) and none belonged to the B group. The conifer sequences were assigned to 7 of the 22 subgroups previously defined based upon Arabidopsis sequences [18]. Four of the conifer sequences (PgMYB2 and 4; PtMYB2 and 4) clustered with Arabidopsis sequences that do not fit into a defined subgroup. Several pairs of spruce and pine sequences clustered closely together with short branch-lengths indicative of a high degree of homology (Fig. 2). Indeed, pair-wise optimal alignments with the Clustal W algorithm of the pine and spruce pairs 1, 2, 3, 4, 7 and 8 gave amino acid identities from 95% to 100% for the DBD and of 79%–93% for the complete coding sequence (Table 3), suggesting that they are putative orthologous pairs. By comparison, PtMYB14 was less homologous to its neighbouring spruce sequences PgMYB5, 10 and 13 (60% to 67% homologous for the full CDS).
Figure 2.
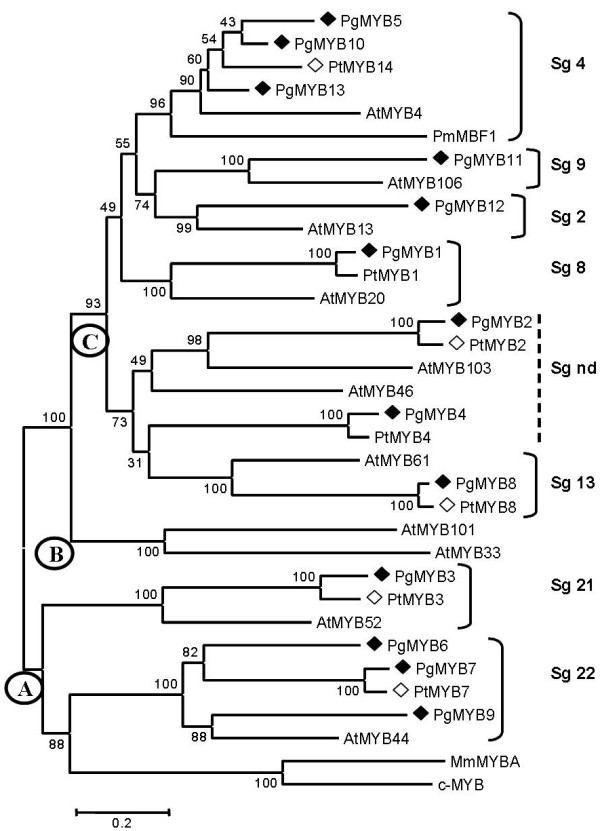
Phylogenetic tree of gymnosperm and angiosperm R2R3-MYB proteins. This neighbour-joining (1000 Bootstraps) tree was based on the Clustal W alignment of the complete coding sequences of 13 spruce and five pine MYB proteins identified in this study (represented by filled and empty lozenges, respectively). The bar indicates an evolutionary distance of 0.2%. Arabidopsis proteins were chosen as landmarks representing the three main groups (circles A, B and C) and subgroups (Sg next to bracket; nd, not determined) defined by Romero et al. [19] and Kranz et al. [18]. Human c-MYB [GenBank: P10242] and Mus musculus MmMYBA [GenBank: X82327] were not used as out groups but as landmarks. The accession numbers of the Arabidopsis genes are given in Methods. Other abbreviations are in Figure 1.
Table 3.
Pair-wise sequence amino acids identities of the DBD and full CDS of closest spruce and pine homologs
| DNA Binding Domains | Full coding sequences | |||
| Amino acids percentage | Identity | Similarity | Identity | Similarity |
| PgMYB1/PtMYB1 | 99,1 | 100 | 87,1 | 91,3 |
| PgMYB2/PtMYB2 | 100 | 100 | 88 | 91,6 |
| PgMYB3/PtMYB3 | 94,6 | 95,4 | 79,2 | 82,3 |
| PgMYB4/PtMYB4 | 96,6 | 97,5 | 84,1 | 88,6 |
| PgMYB7/PtMYB7 | 94,8 | 96,5 | 90,4 | 93,6 |
| PgMYB8/PtMYB8 | 98,3 | 100 | 93,1 | 95,5 |
| PgMYB5/PtMYB14 | 88 | 94,8 | 67,3 | 77,4 |
| PgMYB10/PtMYB14 | 87,8 | 95,6 | 64,3 | 73 |
| PgMYB13/PtMYB14 | 82,7 | 92,2 | 60 | 70 |
Percent similarity calculations were performed using pairwise optimal alignment with Bioedit software (Clustal W, matrix blosum62).
We also analysed the number, size and sequences of introns in PCR-amplified genomic DNA, as a complement to the phylogenetic analysis based on the coding sequences. In angiosperm R2R3-MYBs the introns are located in the Myb DBD, therefore we sequenced this specific region in genomic DNAs of the 13 spruce R2R3-MYBs, isolated by PCR amplification with gene specific primer pairs spanning each gene's coding region (Additional file 1). Most of the gDNA sequences were identical to the cDNAs, ranging from 100% to 99.3% in amino acid identities (data not shown), due to a few variations in the predicted amino acid sequences. The sequences were also verified for the lack of non-sense mutations (stop codons or frameshifts). As observed in angiosperms, we found spruce MYB genes with one (I), two (I, II) or no introns (Table 2).
Table 2.
Length of spruce MYB coding sequences and introns with their predicted splice junctions
| Length (bp) | Intron I (phase 1) | Intron II (phase 2) | |||||
| Coding sequence | Intron I | Intron II | 5'Splice site | 3'Splice site | 5'Splice site | 3'Splice site | |
| Pg MYB 1 | 999 | 83 | 101 | CCG:GTAAAT | TTGCAG:GTC | TAG:GTATAT | CACCAG:GTG |
| Pg MYB 2 | 1347 | 501 | 88 | CAG:GTACTC | TGACAG:GTC | CAG:GTTTGT | GTGCAG:GTG |
| Pg MYB 3 | 924 | 1427 | none | CAG:GTAAAG | ATGCAG:GGA | none | none |
| Pg MYB 4 | 1005 | 194 | 267 | CTG:GTAAGC | GTACAG:GTC | CAG:GTTTTT | GCGCAG:GTG |
| Pg MYB 5 | 774 | 191 | 186 | CAG:GTTGAA | TTGCAG:GGC | CAA:GTATGT | GCGCAG:GTG |
| Pg MYB 6 | 1053 | none | none | none | none | none | none |
| Pg MYB 7 | 1122 | none | none | none | none | none | none |
| Pg MYB 8 | 1581 | 97 | 90 | CTG:GTAAAG | TCGCAG:GCC | CAG:GTAATG | ACACAG:GTG |
| Pg MYB 9 | 1251 | none | none | none | none | none | none |
| Pg MYB 10 | 633 | 94 | 187 | CAG:GTTTCT | ATGCAG:GGC | CAA:GTATGT | GTGCAG:GTG |
| Pg MYB 11 | 1500 | 644 | 132 | CAG:GTATTT | ATGCAG:GAC | CAA:GTAAGG | TTACAG:ATG |
| Pg MYB 12 | 1110 | 94 | 294 | CAG:GTCACT | TTGCAG:GGC | CAG:GTGAGT | ATGTAG:ATG |
| Pg MYB 13 | 591 | 94 | 139 | CAG:GTTTCT | ATGCAG:GGC | CAA:GTATGT | GTGCAG:GTG |
Coding sequences indicate the length in nucleotides from the translation start codon to the stop codon. Introns I and II represent the first and second introns, respectively. Intron phase refers to the position in a codon where the intron is inserted: after the first nucleotide (phase 1) or after the second nucleotide (phase 2) of a codon. Italic nucleotide pairs GT and AG represent the beginning and the end of the introns, respectively; and underlined nucleotides are conserved splice-site sequences.
Similarity was found between the spruce MYBs in terms of intron position, phases and, in some cases, between intron sequences, but the number and length intron was quite variable. Generally, the second intron (II) was longer than the first (I) except in PgMYB11 where intron I was five times longer than intron II. The spruce sequences belonging to group A MYBs fell into two subfamilies with distinct gene structures, i.e. with one intron (Sg21) and one without introns (Sg22). The group C sequences all had one or two introns, as in Arabidopsis [30]. The intron I occurred before the GL amino acid pair in repeat 2 and the intron II occurred after the GN amino acid pair in repeat 3 (Fig. 1), as found in the majority of Arabidopsis R2R3-MYB genes[30]. Only PgMYB3 had a different intron I site, named Ib, located before the GKS amino acids. Moreover, the phase (1 or 2) of insertion was consistent, and the end sequences (GT in 5' and AG in 3') were conserved among the sequences we analysed (Table 2). Phylogenetically close sequences, like PgMYB5, 10 and 13, had similar 5' and 3' splice junctions for both introns (Table 2). The intron of these three genes also showed strong nucleotide sequence conservation, although the first intron of PgMYB5 was much longer due to a 20-nucleotide triplicated sequence (not shown).
Sequence analysis of conserved regions in the C-terminal of P. glauca MYBs
The coding regions of the spruce PgMYB sequences ranged widely in length, encoding between 196–526 amino acid residues depending on the length of the C-terminal region (Table 1). We used the predicted C-terminal coding regions of the spruce MYB proteins to search for conserved sequences, reasoning that such motifs might be important for the function or post-translational regulation of MYB. We used the MEME motif-detection software to analyse the C-terminal region of spruce MYBs using a set of protein sequences selected for their high degree of similarity to each of the spruce MYBs (Table 1, Additional files 2 and 3). Our approach incorporated a large diversity of sequences; it identified a total of 20 different motifs (A-T) in the spruce MYBs, including nine new unpublished motifs (Table 1) and 11 that were reported previously [18,19,30]. The probability scores for each of the motifs identified in this study ranged from 2.39e-09 to 1.79e-18. The lowest previously published score for such motif was 6.79e-12 (motif S in PgMYB11) [30]. We detected between zero (in PgMYB4) and five (in PgMYB9) motifs per protein in the predicated spruce MYB sequences. The large number of conifer sequences enabled us to detect three amino acids regions, I, K and P, that appeared to be specific to gymnosperms (in PgMYB6, 7 and 9). Other motifs, such as F and G, were shared between gymnosperm and angiosperm sequences. Four of the conserved amino acid sequences (A, F, L and Q) shared the central core residues GIDPxTH but displayed differences in neighbouring amino acids between the consensus sequences of subgroups 4, 8, 9 and 13 defined by Kranz et al. [18].
Expression of P. glauca MYB genes in tissues of young and mature trees
We surveyed the abundance of each of the 13 PgMYB gene transcripts by Q-RTPCR, in mature (33-year-old trees) and young (3-year-old) green-house-grown trees to determine their tissue distribution during normal development. Six different organs and differentiating tissues (the young needles; the periderm, phloem and xylem from the stems; and the periderm with phloem, or bark, and xylem from the roots) were collected from two different mature trees (Fig. 3). For tissue comparisons, we calculated the number initial MYB RNA molecules per ng of total RNA. Spruce PgMYBs 2, 4 and 8 were expressed preferentially in differentiating xylem from stem and root. Other MYBs were abundant in the needles along with one to two other tissues from the stem or the roots or both, Some MYB mRNAs also appeared to have rather ubiquitous profiles or low abundance transcripts. The RNA abundance of lignin biosynthesis enzymes PAL, 4CL, CCoAOMT and CAD were also determined in the same tissue samples. The lignin enzymes RNAs all gave very similar profiles, and they were most abundant in differentiating xylem (only 4CL is shown; Fig. 3).
Figure 3.
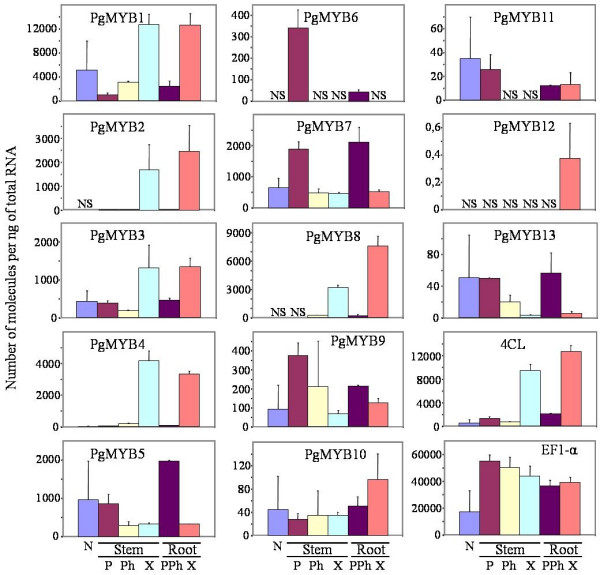
Transcript abundance for 13 spruce MYB genes and 4CL in various organs and tissues. Transcript abundance was determined by Q-RTPCR of six tissues from two different 33-year-old trees (number of molecules per ng of total RNA, see methods). The transcript level of an elongation factor (EF1-α) gene was used as an RNA control. N, needles; Stem tissues: P, periderm; Ph, differentiating phloem; X, differentiating xylem; Root tissues: PPh, root periderm with differentiating phloem; X, root differentiating xylem. Data are based on three technical repetitions per tree, i.e. six measurements per data point. Vertical bars represent the standard error. 4CL: 4-coumarate: CoA ligase. NS, no PCR product detected.
We also compared the abundance of the different MYB transcripts in the differentiating secondary xylem and in the elongating apical leader of young spruce trees (Fig. 4). The cell wall-related genes PAL, 4CL, CAD, CCoAOMT and an arabinogalactan protein (AGP) were included in this analysis. For these within tissue comparisons, the data were normalized against the EF1-α transcript levels. Again, the spruce MYB transcripts 2, 4 and 8 were clearly the most abundant among the MYBs detected in the secondary xylem, consistent with the data from the mature trees. In the apical leader, the relative abundance of the MYB transcripts was quite different than in the secondary xylem, except that PgMYB4 transcripts remained very abundant. Some MYB genes that were weakly expressed or not detectable in secondary xylem were among the most highly expressed in apical stem (PgMYB6, 7 and 11; Fig. 4a).
Figure 4.
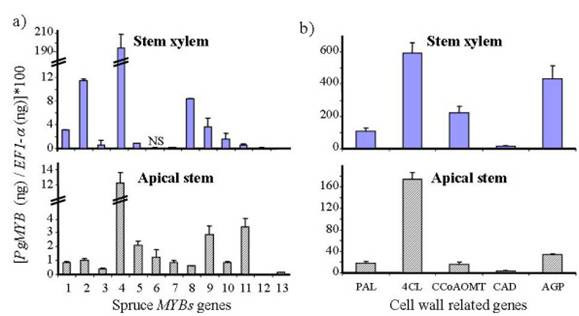
Transcript abundance for MYB genes and secondary cell-wall-related genes in differentiating secondary xylem and in primary growth (new flush) of spruce seedlings. Transcript abundance was determined as in Figure 4 for, a) 13 spruce MYB genes, and b) five cell-wall-related genes in differentiating secondary xylem from stem and in the elongating terminal leader (apical stem) from 3-year-old spruce seedlings. The standard error (bars) was calculated from three biological replicates and two independent technical repetitions (i.e. six independent measurements). PAL, phenylalanine ammonia lyase; 4CL, 4-coumarate: CoA ligase; CCOaOMT, caffeoyl-CoA 3-O-methyltransferase; AGP, arabinogalactan protein; CAD, cinnamyl alcohol dehydrogenase. NS, no PCR product detected.
Spruce MYB genes are differentially expressed in compression wood
We followed the expression of the 13 spruce MYB genes and five cell-wall-related genes during the early phases of compression wood formation, in order to explore further the potential involvement of MYBs in wood formation and lignin biosynthesis. Gymnosperm trees form a type of reaction wood (known as compression wood) on the lower side of a bent or leaning stem, or in branches. Compression wood is enriched in lignin and contains lignins that are more condensed. Therefore compression wood formation requires the modulation of lignin biosynthesis, which we hypothesized to involve such gene sequences as R2R3-MYBs. We induced the formation of compression wood in actively growing 3-year-old spruces by maintaining at a 45° angle (relative to vertical) (Fig. 5). After 21 days of growth in this leaning position, characteristic compression wood was well developed on the lower side of the stems (Fig. 5a). We chose to monitor transcript abundance over a 76-hour period immediately after induction, and found that several transcripts accumulated between 28 and 76 hours (Fig. 5c). The transcripts of PgMYB2, 4 and 8 clearly increased in the xylem forming compression wood compared to the opposite wood and compared to the vertical trees (0 hour time point). The transcripts of PgMYB9, 11 and 13 RNA were slightly increased and the seven others did not fluctuate significantly. By contrast, no significant variation in spruce MYB RNA abundance was observed in the opposite wood, which is found on the upper side of the stem (opposite to the the compression wood). Transcripts for PAL, 4CL, CCoAOMT and CAD lignin biosynthesis enzymes as well as the AGP also increased within the same time-frame as the MYB transcripts (Fig. 5b). In the opposite wood, only CCoAOMT RNA transcripts decreased. No significant variation in transcript abundance was observed for the spruce MYB or lignin genes in the terminal shoots of the same seedlings (data not shown).
Figure 5.
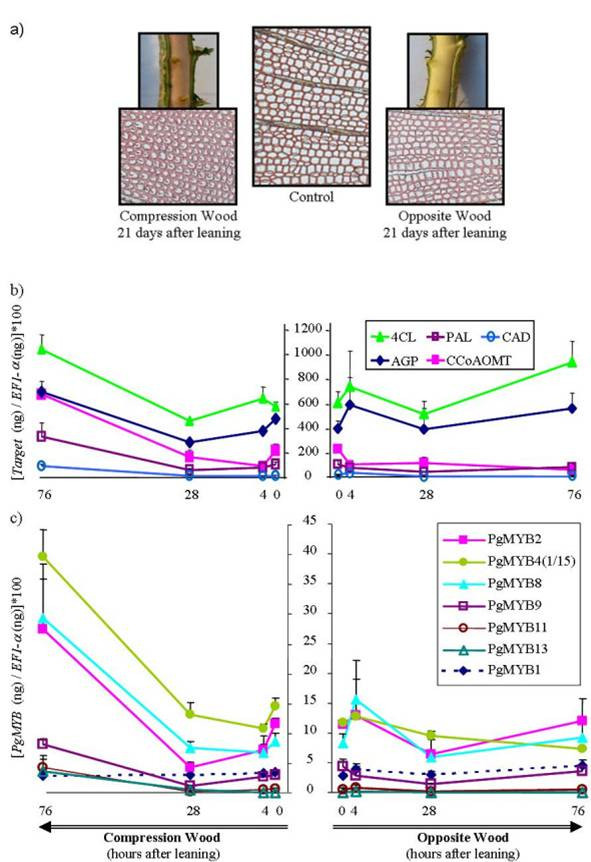
Transcript accumulation for MYB genes and secondary cell-wall-related genes in differentiating compression wood and opposite wood. a) Compression wood and opposite wood formed in a leaning spruce seedling after 21 days of treatment, compared to the control from vertical seedling. Exposed wood (compression wood is light brown) and wood cross-sections (10 μm thick) were stained by the safranin-orange procedure [53] (magnification, ×40). Steady-state mRNA levels were determined as in Figures 4 and 5 for cell-wall-related genes (b) and for several PgMYB genes (c) in the compression wood (left panels) and opposite side wood (right panels) of spruce seedlings leaning at a 45° angle from vertical. Continuous lines indicate genes with significant variation, and standard error bars are shown three trees (biological replicates) with two independent technical repetitions). Discontinuous lines indicate examples of gene transcripts that do not fluctuate in abundance. The zero time point represents vertical control trees only. PgMYB4 (1/15) means that mRNA level is divided by 15.
Discussion
In this paper, we report the complete coding sequences of 18 conifer gene sequences that share the characteristic features of the R2R3-MYB gene family. Thirteen sequences were from P. glauca (white spruce; PgMYBs) and five from P. taeda L. (loblolly pine; PtMYBs). We characterised the full-length cDNA sequences, as well as the spruce exon-intron structure. We assigned the conifer sequences to several phylogenetic clades of the R2R3-MYB family and identified conserved motifs within them based on predicted amino acid sequences. The steady-state mRNA levels of spruce MYBs were surveyed in several tissues to identify those genes that are preferentially expressed in wood-forming tissues. Furthermore, we identified PgMYBs whose transcript levels are upregulated, along with those of an AGP and enzymes of lignin biosynthesis, during the induction of compression wood in young spruce trees.
Sequence conservation and identification of amino acid motifs in spruce R2R3-MYBs
Our data show that the DBDs of conifer MYBs are highly conserved, whereas the C-terminal region are highly variable, as shown in prior studies of other plant MYBs. The predicted amino acid sequences of some of the spruce MYB DBDs contain a motif for interaction with bHLH proteins and/or with calmodulin. We identified twenty amino acid motifs in the variable C-terminal region, of which nine were previously unreported. The amino acid motifs in the DBD and in the C-terminal region are useful to better characterise the spruce R2R3-MYB sequences belonging to each phylogenetic clade.
The R2R3-MYBs are specific to plants and are subdivided into three major groups according to their binding affinities [19]. The more than 120 Arabidopsis sequences were placed into 22 subgroups based on their overall amino acid sequences and C-terminal motifs [18]. Amino acid motifs are conserved among members of several of the 22 phylogenetic clades or subgroups [18,30,31], including the bHLH-interaction and calmodulin-binding motifs in the DNA-binding domain [28,29], and the repression domain pdLNLD/ELxiG/S in the C-terminus [7]. In our study, the spruce group C sequences were dispersed among seven phylogenetics clades, five of which were previously defined as distinct subgroups by Kranz et al[18]. All the spruce members of subgroup 4 harboured the bHLH-interaction motif as well as the C-terminal motifs F and G, except for PmMBF1 [12], which lacked the motif G (pdLNLD/ELxiG/S) described by Kranz et al[18]. The bHLH-interaction motif identified by Zimmermann et al. [28] is required for MYB proteins to transactivate some of the phenylpropanoid and anthocyanin genes through protein-protein interactions [32]. The G motif in the C-terminus has been linked to transcriptional repression of the cinnamate 4-hydroxylase (C4H) gene by AtMYB4 [7]. Several genes in group C also encoded a conserved GIDP sequence located after the end of the DBD, suggesting a conserved molecular function for this motif. The DBDs of PgMYB2, 4 and 8, which were upregulated during compression wood formation, harboured a motif similar to the calmodulin-interaction site of AtMYB2 [29], suggesting a potential link with the calcium signalling pathway implicated in the regulation of secondary wall formation [33]. No conserved regions were detected in the C-terminal region of PgMYB4 and its closest homolog PtMYB4, even though experimental evidences indicate that PtMYB4 is a regulator of lignin synthesis enzymes [10], as is the case for the closely related EgMYB2 [9]. The presence of a regulator motif in PgMYB4 may have escaped our analysis because the parameters were set to detect motifs ranging from 5–15 amino acids in length; motifs of less than five amino acids or scattered in several small modules may thus remain undetected.
Spruce MYBs were relatively under-represented in group A, where they fell into subgroups 21 and 22. In our analysis, spruce group A MYBs contained six of the nine newly identified C-terminal consensus amino acid sequences. Three of these motifs were specific to conifers assigned to subgroup 22: motifs I, K and P found in PgMYB6, 7 and 9. The motifs might be involved in protein or DNA interactions; however, it remains to be seen whether they play a role in protein structure or function.
Spruce MYB phylogeny and evolution
There are very few reports from which to estimate the number of R2R3-MYB genes in gymnosperms or to gain insights into the molecular evolution of this protein family [10-12,34,35]. According to the phylogenetic relationship with other MYB genes in angiosperms and gymnosperms, the spruce MYB sequences described here belong to nine different MYB clades distributed between group A and group C described by Romero et al. [19]. None of the conifer sequences identified in this study and none of the reported gymnosperm R2R3-MYBs were assigned to the B group [19]. We may hypothesize that group B sequences are present only in angiosperms, however, more gene discovery work is needed to draw conclusions since only four of the 125 Arabidopsis MYB genes belong to this group B [19,31].
Despite recent large-scale gene discovery initiatives for conifers like pine and spruce (e.g. [15,16]), only a few regulatory gene families have been characterised in any conifer species. The R2R3-MYBs family has evolved and expanded very rapidly through numerous gene duplications in Angiosperms [36]. Given the very distant separation of gymnosperms and angiosperms (approx. 300 million years), we were interested in assessing whether a similar gene family evolution would be present in both taxonomic groups. In other words, is the R2R3-MYB gene family structure similar in these two groups? In the knox-I gene family of conifer trees, the structure and number of genes was shown to be very different from than of angiosperms, in a recent study investigating evolution of the family in great detail [17]. Several of the angiosperm clades are missing in conifers which appear to have undergone several recent gene duplications with relatively low sequence divergence levels. Our work provides a clear indication that the conifer MYB family structure is not all that divergent from that of the angiosperms, in contrast to the Knox-I report, suggesting that the basic family structure predates the gymnosperm – angiosperm split. In maize, several subgroups of R2R3-MYB genes have expanded within the past 50 million years [36,37]. Consistent with this, our analysis of coding sequences and introns in spruce MYB genes also suggests more recent gene duplications in, at least in some of the clades. For example, PgMYB5, 10 and 13 have high levels of nucleotide sequence similarity in coding sequence as well as introns I and II.
Further investigation is needed to discover the full complement of conifer MYB sequences. By comparison to the angiosperms, we predict that the set of sequences described here represents a fraction of the conifer R2R3-MYB family. Identification of new sequences would complete the evolutionary picture of this conspicuous family of regulators and help to determine its position in the evolution of plant lineages.
Potential involvement of the spruce R2R3-MYBs in the lignification of woody tissue
The spruce and pine sequences we analysed represent diverse subgroups of the R2R3-MYB family. Thus, we hypothesized that they could play diverse roles in metabolism and development. The involvement of specific R2R3-MYB gene products in lignin biosynthesis and/or wood formation is suggested by their expression profiles and by their sequence homology with genes from pine ([10,11]) and in other species whose functions have been previously tested. The AC cis-regulatory elements, for example, which are found in many promoters of phenylpropanoid and lignin biosynthesis genes, play an important role in gene regulation in lignifying xylem cells, thus linking R2R3-MYB genes with lignin biosynthesis. AC elements have been implicated in the transcriptional regulation of PAL in bean [38], 4CL in parsley [39], CCR and CAD in Eucalyptus [40,41].
We compared the abundance of the 13 different spruce MYB mRNAs in selected tissues and organs that develop a secondary vasculature in mature spruce, and in the primary stems and differentiating secondary xylem in young trees. PgMYB2, 4 and 8, all of which belong to the same phylogenetic clade, were expressed preferentially in the secondary differentiating xylem of both juvenile plants and mature trees. Interestingly, all three genes were also expressed preferentially in xylem tissues isolated from large roots. By comparison, 4CL had a very similar transcript profile in prospected tissues. The other MYB genes had various patterns of expression including phloem-preferential and ubiquitous patterns.
We also compared the RNA levels in differentiating secondary xylem during the induction of compression wood in spruce seedlings. Compression wood development in conifers that are leaning or bent is characterised by the formation of thicker cell walls, increased lignin content and the deposition of more condensed lignin polymers, among other features [42]. The plasticity of lignin biosynthesis and cell wall architecture observed in compression wood have been linked to the fluctuation in abundance of several gene transcripts and proteins [43,44]. Although the transcriptional regulators that orchestrate this plasticity are unknown, they might include MYB transcription factors due to their implication in xylem differentiation and in lignin biosynthesis. The three PgMYBs (2, 4 and 8) that were preferentially expressed in xylem are likely candidates because they were also upregulated on the compression wood-forming side (downward side) of the stem but remained relatively constant on the opposite side. The time-course and relative magnitude of the changes in transcript levels of the PgMYBs 2, 4 and 8 were quite similar to those seen for genes encoding lignin biosynthesis enzymes and an AGP surveyed in the same samples. Three other PgMYBs (9, 11 and 13) also showed an increase in transcript abundance in secondary xylem upon induction of compression wood. In the control trees, the mRNA for PgMYB11 was one of the highest we examined in the apical portion of the stem but it was low in secondary xylem. These observations might imply a role for PgMYB11 in processes that are common to primary stem growth and compression wood formation. The MIXTA gene from Antirrhinum majus, which belongs to the same phylogenetic subgroup, is involved in cellular development [45] and may provide clues to the role of PgMYB11 in conifers.
The expression profiles of a few of the spruce MYB genes are consistent with previous reports describing the putative function of homologous genes. For example, spruce PgMYB4 is a close homolog of PtMYB4 [10], which induced ectopic lignification when overexpressed in transgenic tobacco. A putative role for PgMYB4 in lignification is consistent with our data showing a higher mRNA level in compression wood, characterised by increased lignin deposition. The gene PgMYB8 (subgroup 13) showed strong similarity with AtMYB61, which is expressed in xylem tissues of Arabidopsis and was shown to play an important role in regulating lignification [8]. AtMYB61 is also expressed in developing seeds, where it regulates the extrusion of seed coat-derived rhamnogalacturonan mucilage [23]. The accumulation of PgMYB8 transcripts in compression wood is consistent with the ectopic lignification resulting from the constitutive overexpression of AtMYB61 in Arabidopsis [8]. By contrast, AtMYB103, the most similar Arabidopsis sequence to PgMYB2, is not expressed in the stem but is involved in trichome and tapetum development [46], suggesting a putative role in xylem differentiation other than lignin biosynthesis.
The spruce sequence PgMYB1 has a close pine homolog, PtMYB1, that has been linked to lignin biosynthesis [11], however the spruce sequence was not expressed preferentially in secondary xylem (it was also expressed in needles, phloem and the shoot apex) nor was it induced during compression wood formation. It was demonstrated that PtMYB1 is able to bind the AC-I and AC-II elements (PAL-Box) [11]. Recently, Gomez-Maldonaldo et al. [13] showed that pine MYB1 and MYB4 bind to glutamine synthetase AC elements and that MYBs are linked to several metabolic pathways by shared cis-acting elements. Based on these observations, it appears that the MYB1 genes of pine and spruce may regulate phenylpropanoid metabolism as well as nitrogen assimilation in various plant tissues.
Conclusion
Through a systematic survey of EST sequence data followed by full length sequencing, we characterised 18 conifer R2R3-MYB gene sequences (13 from P. glauca, white spruce; 5 from P. taeda, loblolly pine). Three R2R3-MYBs from spruce, namely MYB 2, 4 and 8 were shown to be expressed preferentially in secondary xylem. We also found that transcript levels of six PgMYB genes (including the MYB 2, 4 and 8 genes), were upregulated in differentiating secondary xylem from young trees during the induction of compression wood along with cell-wall-related genes. Our study highlights a small set of spruce MYB transcription factors that could be good candidate genes for marker development studies. Gain-of-function/loss-of-function studies using transgenic plants are also needed to delineate the roles of these different MYBs. Such studies are expected to lead to greatly lacking insights into the regulation of wood formation in conifers.
Methods
Plant material and RNA isolation
Several tissues were isolated from two 33-year-old P. glauca trees felled in July 2003, from a progeny trial established near Quebec City (Canada). All tissues were frozen in liquid nitrogen immediately upon removal from the tree and stored at -80°C until further use. We collected newly formed needles from the upper crown. Differentiating secondary xylem and phloem, as well as bark tissues were collected from three 30–40 cm bolts taken from the lower third of the main stem. These vascular tissues were scraped with a scalpel immediately after peeling the bark. Tissues scrapped from the exposed inner side of the bark and from surface of the exposed wood were labelled as differentiating secondary phloem and xylem, respectively. Similarly, differentiating xylem and bark (including phloem) were collected from large roots located in a one-meter radius from the base of the stem. Samples from each tree and each tissue were kept separate for RNA extraction and gene expression studies.
A gravitropic treatment to induce compression wood formation was performed on 3-year-old spruce seedling stock. The seedlings were transferred to 3 L pots one month before the experiment, grown in a greenhouse with 16 hours light per day, and fertilised weekly with 20 g/L N-P-K. A randomised design of 24 young trees was established in which 12 trees were maintained at 45° angle by leaning the pots and tying the plants to stakes (also at 45°); 12 seedlings were grown in the normal vertical position. Destructive tissue samplings were carried out 4, 28 and 76 hours after the beginning of the treatment. The average diameter of the plants near the base was 7.2 +/- 0.61 mm, their average height was 60.63 +/- 7.25 cm, and the terminal leader was 19.83 +/- 2.91 cm. For each time point, four vertical and four leaning trees were harvested mid-morning in a randomised order, and three randomly selected trees were used for gene expression analyses. Secondary xylem was collected as described above from the two sides of the main stem of the seedlings; the lower and upper sides representing compression wood and opposite side wood, respectively, for the leaning trees, or left and right side for vertical tree. The whole terminal leader was also collected from each plant. Total RNAs were isolated from tissues described above and ground in liquid nitrogen with a pestle and mortar, except for the gravitropic treatment where a Mixer Mill MM300 engine (Retsch) was used to grind in Microtubes (Eppendorf). The RNAs were extracted from each tissue sample and each tree or seedling separately, following the procedure of Chang et al. [47], in Oakridge tubes or in Microtubes. RNA concentration and quality were determined with a bioAnalyser (model 2100, Agilent Technologies; RNA 6000 Nano Assay kit).
DNA cloning and accession numbers
Previous reports described two complete coding sequences of R2R3-MYB genes from pine (PtMYB1 and PtMYB4) [10,11] and one from spruce [12] as well as several partial sequences [35] expressed in xylem tissues of pine. We isolated partial spruce cDNA clones representing putative orthologues of the pine MYB genes by PCR amplification, and identified several partial and putative full-length sequences among the spruce EST sequences data of the ARBOREA project derived from 17 different cDNA libraries[48] For each of the partial spruce and pine gene sequences, we obtained complete coding sequence and UTRs by using 3' RACE, 5' RACE or both cloning methods on spruce or pine mRNA from needles or xylem (SMART RACE cDNA Amplification Kit, Invitrogen, Carlsbad, CA). DNA was cloned in pCR2.1 with the TA cloning Kit (Invitrogen, Carlsbad, CA) and sequenced. The sequence analyses presented hereafter are based upon cDNA clones containing the complete coding sequences of the MYB, which were isolated as a single fragment from reverse transcribed RNA, with gene specific primer pairs for each of the 13 sequences from spruce and five sequences from pine. The numbering of pine MYB genes (PtMYB1-8 and PtMYB14; no PtMYB5 and 6 reported) is in accordance with Patzlaff et al., [10,11]; the numbering of spruce genes was established on the basis of putative orthology with the pine sequences.
In addition, we isolated the 13 corresponding sequences from spruce genomic DNA (gDNA). Genomic DNA was extracted from needles of white spruce using the Genomic-Tip Kit (Qiagen, Mississauga, Ontario). The entire coding region with introns was isolated by PCR amplification with gene specific primer pairs spanning each gene's coding region (Additional file 1) and cloned in pCR2.1 with the TA cloning Kit (Invitrogen, Carlsbad, CA). The gDNA was from Picea glauca genotype Pg653 and so did most of the cDNA clones (although a few came from wild Picea glauca genotypes). Each clone was sequenced at least through the MYB DBD in order to determine the number of introns present in this region. Some nucleotide differences were observed between cDNA and gDNA sequences due to the genotypic variation, but no non-sense mutation were detected. The genomic sequences of PgMYB 3, 6, 7 and 11 showed 1 to 3 non synonymous substitutions giving no less than 99.2% amino acids identity; however, we do not find nucleotide mismatches in spruce MYB 2, 4, 8, 9 and 12.
The 13 MYB genes from spruce and five MYB genes from pine have the following accession numbers: PgMYB1 [GenBank: DQ399073], PgMYB2 [GenBank: DQ399072], PgMYB3 [GenBank: DQ399071], PgMYB4 [GenBank: DQ399070], PgMYB5 [GenBank: DQ399069], PgMYB6 [GenBank: DQ399068], PgMYB7 [GenBank: DQ399067], PgMYB8 [GenBank: DQ399066], PgMYB9 [GenBank: DQ399065], PgMYB10 [GenBank: DQ399064], PgMYB11 [GenBank: DQ399063], PgMYB12 [GenBank: DQ399062], PgMYB13 [GenBank: DQ399061] and PtMYB2 [GenBank: DQ399060], PtMYB3 [GenBank: DQ399059], PtMYB7 [GenBank: DQ399058], PtMYB8 [GenBank: DQ399057], PtMYB14 [GenBank: DQ399056].
The nucleotides sequences of candidate genes involved in wood formation come from the spruce EST assembly directory number 8 (dir8) of the ARBOREA project[48] Their percentage amino acid sequence similarity with other species is given in brackets. They are:
phenylalanine ammonia lyase (PAL) [dir8: contig10199] partial coding sequence (cds), 85% to Pinus taeda [GenBank: U39792];
4-coumarate: CoA ligase (4CL) [dir8: contig10433] partial cds, 86% to Pinus taeda [GenBank: U39405];
caffeoyl-CoA 3-O-methyltransferase (CCOaOMT) [dir8: contig5884] complete cds, 92 % to Pinus taeda [GenBank: AF036095];
arabinogalactan protein (AGP) [dir8: contig10745] complete cds, 75 % to Pinus taeda [GenBank: U09556];
cinnamyl alcohol dehydrogenase (CAD) [dir8: contig9065] complete cds, 95 % to Pinus radiata [GenBank: AF060491],
and the housekeeping gene elongation factor alpha (EF1-α) [dir8: contig10829] complete cds, 99% to Picea abies [GenBank: AJ132534].
Sequence analyses and phylogenetic studies
Nucleotide and amino acid sequence alignments were obtained with Clustal W [49] using BioEdit software version 6.0.7 and BoxShade 3.21 to highlight sequences. Delineation of introns was achieved by aligning the cDNA and genomic nucleotide sequences of the PgMYB from the start codon to stop codon on the basis that introns begin with GT and end with AG dinucleotides. Phylogenetic studies were performed with the 13 predicted MYB protein sequences from white spruce, one sequence from black spruce (PmMBF1, [12]), and seven loblolly pine sequences including previously reported PtMYB1 and 4 [10,11]. We also included 11 diverse Arabidopsis MYB sequences and two R1R2R3-MYB genes from human and mouse as landmarks to classify the MYBs according to previous reports [18,19,30]. We constructed a neighbour-joining tree based on a Clustal W amino acid alignment generated with the Mega 2.0 method [50] and using 1000 bootstraps to estimate the node strength (parameters are Poisson correction and pair-wise deletion as described in [30]).
The accession numbers of the Arabidopsis genes analysed are: AtMYB13 [GenBank: At1g06180], PtMYB1 [GenBank: AY356372], AtMYB4 [GenBank: At4g38620], AtMYB103 [GenBank: At1g63910], AtMYB46 [GenBank: At5g12870], PtMYB4 [GenBank: AY356371], AtMYB61 [GenBank: At1g09540], AtMYB52 [GenBank: At1g17950], AtMYB44 [GenBank: At5g67300], AtMYB20 [GenBank: At1g66230], AtMYB101 [GenBank: At2g32460], AtMYB106 [GenBank: At3g01140], AtMYB33 [GenBank: At5g06100], PmMBF1 [GenBank: U39448]. The accession numbers of the conifer MYB genes are as above.
MEME analysis software [51] was used to identify amino acid regions conserved between several members of a subgroup of sequences (according to Kranz et al. [18]) containing one or more spruce MYBs. The parameters setting was the number of motifs to find: 5; minimum width of motif: 5 and maximum: 15. We used complete protein sequences of MYBs from 12 conifer species and from 14 angiosperm species (Additional file 2). We also included 10 partial conifer sequences that encompassed the C-terminal region [34] and were closely related to PgMYB6, 7 and 9 (Additional file 3).
Analysis of transcript accumulation by Q-RTPCR
To analyse the transcript abundance of PgMYBs, first-strand cDNA was synthesised starting from one microgram of RNA treated with amplification grade DNAse I (Invitrogen) and purified on an RNeasy column (Qiagen), using oligo(dT) primers and SuperScript II RT (Invitrogen) according to the manufacturer's instructions. The resulting cDNA was diluted 1:5 with sterile RNAse-free water and stored at -20°C; the same cDNA was used for all Q-RTPCR (quantitative reverse transcription PCR) assays for any given tissue sample. Steady-state mRNA levels were determined on cDNA by quantitative RT-PCR using Opticon Monitor 2 fluorescence detection system (MJ research) and DyNAmo SYBR Green QPCR kit (Finnzymes Oy, Espoo, Finland). Gene primer pairs were designed using the Primer 3 software [52] to anneal near the 3' end of each transcript (usually in 3' UTR) to ensure primer specificity. The forward and reverse primers were as follows (amplicon length indicated in brackets):
PgMYB1: 5'-gattgtacattaacccagtaa-3' and 5'-taaaccatgtggtatctgtta-3' (148 bp);
PgMYB2: 5'-tgggtattctaggtatttcc-3' and 5'-attaggtaagtatgcaggg-3' (99 bp);
PgMYB3: 5'-agatcacggacccagatcaac-3' and 5'-gagcgaacgacctccttcag-3' (147 bp);
PgMYB4: 5'-gcagtttgagtttgagtgtg-3' and 5'-ctggagcatagatttgatga-3' (162 bp);
PgMYB5: 5'-aattctggcagcgaactg-3' and 5'-aatgcttcgtggtggaatc-3' (173 bp);
PgMYB6: 5'-tttccttccttcatttcaac-3' and 5'-taaatttgggtttctgttgc-3' (108 bp);
PgMYB7: 5'-tcgagttgcacatcaggag-3' and 5'-gagtgtggatggcaaacag-3' (156 bp);
PgMYB8: 5'-ggtggactcagttgtaataa-3' and 5'-gtatctcacctatttacagatca-3' (101 bp);
PgMYB9: 5'-gaaattcgagaaacatggtg-3' and 5'-aaacgacagaaatcgagaac-3' (149 bp);
PgMYB10: 5'-gctgtattttaacatttcatgg-3' and 5'-acaacaatctttctttttctcc-3' (133 bp);
PgMYB11: 5'-cccagcttatgactggaag-3' and 5'-tacagaacaaccatgcagac-3' (157 bp);
PgMYB12: 5'-caggtgacttaactctattccag-3' and 5'-tcacatagaacaggcatgg-3' (143 bp);
PgMYB13: 5'-aaattacagctagagtgagagg-3' and 5'-aacttgaaccgtacacgac-3' (86 bp) and
EF1-α (elongation factor alpha): 5'-aactggagaaggaacccaag-3' and 5'-aacgacccaatggaggatac-3', (114 bp). Forward and reverse primer pairs used for the Q-RTPCR of the wood formation-related genes are:
PAL: 5'-tggatttgcatcctactg-3' and 5'-tccatcttcaactataggac-3' (103 bp);
4CL: 5'-cattcctcaaaagcatgaagag-3' and 5'-atcgcatccacaaagtacac-3' (150 bp);
CCOaOMT: 5'-attgagatcagccaaatcc-3' and 5'-gcgctctccctataatcag-3' (124 bp);
AGP: 5'-gcgtccattgttttaatgtag-3' and 5'-tgtatttatccctctgtctgc-3' (181 bp) and
CAD: 5'-ctggactacatcaatactgc-3' and 5'-gatttactcattctgcacg-3' (141 bp).
The PCR reaction mixture (20 μL) consisted of 10 μL DyNAmo SYBR Green qPCR mix, 1 μL primers (0.25 μM forward and 0.25 μM reverse), 1 μL of cDNA and 8 μL of RNase-free water. The cycling conditions were 95°C for 2 min, then 35 cycles of 95°C for 10 sec, 55°C for 10 sec, 72°C for 8 sec, followed by a plate reading. The melting curve readings were carried out every 0.2°C from 65 to 95°C, holding 1 sec at each temperature. Standard curves were established for each spruce MYB and cell-wall-related cDNAs and were used to determine RNA abundance in each sample. The standards consisted of serial dilutions of PCR amplicons prepared from each cDNA, cloned in pCR2.1, with M13 reverse and forward primers. Amplicon standards were gel purified (Qiagen), and product length and concentration were verified using a bioAnalyser (model 2100 Agilent Technologies, DNA 1000 LabChip kit). The standard curves were determined from duplicate reactions from the dilutions series of each amplicon. Raw data were converted with the following parameters: no blank subtraction, subtract baseline and average over cycle range according each case. We calculated the number of transcript molecules per ng of total RNA [(DNA quantity quantified in g * DNA base pair mass per gram of DNA)/M13 Reverse-Forward amplified fragment length in bp]. For within tissue comparisons that were carried on differentiation xylem (including compression wood induction) and apical shoots young trees, the RNA abundance was normalised using the abundance of RNA of the elongation factor alpha (EF1-α), as an endogenous control (calculated as the following ratio: target gene (ng)/EF1-α (ng)).
Abbreviations
Q-RTPCR, quantitative reverse transcriptase polymerase chain reaction; EST, expressed sequence tag; RACE, rapid amplification of cDNA-ends.
Authors' contributions
FB carried out the experimental work and the data analysis, participated in the design of the study and drafted the manuscript. JGP and JM participated in the design of the study and in manuscript preparation. All authors read and approved the final manuscript.
Supplementary Material
Primers sequences of spruce MYBs used for genomic amplification and sequencing. The table provides the nucleotides sequences of the primers with respective Tm values and amplified genomics DNA length, for each spruce MYB genes.
Phylogenetic tree of MYBs from spruce, pine and nearest sequences from other species. The figure shows the phylogenetic relationship between conifers MYBs and others MYBs on the basis of their complete amino acids sequences. Protein sequences from each clade incorporating spruce MYBs was used to search for conserved amino acid motifs.
Conifer MYB phylogeny based on partial sequences using a 31 amino acids region. The figure shows the phylogenetic relationship between several conifers MYBs on the basis of a 31 amino acid region in the third MYB domain repeat. Due to the availability of many partial sequences, this short region leads to the addition of several conifer sequences.
Acknowledgments
Acknowledgements
We are grateful to R.R Sederoff (North Carolina State University, Raleigh, NC) for kindly providing clones of Pinus taeda. We acknowledge the technical assistance of S. Blais for cloning of pine sequences and F. Boileau for tissue and RNA isolation. The authors thank C. Bomal for critical reading of the manuscript. Funding from Genome Canada and Génome Québec to JM for the ARBOREA project supported this research.
Contributor Information
Frank Bedon, Email: frank.bedon@rsvs.ulaval.ca.
Jacqueline Grima-Pettenati, Email: grima@scsv.ups-tlse.fr.
John Mackay, Email: jmackay@rsvs.ulaval.ca.
References
- Rogers LA, Campbell MM. The genetic control of lignin deposition during plant growth and development. New Phytol. 2004;164:17–30. doi: 10.1111/j.1469-8137.2004.01143.x. [DOI] [PubMed] [Google Scholar]
- Chaffey N. Why is there so little research into the cell biology of the secondary vascular system of trees? New Phytol. 2002;153:213–223. doi: 10.1046/j.0028-646X.2001.00311.x. [DOI] [Google Scholar]
- Scarpella E, Meijer AH. Pattern formation in the vascular system of monocot and dicot plant species. New Phytol. 2004;164:209–242. doi: 10.1111/j.1469-8137.2004.01191.x. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, -Z. C, Jiang. Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, -L. Yu G. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. The AmMYB308 and AmMYB330 Transcription Factors from Antirrhinum Regulate Phenylpropanoid and Lignin Biosynthesis in Transgenic Tobacco. Plant Cell. 1998;10:135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LJ, Perazza DE, Juda L, Campbell MM. Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 2004;37:239–250. doi: 10.1046/j.1365-313x.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- Goicoechea M, Lacombe E, Legay S, Mihaljevic S, Rech P, Jauneau A, Lapierre C, Pollet B, Verhaegen D, Chaubet-Gigot N, Grima-Pettenati J. EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 2005;43:553–567. doi: 10.1111/j.1365-313X.2005.02480.x. [DOI] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, Campbell MM. Characterisation of a pine MYB that regulates lignification. Plant J. 2003;36:743–754. doi: 10.1046/j.1365-313X.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- Patzlaff A, Newman LJ, Dubos C, Whetten RW, Smith C, McInnis S, Bevan MW, Sederoff RR, Campbell MM. Characterisation of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol Biol. 2003;53:597–608. doi: 10.1023/B:PLAN.0000019066.07933.d6. [DOI] [PubMed] [Google Scholar]
- Xue B, Charest PJ, Devantier Y, Rutledge RG. Characterization of a MYBR2R3 gene from black spruce (Picea mariana) that shares functional conservation with maize C1. Mol Genet Genomics. 2003;270:78–86. doi: 10.1007/s00438-003-0898-z. [DOI] [PubMed] [Google Scholar]
- Gomez-Maldonado J, Avila C, Torre F, Canas R, Canovas FM, Campbell MM. Functional interactions between a glutamine synthetase promoter and MYB proteins. Plant J. 2004;39:513–526. doi: 10.1111/j.1365-313X.2004.02153.x. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. LIGNIN BIOSYNTHESIS. Annu Rev Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Kirst M, Johnson AF, Baucom C, Ulrich E, Hubbard K, Staggs R, Paule C, Retzel E, Whetten R, Sederoff R. Apparent homology of expressed genes from wood-forming tissues of loblolly pine (Pinus taeda L.) with Arabidopsis thaliana. PNAS. 2003;100:7383–7388. doi: 10.1073/pnas.1132171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy N, Laroche J, Bousquet J, Mackay J. Large-scale statistical analysis of secondary xylem ESTs in pine. Plant Mol Biol. 2005;57:203–224. doi: 10.1007/s11103-004-6969-7. [DOI] [PubMed] [Google Scholar]
- Guillet-Claude C, Isabel N, Pelgas B, Bousquet J. The evolutionary implications of knox-I gene duplications in conifers: correlated evidence from phylogeny, gene mapping, and analysis of functional divergence. Mol Biol Evol. 2004;21:2232–2245. doi: 10.1093/molbev/msh235. [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, Smeekens S, Tonelli C, Paz-Ares J, Weisshaar B. Towards functional characterisation of the members of theR2R3-MYBgene family fromArabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. More than 80R2R3 MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313X.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- Lipsick JS. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- Ogata K, Hojo H, Aimoto S, Nakai T, Nakamura H, Sarai A, Ishii S, Nishimura Y. Solution Structure of a DNA-Binding Unit of Myb: A Helix-Turn-Helix-Related Motif With Conserved Tryptophans Forming a Hydrophobic Core. Proc Natl Acad Sci USA. 1992;89:6428–6432. doi: 10.1073/pnas.89.14.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41:577–585. doi: 10.1023/A:1006319732410. [DOI] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. MYB61 Is Required for Mucilage Deposition and Extrusion in the Arabidopsis Seed Coat. Plant Cell. 2001;13:2777–2791. doi: 10.1105/tpc.13.12.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylides C, Roby D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA. 2002;99:10179–10184. doi: 10.1073/pnas.152047199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr op Plant Biol. 2003;6:74–78. doi: 10.1016/S136952660200002X. [DOI] [PubMed] [Google Scholar]
- Neale DB, Savolainen O. Association genetics of complex traits in conifers. Trends Plant Sci. 2004;9:325–330. doi: 10.1016/j.tplants.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Avila J, Nieto C, Canas L, Benito MJ, Paz-Ares J. Petunia hybrida genes related to the maize regulatory C1 gene and to animal myb proto-oncogenes. Plant J. 1993;3:553–562. doi: 10.1046/j.1365-313X.1993.03040553.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
- Yoo JH, Park CY, Kim JC, Do Heo W, Cheong MS, Park HC, Kim MC, Moon BC, Choi MS, Kang YH, Lee JH, Kim HS, Lee SM, Yoon HW, Lim CO, Yun DJ, Lee SY, Chung WS, Cho MJ. Direct Interaction of a Divergent CaM Isoform and the Transcription Factor, MYB2, Enhances Salt Tolerance in Arabidopsis. J Biol Chem. 2005;280:3697–3706. doi: 10.1074/jbc.M408237200. [DOI] [PubMed] [Google Scholar]
- Jiang C, Gu X, Peterson T. Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol. 2004;5:R46. doi: 10.1186/gb-2004-5-7-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr op Plant Biol. 2001;4:447–456. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Development. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Fukuda H. Involvment of calmodulin and calmodulin binding proteins in the differentiation of tracheray elements in Zinnia cells. Planta. 1994;194:388–394. doi: 10.1007/BF00197540. [DOI] [Google Scholar]
- Kusumi J, Tsumura Y, Yoshimaru H, Tachida H. Molecular Evolution of Nuclear Genes in Cupressacea, a Group of Conifer Trees. Mol Biol Evol. 2002;19:736–747. doi: 10.1093/oxfordjournals.molbev.a004132. [DOI] [PubMed] [Google Scholar]
- Mackay J, Bérubé H, Regan S, Séguin A. Functional genomics in forest tress: Application to the investigation of defense mechanisms and wood formation. Plantation Forest Biotechnology for the 21st century: Research Signpost. 2004. p. 446p.
- Rabinowicz PD, Braun EL, Wolfe AD, Bowen B, Grotewold E. Maize R2R3 Myb Genes: Sequence Analysis Reveals Amplification in the Higher Plants. Genetics. 1999;153:427–444. doi: 10.1093/genetics/153.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun EL, Grotewold E. Newly Discovered Plant c-myb-Like Genes Rewrite the Evolution of the Plant myb Gene Family. Plant Physiol. 1999;121:21–24. doi: 10.1104/pp.121.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A, Liang X, Pintor-Toro JA, Dixon RA, Lamb CJ. cis-Element Combinations Determine Phenylalanine Ammonia-Lyase Gene Tissue-Specific Expression Patterns. Plant Cell. 1992;4:263–271. doi: 10.1105/tpc.4.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauffe KD, Lee SP, Subramaniam R, Douglas CJ. Combinatorial interactions between positive and negative cis-acting elements control spatial patterns of 4CL-1 expression in transgenic tobacco. Plant J. 1993;4:235–253. doi: 10.1046/j.1365-313X.1993.04020235.x. [DOI] [PubMed] [Google Scholar]
- Lauvergeat V, Rech P, Jauneau A, Guez C, Coutos-Thevenot P, Grima-Pettenati J. The vascular expression pattern directed by the Eucalyptus gunnii cinnamyl alcohol dehydrogenase EgCAD2 promoter is conserved among woody and herbaceous plant species. Plant Mol Biol. 2002;50:497–509. doi: 10.1023/A:1019817913604. [DOI] [PubMed] [Google Scholar]
- Lacombe E, Van Doorsselaere J, Boerjan W, Boudet AM, Grima-Pettenati J. Characterization of cis-elements required for vascular expression of the Cinnamoyl CoA Reductase gene and for protein-DNA complex formation. Plant J. 2000;23:663–676. doi: 10.1046/j.1365-313x.2000.00838.x. [DOI] [PubMed] [Google Scholar]
- Timell TE. Compression wood in gymnosperms. Springer, Berlin Heidelberg New York. 1986.
- Plomion C, Leprovost G, Stokes A. Wood Formation in Trees. Plant Physiol. 2001;127:1513–1523. doi: 10.1104/pp.127.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gion JM, Lalanne C, Le Provost G, Ferry-Dumazet H, Paiva J, Chaumeil P, Frigerio JM, Brach J, Barré A, de Daruvar A, Claverol S, Bonneu M, Sommerer S, Negroni L, Plomion C. The proteome of maritime pine wood forming tissue. Proteomics. 2005;5:3731–3751. doi: 10.1002/pmic.200401197. [DOI] [PubMed] [Google Scholar]
- Noda K, Glover BJ, Linstead P, Martin C. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature. 1994;369:661–664. doi: 10.1038/369661a0. [DOI] [PubMed] [Google Scholar]
- Higginson T, Li SF, Parish RW. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 2003;35:177–192. doi: 10.1046/j.1365-313X.2003.01791.x. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Pavy N, Johnson JJ, Crow JA, Paule C, Kunau T, MacKay J, Retzel EF. ForestTreeDB: a database dedicated to the mining of tree transcriptomes. Nucleic Acids Res. 2007;35:D888–894. doi: 10.1093/nar/gkl882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. S Krawetz and S Misener (Eds), Bioinformatics Methods and Protocols: Methods in Molecular Biology Humana Press, Totowa, NJ. 2000. pp. 365–386. [DOI] [PubMed]
- Sharman BC. Tannic acid and iron alum with safranin and orange G in studies of the shoot apex. Stain Techn. 1943. pp. 105–111.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers sequences of spruce MYBs used for genomic amplification and sequencing. The table provides the nucleotides sequences of the primers with respective Tm values and amplified genomics DNA length, for each spruce MYB genes.
Phylogenetic tree of MYBs from spruce, pine and nearest sequences from other species. The figure shows the phylogenetic relationship between conifers MYBs and others MYBs on the basis of their complete amino acids sequences. Protein sequences from each clade incorporating spruce MYBs was used to search for conserved amino acid motifs.
Conifer MYB phylogeny based on partial sequences using a 31 amino acids region. The figure shows the phylogenetic relationship between several conifers MYBs on the basis of a 31 amino acid region in the third MYB domain repeat. Due to the availability of many partial sequences, this short region leads to the addition of several conifer sequences.


