Abstract
We have identified a novel mature human B-cell subpopulation in the human tonsil that has characteristics of both naive B cells and germinal center B cells including the expression of activation-induced cytidine deaminase (AID), which is essential for the process of immunoglobulin somatic hypermutation and class-switch recombination. These cells are clearly somatically hypermutated, albeit modestly. Their phenotype (IgD+CD38−CD23−FSChiCD71+) is unique and suggests they may be intermediate between both naive and germinal center cells. Morphologically they are also distinct from other B-cell subpopulations. The evidence presented suggests these cells may be the founder cells of the germinal center reaction (a pro-GC cell) and may be the normal counterpart of the mantle cell lymphoma cell.
Introduction
Peripheral B cells are widely recognized to be broadly divided into naive, germinal center, memory, and plasma cell populations. Over a decade ago, knowledge available through immunohistochemical staining of human lymphoid tissue was applied to the separation of B cells from peripheral lymphoid tissue into these and other subpopulations using flow cytometry.1,2 Naive B cells are recognized as IgD+IgM+CD38−CD27− cells that can be further divided by expression of CD23 into Bm1 (Bm = “B mature”; CD23−) and Bm2 (CD23+) cells. With rare exception, Ig V genes isolated from such cells are unmutated. Germinal center cells are IgD−CD38+CD27− and can be divided into Bm3 (CD77+: centroblasts) and Bm4 (CD77−: centrocytes) populations. With few exceptions, cells isolated from germinal centers contain highly mutated IgV genes. While labeled numerically, it has not been conclusively established which subpopulation follows another beyond the progression of naive B cells to become germinal center B cells before finally differentiating into an effector B cell (memory or plasma cell)
With advances in flow cytometry, the number of subpopulations that can be sorted from peripheral lymphoid tissue has expanded. Pre-GC cells (IgD+IgM+CD38+), δ-class–switched cells (IgD+IgM−CD38+CD27−),3 plasmablasts (IgD−CD38++CD27+), memory cells (Bm5) (IgD−CD38−CD27+), and marginal zone T-independent cells (IgD+IgM+CD38−CD27+),4 among others, have all been identified as distinct subpopulations of B cells in human tonsil. All of these subpopulations of cells can now be isolated from a single tonsil using 6-color flow cytometry of isolated B cells. This subdivision of peripheral B cells continues to allow greater understanding of not only their function but also of the origin, prognosis, and treatment targets for human B-cell lymphomas and leukemias.
The process of affinity maturation is central to the development of high-affinity antibodies. Somatic hypermutation, critical to this process, requires the presence of activation-induced cytidine deaminase (AID).5,6 AID is also essential for class-switch recombination.7–9 While absent in naive B cells, AID expression has been repeatedly associated with mutated germinal center B cells.10,11 The nucleic acid upon which AID acts is still a matter of debate between camps holding to the RNA hypothesis (indicating AID would itself not be the Ig somatic hypermutator but work on the transcript for another protein—reviewed in Tasuku Honjo12) or the DNA hypothesis (AID acting directly to mutate Ig genes—the prevailing view).13–16 Factors surrounding its activation and regulation have yet to be fully elucidated. Identifying the initial cell population that expresses AID is therefore critical to a further understanding of the function of this molecule. Here, we describe what appears to be the earliest B-cell population in which AID is expressed—an activated B-cell population with many of the cell surface and molecular Ig characteristics of a naive B cell and yet with important similarities to the cells in the germinal center.
Materials and methods
Tonsil tissue
Tonsils were obtained from the Children's Hospital of Oklahoma in Oklahoma City, OK. The institutional review boards of the University of Oklahoma Health Sciences Center and the Oklahoma Medical Research Foundation (OMRF) approved the use of human tissue. Protocols and informed consent procedures were provided according to Declaration of Helsinki guidelines.
Preparation of human cells
B lymphocytes were obtained from tissue removed during routine tonsillectomy. Total cells were collected and diluted with Hanks balanced salt solution (Mediatech, Norcross, GA), and Lymphocyte Separation Media (Mediatech) was used as a density gradient to separate the mononuclear cells. After separation, the cells were washed with magnetic-activated cell sorter (MACs) buffer (1 × PBS and 0.5% BSA) and counted. They were then enriched for B cells according to the protocol in the MACs B Cell Isolation Kit (Miltenyi Biotec, Auburn, CA).
After separation, the B lymphocytes were stained with various antibodies for flow cytometry using a MoFlo (DakoCytomation, Fort Collins, CO) cell sorter. Enriched B cells were sorted into the following populations: IgD+/CD38−/CD23−/IgM+/CD27−/FSClo (Bm1), IgD+/CD38−/CD23+/IgM+/CD27−/FSClo (Bm2), IgD+/CD38−/IgM+/CD27−/CD23−/FSChi (pro-GC), IgD−/CD38+/IgM−/CD27−/CD77+ (Bm3), IgD−/CD38+/IgM−/CD27−/CD77− (Bm4), IgD−/CD38−/CD27+/IgM+/− (Bm5; memory), or IgD−/CD38++/CD27+/IgM+/− (plasmablast; PB) cells.
Antibodies for flow cytometry
Antibodies were either biotinylated or conjugated for sorting with fluorescein isothiocyanate (FITC), phycoerythrin (PE), allophycocyanin (APC), tricolor (TC), APC-cyanine (APC-Cy5.5), or PE–Texas Red (ECD) and were as follows: anti-CD77 (clone 5B5), anti-CD5 (L17F12), anti-CD10 (H110a), anti-CD39 (TU66), anti-CD40 (5C3), and anti-CD95 (DX2) (all from Becton Dickinson, San Jose, CA); anti-CD23 (clone 9P25; Beckman Coulter, Fullerton, CA); anti-IgM (clone SA-DA4; Southern Biotech, Birmingham, AL); anti-IgD (clone IA6-2; Becton Dickinson); anti-CD38 (clone HIT 2), anti-CD19 (clone SJ25-C1), anti-CD27 (clone CLB-27/1), anti-CD4 (clone S3.5), anti-CD71 (clone T56/14), anti-CD20 (H147), anti-CD21 (BU32), anti-CD44 (MEM-85), anti-CD69 (CH/4), anti-CD80 (MEM-233), anti-CD86 (BU63), and anti–HLA-DR (TU36) (all from Caltag Laboratories, Burlingame, CA). The biotinylated antibodies were stained with a secondary step using Streptavidin Red 613 (Caltag Laboratories).
Cloning and sequencing of Ig VH4 gene segment cDNAs
After collection of pro-GC or specifically AID-expressing pro-GC cells, they were centrifuged and lysed using lysis buffer and prepared for isolation of RNA according to the protocol for the RNAqueous Micro RNA Isolation Kit (Ambion, Austin, TX). RNA was converted to cDNA by reverse transcription and amplified by polymerase chain reaction (PCR) using gene-specific primers,17 according to the protocol for the Qiagen OneStep RT-PCR kit (Qiagen, Valencia, CA). Products of RT-PCR were extracted from agarose gel using a QIAquick Gel Extraction Kit (Qiagen), ligated, and cloned using the Qiagen PCR Cloning Kit (Qiagen). Plasmids from individual bacterial colonies were purified using a QIAprep Spin Miniprep Kit (Qiagen). The purified plasmid DNAs were submitted to the OMRF DNA Sequencing Facility.
Giemsa staining
Sorted B-cell populations (Bm1, Bm2, pro-GC [CD71+], Bm3, Bm4, Bm5, and PB) were cytocentrifuged for 5 minutes at 400 rpm onto microscope slides, air dried, and fixed in methanol for 5 to 7 minutes at room temperature. The slides were incubated with Giemsa stain (Sigma-Aldrich, St Louis, MO) diluted 1:20 with distilled water for 30 minutes to 1 hour at room temperature, and then washed with distilled water. Images were captured using a Zeiss Standard light microscope (Carl Zeiss, Heidelberg, Germany) with a 40× plan objective using a Canon Digital Rebel XT camera back (Canon, Lake Success, NY). Brightness and contrast were globally adjusted to the same level for each image using Adobe Photoshop software (Adobe Systems, San Jose, CA).
Cell cycle fractionation with Hoechst and pyronin Y
After B-cell isolation, cells were enriched with IgD by magnetic cell sorting and further stained to isolate populations for cell cycle analysis. IgD+ cells were stained with anti–human CD38, anti–human CD27, and anti–human CD23 to obtain Bm1, Bm2, and pro-GC (without CD71 preselection). IgD− cells were sorted into germinal center (GC), Bm3, and Bm4 populations.
A combination of Hoechst 33342 (Hst) and pyronin Y (PY) was used for the differential staining of cellular RNA and DNA as described elsewhere.18 Briefly, the isolated B-cell populations were fixed in 70% ethanol overnight, then resuspended in a solution of 2 μg/mL Hst (Molecular Probes, Eugene, OR) and 4 μg/mL PY (Polysciences, Warrington, PA) and measured by flow cytometry on a MoFlo equipped with UV laser (DakoCytomation). Since RNA staining with PY yields a continuous histogram without demarcation between positive and negative cells,19 an arbitrary analysis window comprising about 90% of Bm1 and Bm2 subpopulations displaying minimal PY staining was used to designate the G0 fraction in all experiments.
B-cell–T-cell cocultures
The sort-purified B-cell populations Bm1, Bm2, and pro-GC (CD71+) were cultured with autologous irradiated T cells that were CD3+/CD4+. A 96-well round-bottom tissue-culture plate was first coated with an anti–mouse IgG/PBS mix, and then diluted 1:5000 for 2 hours at 37°C. After 2 hours, the plate was washed and coated with a 1:5 dilution of OKT3 cell supernatant in RPMI (with 10% FCS, antibiotic-antimycotic, sodium pyruvate, and l-glycine) for 2 hours to overnight at 37°C. The wells were filled with 100 μL RPMI, and B cells at 1.0 × 104/well were added with 1.0 × 105 at 10 μL/well of irradiated T cells (irradiated at 40 gray) to a total final volume of 200 μL. The control wells contained only B cells with no T cells. Diluted (1:100) anti-IgM Fab was added at 2 μL/well to all wells except the control wells. The cells were incubated at 37°C for 1 week. After 1 week, the similar cell populations were pooled and analyzed using a FACs Calibur with anti–human CD27, anti–human CD23, anti–human CD38, anti–human IgD, and anti–human CD71.
Western blot
One hundred thousand cells were isolated by flow cytometry from each population and lysed with mild detergent (1% Igepal CA-630, 0.4% sodium deoxycholate, 10 mM Tris-HCl [pH = 8.0], 0.1% SDS, 5 mM EDTA) for 5 minutes on ice. Lysis material was centrifuged for 5 minutes at 14 000 rpm and cytoplasmic supernatants were transferred to clean tubes. Nuclear pellets were resuspended in detergent solution. AID (sc-14680 goat polyclonal antibody; Santa Cruz Biotechnology, Santa Cruz, CA) was added to nuclear and cytoplasmic fractions and immunoprecipitated for 1 to 2 hours at 4°C with constant tumbling. Pansorbin was added to each sample and tumbled for 1 hour at 4°C. Samples were sedimented and pellets washed with detergent solution, then suspended in 2 × sample buffer, mixed, and boiled. Samples were run on a 12% acrylamide gel and transferred to PVDF membrane (Millipore Immobilon-P; Millipore, Billerica, MA). Membranes were blocked for one hour in 1% BSA in TTBS buffer and incubated in primary antibody (AID mAb, mouse, no. 4975 [Cell Signaling Technology, Danvers, MA] 1:1000 in TTBS) overnight at 4°C with gentle shaking. Membranes were then incubated in secondary antibody (biotin antimouse [Vector Labs, Burlingame, CA] 1:1000 in TTBS) for 1 hour at room temperature. Membranes were then incubated with HRP-Streptavidin (Vector Labs) 1:10 000 in TTBS for 30 minutes at room temperature. Enhanced chemiluminescence (ECL) reagents (Amersham, Piscataway, NJ) were applied to the membrane and visualized using a LumiImager (Roche, Indianapolis, IN). Analysis was performed using IPLab software (Scanalytics, Rockville, MD).
Immunohistochemistry
Human tonsil tissue was fixed with 4% paraformaldehyde for 1.5 hours, rinsed, and incubated at 4°C overnight in 30% sucrose. Tissue was embedded in OCT mounting media (Bayer, Pittsburgh, PA), and 5-μm sections were mounted on charged glass slides. Tissue was blocked with normal donkey serum and stained with mouse anti–human IgD (LE 2) (Serotec, Oxford, United Kingdom), rabbit anti–human CD38 (Anaspec, San Jose, CA), and goat anti–human AID (C-20) (Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies were as follows: rhodamine-conjugated donkey anti–mouse IgG, Cy5-conjugated donkey anti–rabbit IgG (both Jackson ImmunoResearch, West Grove, PA), and Alexa Flour 488 donkey anti–goat IgG (Molecular Probes). DAPI counterstain was used to visualize nuclei. For imaging a Zeiss LSM 510 confocal with META, C-apochromat 40× (NA 1.2) water immersion objective using PMT detectors was used. Adjustments for color balance were globally applied to images using Aperature Software (Apple Computer, Sao Palo, CA).
Results
IgD+CD38−CD23−FSChi cells represent a novel human B-cell subset
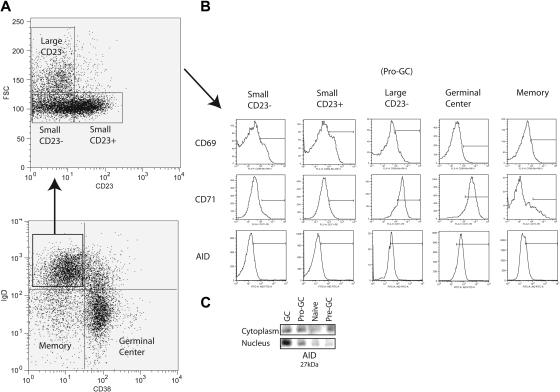
Peripheral mature naive B cells (IgD+CD38−) were originally divided into 2 subpopulations by the expression of CD23 (Bm1 and Bm2)1,2 (Figure 1A). In this study, in conjunction with CD23, we used forward light scatter (a measure of cell size) and the activation marker CD71 in an attempt to further subdivide this population of B cells in the human tonsil. We identified among IgD+CD38− cells a population that was CD23− but FSChi (“large”) instead of FSClo and was mostly CD71hi (85% of IgD+CD38−CD23−FSChi cells) (Figure 1A-B). For reasons expanded upon in “Discussion,” we refer to these as pro-GC cells. This population among multiple sorts averages in size to be 0.2% to 2% of total tonsillar B lymphocytes. Previously, these cells would have been classified in the Bm1 subpopulation as they are IgD+CD38−CD23−. We saw them as distinct since Bm1 (and Bm2) cells are FSClo (“small”) and CD71lo. Germinal center cells are known to express CD71, and most have the same mean fluorescence intensity as this new subpopulation. These cells were routinely found in the tonsil but generally were not found in peripheral blood or bone marrow (data not shown).
Figure 1.
Isolation of a novel B-cell subpopulation from tonsillar B lymphocytes. (A) IgD+CD38− B cells (lymphocyte gate not shown) are further separated into Bm1 (small CD23−) and Bm2 (small CD23+) naive B-cell subpopulations along with a novel subpopulation (large CD23−) using CD23 expression and forward scatter (FSC). (B) CD69, CD71, and AID expression is shown for each IgD+CD38− subpopulation as well as germinal center and memory B cells. Bars are set to indicate the mean fluorescence intensity range at which populations were considered positive. (C) Immunoprecipitation and Western blot were performed on 100 000 cells each from naive (Bm1 and Bm2), pro-GC, pre-GC (IgD+CD38+), and GC (Bm3 and Bm4) subpopulations with polyclonal (IP) and monoclonal (blot) antibodies to AID, which migrates at 27 kDa.
Phenotypic analysis of a novel B-cell subpopulation
In order to further characterize these cells, we examined their surface phenotype (Table 1). By definition, they are IgD+ (like naive and pre-GC cells but unlike GC cells) CD38− (like naive and traditional memory cells but unlike pre-GC cells), CD23− (unlike Bm2), and CD71+ (like pre-GC and GC cells). In addition, we found that similar to Bm1 and Bm2 cells, they were CD10−. As in GC cells, CD95 (Fas ligand) was expressed along with variable expression of CD77 and CD5. Unlike memory B cells, IgD+CD38−CD23−FSChiCD71+ cells were CD27−. These data distinguish this population from previously characterized human B cells.
Table 1.
Phenotype of 5 tonsillar B-cell subpopulations
| Marker | Population |
||||
|---|---|---|---|---|---|
| Naive | Pro-GC | Pre-GC | GC | Memory | |
| CD5 | − | −/+ | − | +/− | −/+ |
| CD10 | − | − | + | + | − |
| CD19 | + | + | + | + | + |
| CD20 | + | + | + | ++ | ++ |
| CD21 | +/− | + | −/+ | + | + |
| CD23 | −/+ | − | |||
| CD27 | − | − | − | − | + |
| CD38 | − | − | + | + | − |
| CD39 | − | +/− | − | − | −/+ |
| CD40 | + | ++ | + | + | + |
| CD44 | + | + | + | + | + |
| CD69 | −/+ | − | − | − | |
| CD71 | − | + | + | + | |
| CD77 | − | −/+ | +/− | ++/− | −/+ |
| CD80 | + | +++ | ++ | ++ | + |
| CD86 | + | ++ | +++ | ++ | + |
| CD95 | − | + | + | + | − |
| HLA-DR | + | + | + | ++ | + |
| IgD | + | + | + | − | − |
| IgM | −/+ | −/+ | −/+ | − | − |
Relative MFIs of different levels are indicated by multiple + signs. The order of + or − signs indicates predominant population.
IgD+CD38−CD23−FSChiCD71+ cells have Ig transcripts that are minimally mutated
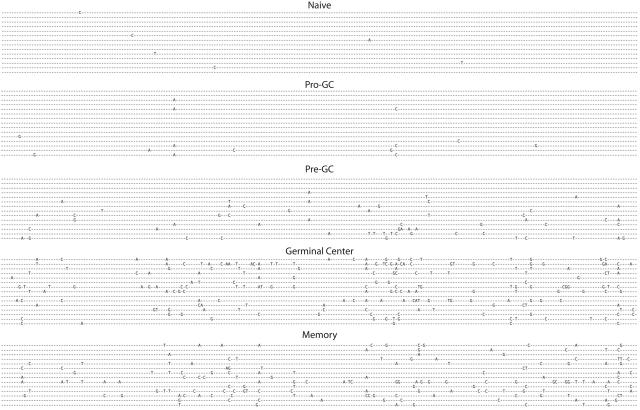
Having isolated an apparently unique population of human B cells from what would previously have been considered a “naive” pool, we sequenced 35 unique transcripts from these cells and compared them with naive, pre-GC (IgD+IgM+CD38−), germinal center, and memory cells all from tonsil tissue as shown in Figure 2 and Table 2. The sine qua non of a naive B cell is the absence of somatic mutation, however this new cell population had significantly greater numbers of mutations per transcript (0.4 versus 1.0), yet fewer than pre-GC cells, which average 3.9 mutations per sequence. This is, of course, substantially lower than the level of mutations present in germinal center cells, which average 14.7 mutations per transcript. IgD+CD38−CD23−FSChiCD71+ cells had a replacement-to-silent ratio of 1.9 to 1, in line with all other mature B-cell populations that are mutating, with the exception of GC cells, where it is substantially higher (3.9).
Figure 2.
Somatic hypermutation is present in pro-GC cells. VH regions from immunoglobulin sequences are shown from naive, pro-GC, pre-GC, germinal center, and memory cells. VH4-34 gene segment transcripts are shown. All sequences except for those from GC and memory subpopulations were IgM+. Otherwise, IgG was used.
Table 2.
Somatic hypermutation in B-cell subpopulations
| Name | Average SHM/sequence | R/S ratio |
|---|---|---|
| Naive*† | 0.7 | 1.8 |
| Naive without marginal zone and pro-GC cells†‡ | 0.4 | 1 |
| Pro-GC | 1.0 | 1.9 |
| Pre-GC‡ | 3.9 | 1.9 |
| Germinal center‡ | 14.7 | 3.9 |
| Memory‡ | 14.3 | 1.8 |
Here, naive cells represent a population contaminated by marginal zone T-independent B cells (IgM+IgD+CD27+CD38−) and pro-GC cells artificially increasing the mutation rate and R/S ratio.
Top 10% of mutated cells representing marginal zone and pro-GC cells have been removed.
‡Sequences from Zheng et al.3
IgD+CD38−CD23−FSChi cells are primarily in the G1 phase of the cell cycle
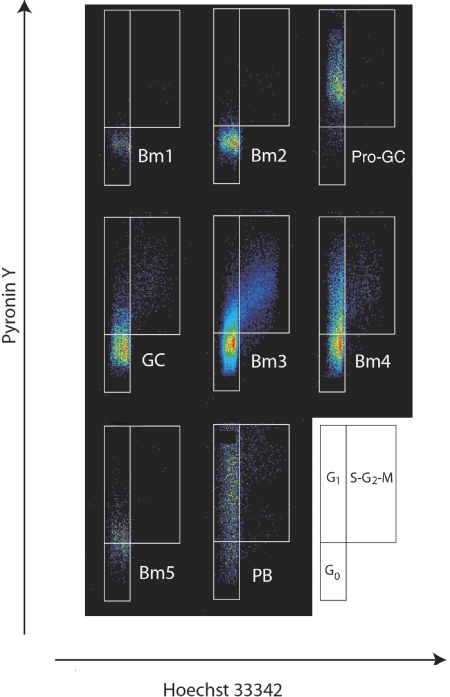
Next, we addressed the cell cycle stage of this new population of cells in order to understand their dynamics. Using flow cytometry, we collected cells from each major subpopulation (Bm1, Bm2, Bm3, Bm4, Bm5, and plasmablasts) in addition to IgD+CD38−CD23−FSChi cells. Consistent with their CD71 expression, an analysis of DNA/RNA content revealed that a majority of pro-GC tonsil cells are in a G1 state at any given time. They were then fixed and stained with pyronin Y and Hoechst 33342 dye18–20 (Figure 3). To our knowledge, this technique has not been previously reported for tonsillar B-cell subpopulations. Both Bm1 and Bm2 subpopulations were primarily in the inactive G0 fraction (92% and 94%, respectively), whereas the germinal center cell subpopulation Bm4 was primarily in the early and late G1 stages (45%; for total GC subpopulations was 23%) with some cells in the S-G2-M stage (9%). The Bm3 population had the greatest percentage of cells in the S-G2-M stage (10%) with a noticeable paucity of late G1 cells indicating a relatively fast cell cycle kinetic. Memory B cells were evenly divided between G0 and G1 stages of the cell cycle. Unlike the naive B-cell subpopulations, this new subpopulation of cells was almost entirely (83%) in the G1 stage of the cell cycle (primarily early G1).
Figure 3.
Cell cycle analysis of human B-cell subpopulations reveals IgD+CD38−CD23−FSChi (CD71+) cells are in G1 phase of the cell cycle. B-cell subpopulations were separated as described previously1,2 with the addition of IgD+CD38−CD23−FSChi cells described here. Cell cycle status was determined by flow cytometry of Hoechst 33342– and pyronin Y–stained populations. The diagram at the side illustrates how actively dividing cells can be resolved on the basis of RNA/DNA staining into G1 or S + G2 + M fractions.
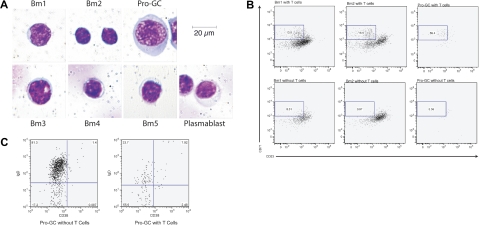
IgD+CD38−CD23−FSChiCD71+ cells are larger and distinctive morphologically
The morphology of these cells in cytospins also distinguishes them among B-cell subpopulations (Figure 4A) in the tonsil. They are large (> 30 μm diameter) with significant amounts of homogenously stained cytoplasm, perinuclear clearing, and sizeable nuclei. Comparatively, naive B cells are small (10-20 μm diameter) with little cytoplasm and a compact nucleus. Germinal center cells are slightly larger (17-25 μm diameter) than naive B cells and generally have more cytoplasm and a slightly larger nucleus.
Figure 4.
Morphology of IgD+CD38−CD23−CD71+ cells is unique among B-cell subpopulation and is generated by T-cell coculture using naive B cells. (A) B-cell subpopulations were prepared by cytospin and Giemsa staining. Cells are magnified 800×. (B) Bm1, Bm2, and IgD+CD38−CD23−CD71+ cells were cultured with cross-linking anti-IgM and with or without OKT3-stimulated, irradiated T cells. CD71+CD23− cells were identified after 7 days. Results are representative of 5 experiments. (C) IgD+CD38−CD23−CD71+ cells from the same cocultures in panel B were evaluated for the loss of IgD expression. Percents represent the percent of cells within the given dot plot.
Coculture of IgD+CD38−CD23−FSChiCD71+ cells with T cells results in the loss of naive and gain of germinal center markers
We used B-cell–T-cell coculture conditions that included irradiated T cells stimulated with OKT3 along with anti-IgM for one week to stimulate IgD+CD38−CD71lo Bm1 and Bm2 cells to produce cells with the same phenotype as pro-GC cells (13.9%-16.9%) compared with the same conditions without T cells (8.51%-0.97%). Sorted pro-GC cells required T cells to maintain their phenotype (59.4% vs 5.56% without T cells after one week in culture) (Figure 4B). Under the same culture conditions, IgD+CD38−CD23−CD71+ (pro-GC) cells also produced the same germinal center–like phenotype (55.9% GC-like compared with 17.2% GC-like without T cells %) (Figure 4C).
The novel B-cell population expresses AID
We used a polyclonal antibody to AID in order to immunoprecipitate the protein and used a monoclonal antibody to identify AID in both nuclear and cytoplasmic fractions by Western blot (Figure 1C). AID migrated at 27 kDa. Naive cells lacked AID expression in both nucleus and cytoplasm. Germinal center cells, however, express AID in both compartments but primarily express AID in the nucleus. Pre-GC cells have a predominance of cytoplasmic expression. The expression of AID in IgD+CD38−CD23−CD71+ cells is the same between cytoplasmic and nuclear compartments. Flow cytometry using a polyclonal antibody showed that approximately 60% to 70% of IgD+CD38−CD23−CD71+ cells were AID+ by an MFI equivalent to germinal center but not naive cells (Figure 1B).
In most cases, only 1 to 2 IgD+CD38−CD23−FSChiCD71+ cells are found in the germinal center
Finally, we wanted to know the distribution of this novel population of cells within tonsillar lymphoid tissue. Because they express AID, we were able to use 4-color immunohistochemistry to identify these cells using IgD (+), CD38 (−), and AID (+) and DAPI (Figure 5A). Using this combination of antibodies, we could also distinguish naive B cells (IgD+CD38−AID−), germinal center B cells (IgD−CD38+AID+), and pre–germinal center cells (IgD+CD38+AID+). About half (48.6%) of germinal centers contained at least one IgD+CD38−AID+ cell. We found that 72.4% of IgD+CD38−AID+ cells were solitary and found within the germinal center (usually small ones) itself or in an interfollicular region but not in the follicular mantle.
Figure 5.
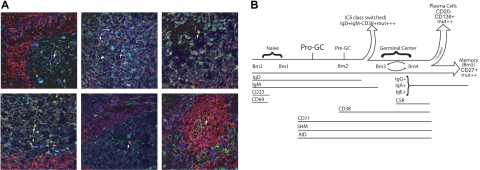
Pro-GC cells that are located in the germinal center. (A) Anti-IgD (red), anti-AID (green), and anti-CD38 (dark blue) were used to stain frozen tonsillar sections. Lymphoid follicles were identified as naive (red), germinal center (green), pre–germinal center (cyan appearance from combination of red, green, and dark blue), and IgD+CD38−AID+ (yellow appearance from combination of red and green). IgD+CD38−AID+ cells are labeled by a white arrow. Six representative sections are shown. (B) Pro-GC cells most likely exist as a transitory population between naive and germinal center cells because of their cell surface and molecular characteristics described here. We propose that pro-GC cells lie at a critical junction in which AID expression and somatic hypermutation (SHM) begin to take place but the cell has not yet matured with a full complement of germinal center surface markers. Because of cell size and the dynamics of CD69 and CD23 expression in the tonsil subpopulations, we place Bm2 prior to Bm1 in this diagram.
Discussion
Over the last decade, our laboratory has sequenced more than 4000 VH transcripts from the B cells of tonsil and peripheral blood and have found consistently that, although the vast majority of naive B cells are completely free of somatic hypermutation, there were some sequences in every pool that were mutated (Table 2). We initially considered these cells contaminated, but were perplexed by the consistency of these findings regardless of the hands of the experimenter. We concluded that there must be another subpopulation of B cells within the IgD+CD38− (naive) population.
Here, we identify a novel tonsillar B-cell subpopulation that has previously not been distinguished from the naive B-cell pool that explains these findings. While these cells share many cell surface markers with naive B cells (IgD+CD38−CD23+/−), they are distinguished by the presence of the activation marker CD71 and are in the early G1 phase of the cell cycle, as opposed to naive B cells, which are exclusively in G0. Additionally, they are larger and, unlike naive B cells, express AID and are just beginning to undergo somatic hypermutation. They do not fulfill the criteria for germinal center cells since they do not share a full complement of late activation surface markers characteristic of Bm3 and Bm4 cells (especially CD38). Consequently, the identification of these cells, which seem to be intermediate between naive and GC cells (Figure 5B), represents an important advance in our understanding of the dynamics of tonsillar B cells and the germinal center reaction.
We isolated IgD+CD38−CD23−CD71+ cells from several patients and, although we found a range in representation among B cells of the tonsil, it averaged close to 0.2%. While this subpopulation of cells is variable among individuals, the characteristics (activated cells with a naive phenotype in the G1 phase of the cell cycle that are unmutated but express AID) were the same. While memory B cells are being recognized as more phenotypically diverse than previously thought,21 this novel subpopulation does not seem to belong to a subset of cells that resembles a memory B cell. Memory B cells are typically highly mutated, often class switched, are usually CD27+,22 and are almost always in the G0 as well as G1 phase of the cell cycle. IgD+CD38−CD23−CD71+ cells do not share these characteristics. Somatic hypermutation in the naive B-cell population can now be attributed to at least 2 previously unrecognized populations that share the IgD+CD38− distinction. One previously described by others are mutated marginal zone B cells that are T-cell independent,4,23 which we suspect to have a unique pattern of mutation in adults,24 and the new population of B cells described here. The removal of these 2 “contaminating” populations is illustrated in the changes to the somatic hypermutation profile of naive cells in Table 2. Somatic hypermutations must be distinguished from PCR error by using calculations of background mutation rates from the constant region sequence obtained with the V region (this corrects for enzyme-specific errors). Taking these corrections into account, pro-GC cells (IgD+CD38−CD23−CD71+ cells) had more mutations than naive B cells, and, in terms of mutation frequency, place them between “true naive” cells and pre-GC cells (Table 2). In addition, replacement-to-silent mutation ratios also aids in distinguishing between PCR error and true somatic hypermutation, since affinity maturation selects preferentially for mutations that change amino acid sequences. Hence, an R/S ratio of 2 replacements to one silent mutation (1.9 for pro-GC) indicates mutations that have been selected as opposed to a ratio of 1 for naive B cells. Thus, these data argue that pro-GC cells are actively mutating (albeit modestly) and are being selected.
We detected AID by Western blot and flow cytometry. We used both monoclonal and polyclonal antibodies and repeatedly detected AID in IgD+CD38−CD23−CD71+ cells and germinal center whole-cell lysates but not in naive B cells. In IgD+CD38−CD23−CD71+ cells, AID expression was the same in cytoplasmic and nuclear compartments. A nuclear export domain is required for AID to shuttle between the nucleus and cytoplasm.25–27 The cleavage of this domain results in the retention of AID within the nucleus and hypermutation of non-Ig targets, but its retention does not prevent somatic hypermutation of Ig genes, it only precludes class-switch recombination.25,26 Since AID expression in the nucleus and cytoplasm of IgD+CD38−CD23−CD71+ cells was nearly identical, the AID protein is likely to traffic between both compartments. This implies that even though the effects of the AID protein are not yet fully manifest as measured by explosive somatic hypermutation, large amounts of AID are present in the developing B cell for the first time. Taken together, we conclude that IgD+CD38−CD23−CD71+ cells are just beginning to express AID and as such should prove useful for the study of the transition of mature naive B cells into true germinal center cells.
IgD+CD38−CD23−CD71+ cells have the phenotypic and molecular characteristics that place them between naive and germinal center cells. Indeed, IgD+CD38−CD23−CD71+ cells are produced in culture from naive B cells and can be stimulated to lose IgD much like true germinal center cells. They are also very modestly somatically hypermutated with an R/S ratio reflecting the beginnings of selection for amino acid changes (these mutations are part of the process of affinity maturation). Functionally, the expression of AID places them at either a stage within the mantle zone about to enter the germinal center or within the germinal center itself. Immunohistochemistry reveals that they (and usually only one cell) are present in the germinal center. Otherwise, individual cells were occasionally located in the interfollicular regions. These observations suggest that IgD+CD38−CD23−CD71+ cells are a candidate to be the founder B cell of the germinal center. While clearly speculative, we propose a model in which a naive B cell is exposed to antigen and, if its receptor is compatible and it receives appropriate T-cell help in the interfollicular region, that it is activated and begins to proliferate and express AID. This cell (we favor the name “pro-GC” cell, indicating its position as a cell prior to pre-GC cells and GC cells) then quickly acquires a germinal center phenotype and the process of affinity maturation begins with other interfollicular cells being pushed to the side in part forming an initial mantle zone. As cells from a maturing mantle zone are found that will recognize the key antigen in that particular germinal center, they are “picked off” and activated, producing more cells with this pro-GC phenotype. The rarity of these cells is perhaps due to their specific phenotype that represents a snapshot of cells in a continuing process but is no less valuable as a way to specifically target the first B-cell subpopulation to express AID.
The division of B-cell subpopulations in peripheral lymphoid tissue has been critical for our understanding of human B-cell malignancies. Prior to our understanding of the dynamics of B-cell subpopulations in tissues such as the lymph node and spleen, the classification of B-cell malignancies was based largely on morphology, location, and in some cases cytogenetics. Since gaining the ability to separate normal human lymphoid tissue into distinct subpopulations and correlate morphology, phenotype, and molecular information about normal cells,1,2,28–30 our understanding of and information about the molecular biology of human B-cell malignancies has grown significantly. The normal counterparts of a large number of leukemias and lymphomas have been described (reviewed in Pascual et al31) but continue to grow as new B-cell subsets are found and as new molecular data refine our understanding of the origin of these cancers.
Chronic lymphocytic leukemia cells contain Ig transcripts that can be mutated or unmutated32–34 and have also been found to variably express AID.35 Similarly, there are reports of both unmutated and, more rarely, mutated forms of mantle cell lymphoma (MCL).36,37 It has not escaped our attention that this B-cell malignancy shares many characteristics to the pro-GC cells described here. Phenotypically, they and MCL cells are IgM+IgD+CD27−CD23− and CD10−.38 While most MCLs are CD5+, there are reports of some that are CD5−39 yet retain the same gene expression profile as those that have this marker. A small number of IgD+CD38−CD23−FSChi cells are CD5+, while most are CD5−. More of the pro-GC cells are CD5+ than either Bm1 or Bm2 populations, and as many as 50% of GC cells were positive for CD5. MCL might simply arise from a subset of pro-GC cells or acquire CD5 immediately subsequent to transformation.
It is of interest that some MCLs express AID.37 While it is well known that a translocation (t(11:14)) of PRAD-1/cyclin D1 and the IgH locus results in constitutive expression of this cell cycle regulator resulting in the pathology of the disease,40,41 the stage at which this expression becomes critical is not yet clear. IgD+CD38−CD23−CD71+ cells may represent a stage in peripheral B-cell development when the initiation of large amounts of transcription of the IgH locus first occurs in the periphery. Increased transcription would result in the disregulation of proliferation and the perpetuation of MCL. These striking similarities (cell surface phenotype, somatic hypermutation, and AID expression) between IgD+CD38−CD23−CD71+ cells and MCL characteristics lead us to speculate that this novel population is a strong candidate for the normal cell counterpart of this lymphoma.
We describe a novel population of B cells that appears to represent cells at the onset of activation that expresses AID while not yet showing evidence of extensive somatic hypermutation (a pro-GC cell). This population may serve as a window into the beginnings of the germinal center reaction and provides a population of cells to target for the further study of expression and regulation of AID. In addition, this population can be used for the identification of immunoglobulin clones that are selected initially in response to a particular pathogen. IgD+CD38−CD23−CD71+ cells are therefore not only a valuable population to study in order to understand the basic process of antigen selection and the onset of somatic hypermutation, but also may serve to be clinically important in furthering our understanding of human B-cell malignancies.
Acknowledgments
This work was supported by COBRE grant number SP20 RR015577-07.
We thank Dr John Houck Jr for his help in acquiring the tissue needed for this study. We thank Viji Dandipani and Jacob Bass of the OMRF Cytometry Facility as well as the technical support and use of the OMRF Imaging Core. In addition, we thank Sheryl Christopherson in the OMRF sequencing core facility and Dr Patrick C. Wilson for the use of his database for somatic hypermutation comparisons. We especially thank Gina Yosten for her technical assistance.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: G.R.K. wrote the paper and designed and performed the experiments included; D.M. performed experiments; R.P. performed experiments for Figure 4; J.D.C. designed experiments, wrote the paper, and provided financial support as principal investigator for this paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: J. Donald Capra, Oklahoma Medical Research Foundation, 825 NE 13th St, Oklahoma City, OK 73104; e-mail: donald-capra@omrf.ouhsc.edu.
References
- 1.Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. J Exp Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu YJ, de Bouteiller O, Arpin C, Durand I, Banchereau J. Five human mature B cell subsets. Adv Exp Med Biol. 1994;355:289–296. doi: 10.1007/978-1-4615-2492-2_49. [DOI] [PubMed] [Google Scholar]
- 3.Zheng NY, Wilson K, Wang X, et al. Human immunoglobulin selection associated with class-switch and possible tolerogenic origins for Cδ class-switched B cells. J Clin Invest. 2004;113:1188–1201. doi: 10.1172/JCI20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weller S, Faili A, Garcia C, et al. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc Natl Acad Sci U S A. 2001;98:1166–1170. doi: 10.1073/pnas.98.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 6.Yoshikawa K, Okazaki IM, Eto T, et al. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 7.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 8.Nagaoka H, Muramatsu M, Yamamura N, Kinoshita K, Honjo T. Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Smu region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J Exp Med. 2002;195:529–534. doi: 10.1084/jem.20012144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okazaki IM, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T. The AID enzyme induces class switch recombination in fibroblasts. Nature. 2002;416:340–345. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- 10.Greeve J, Philipsen A, Krause K, et al. Expression of activation-induced cytidine deaminase in human B-cell non-Hodgkin lymphomas. Blood. 2003;101:3574–3580. doi: 10.1182/blood-2002-08-2424. [DOI] [PubMed] [Google Scholar]
- 11.Smit LA, Bende RJ, Aten J, Guikema JE, Aarts WM, van Noesel CJ. Expression of activation-induced cytidine deaminase is confined to B-cell non-Hodgkin's lymphomas of germinal-center phenotype. Cancer Res. 2003;63:3894–3898. [PubMed] [Google Scholar]
- 12.Honjo T. Does AID need another aid? Nat Immunol. 2002;3:800–801. doi: 10.1038/ni0902-800. [DOI] [PubMed] [Google Scholar]
- 13.Faili A, Aoufouchi S, Gueranger Q, et al. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 14.Reynaud CA, Aoufouchi S, Faili A, Weill JC. What role for AID: mutator, or assembler of the immunoglobulin mutasome? Nat Immunol. 2003;4:631–638. doi: 10.1038/ni0703-631. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Feng J, Komori A, Kim EC, Zan H, Casali P. Immunoglobulin somatic hypermutation: double-strand DNA breaks, AID and error-prone DNA repair. J Clin Immunol. 2003;23:235–246. doi: 10.1023/a:1024571714867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolar GR, Yokota T, Rossi MI, Nath SK, Capra JD. Human fetal, cord blood, and adult lymphocyte progenitors have similar potential for generating B cells with a diverse immunoglobulin repertoire. Blood. 2004;104:2981–2987. doi: 10.1182/blood-2003-11-3961. [DOI] [PubMed] [Google Scholar]
- 18.Darzynkiewicz Z, Kapuscinski J, Traganos F, Crissman HA. Application of pyronin Y(G) in cytochemistry of nucleic acids. Cytometry. 1987;8:138–145. doi: 10.1002/cyto.990080206. [DOI] [PubMed] [Google Scholar]
- 19.Gothot A, Pyatt R, McMahel J, Rice S, Srour EF. Functional heterogeneity of human CD34(+) cells isolated in subcompartments of the G0/G1 phase of the cell cycle. Blood. 1997;90:4384–4393. [PubMed] [Google Scholar]
- 20.Crissman HA, Darzynkiewicz Z, Tobey RA, Steinkamp JA. Correlated measurements of DNA, RNA, and protein in individual cells by flow cytometry. Science. 1985;228:1321–1324. doi: 10.1126/science.2408339. [DOI] [PubMed] [Google Scholar]
- 21.Ehrhardt GR, Hsu JT, Gartland L, et al. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J Exp Med. 2005;202:783–791. doi: 10.1084/jem.20050879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weller S, Braun MC, Tan BK, et al. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104:3647–3654. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolar GR, Mehta D, Wilson PC, Capra JD. Diversity of the Ig repertoire is maintained with age in spite of reduced germinal centre cells in human tonsil lymphoid tissue. Scand J Immunol. 2006;64:314–324. doi: 10.1111/j.1365-3083.2006.01817.x. [DOI] [PubMed] [Google Scholar]
- 25.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–26401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- 27.Ito S, Nagaoka H, Shinkura R, et al. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frazer JK, LeGros J, de BO, et al. Identification and cloning of genes expressed by human tonsillar B lymphocyte subsets. 1997;815:316–318. doi: 10.1111/j.1749-6632.1997.tb52073.x. [DOI] [PubMed] [Google Scholar]
- 29.Liu YJ, Malisan F, de Bouteiller O, et al. Within germinal centers, isotype switching of immunoglobulin genes occurs after the onset of somatic mutation. Immunity. 1996;4:241–250. doi: 10.1016/s1074-7613(00)80432-x. [DOI] [PubMed] [Google Scholar]
- 30.Liu YJ, Banchereau J. Human peripheral B cell subsets. In: Weir D, Blackwell C, Herzenberg L, editors. Handbook of Experimental Immunology. Oxford, United Kingdom: Blackwell Scientific Publishing; 1996. [Google Scholar]
- 31.Pascual V, Liu YJ, Banchereau J. Normal human B cell sub-populations and their malignant counterparts. Bailleres Clin Haematol. 1997;10:525–538. doi: 10.1016/s0950-3536(97)80024-8. [DOI] [PubMed] [Google Scholar]
- 32.Gurrieri C, McGuire P, Zan H, et al. Chronic lymphocytic leukemia B cells can undergo somatic hypermutation and intraclonal immunoglobulin V(H)DJ(H) gene diversification. J Exp Med. 2002;196:629–639. doi: 10.1084/jem.20011693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naylor M, Capra JD. Mutational status of Ig V(H) genes provides clinically valuable information in B-cell chronic lymphocytic leukemia. Blood. 1999;94:1837–1839. [PubMed] [Google Scholar]
- 34.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 35.McCarthy H, Wierda WG, Barron LL, et al. High expression of activation-induced cytidine deaminase (AID) and splice variants is a distinctive feature of poor-prognosis chronic lymphocytic leukemia. Blood. 2003;101:4903–4908. doi: 10.1182/blood-2002-09-2906. [DOI] [PubMed] [Google Scholar]
- 36.Thorselius M, Walsh S, Eriksson I, et al. Somatic hypermutation and V(H) gene usage in mantle cell lymphoma. Eur J Haematol. 2002;68:217–224. doi: 10.1034/j.1600-0609.2002.01662.x. [DOI] [PubMed] [Google Scholar]
- 37.Babbage G, Garand R, Robillard N, Zojer N, Stevenson FK, Sahota SS. Mantle cell lymphoma with t(11;14) and unmutated or mutated VH genes expresses AID and undergoes isotype switch events. Blood. 2004;103:2795–2798. doi: 10.1182/blood-2003-05-1632. [DOI] [PubMed] [Google Scholar]
- 38.Dorfman DM, Pinkus GS. Distinction between small lymphocytic and mantle cell lymphoma by immunoreactivity for CD23. Mod Pathol. 1994;7:326–331. [PubMed] [Google Scholar]
- 39.Liu Z, Dong HY, Gorczyca W, et al. CD5- mantle cell lymphoma. Am J Clin Pathol. 2002;118:216–224. doi: 10.1309/TE56-A43X-29TT-5H8G. [DOI] [PubMed] [Google Scholar]
- 40.Bosch F, Jares P, Campo E, et al. PRAD-1/cyclin D1 gene overexpression in chronic lymphoproliferative disorders: a highly specific marker of mantle cell lymphoma. Blood. 1994;84:2726–2732. [PubMed] [Google Scholar]
- 41.Espinet B, Sole F, Woessner S, et al. Translocation (11;14)(q13;q32) and preferential involvement of chromosomes 1, 2, 9, 13, and 17 in mantle cell lymphoma. Cancer Genet Cytogenet. 1999;111:92–98. doi: 10.1016/s0165-4608(98)00228-3. [DOI] [PubMed] [Google Scholar]