Short abstract
Transcriptional profiling reveals a possible association between schizophrenia and altered miRNA expression
Abstract
Background
microRNAs (miRNAs) are small, noncoding RNA molecules that are now thought to regulate the expression of many mRNAs. They have been implicated in the etiology of a variety of complex diseases, including Tourette's syndrome, Fragile × syndrome, and several types of cancer.
Results
We hypothesized that schizophrenia might be associated with altered miRNA profiles. To investigate this possibility we compared the expression of 264 human miRNAs from postmortem prefrontal cortex tissue of individuals with schizophrenia (n = 13) or schizoaffective disorder (n = 2) to tissue of 21 psychiatrically unaffected individuals using a custom miRNA microarray. Allowing a 5% false discovery rate, we found that 16 miRNAs were differentially expressed in prefrontal cortex of patient subjects, with 15 expressed at lower levels (fold change 0.63 to 0.89) and 1 at a higher level (fold change 1.77) than in the psychiatrically unaffected comparison subjects. The expression levels of 12 selected miRNAs were also determined by quantitative RT-PCR in our lab. For the eight miRNAs distinguished by being expressed at lower microarray levels in schizophrenia samples versus comparison samples, seven were also expressed at lower levels with quantitative RT-PCR.
Conclusion
This study is the first to find altered miRNA profiles in postmortem prefrontal cortex from schizophrenia patients.
Background
Schizophrenia is a common neuropsychiatric disorder affecting one percent of the general population. The personal, familial, and societal costs of the disease are enormous, with chronic symptoms that result in marked functional disability. In fact, approximately three percent of all person-years lived with disability are due to schizophrenia [1].
It is clear that schizophrenia has a strong genetic component, although its genetic basis remains unknown [2]. Consistent with a disease mechanism that involves post-transcriptional dysregulation of gene expression, postmortem studies find altered levels of mRNA and proteins rather than a specific abnormal protein [3]. Postmortem studies also find differences between schizophrenia and unaffected comparison subjects in the relationship of such mRNAs and cognate proteins [4,5].
microRNAs (miRNAs) are a class of noncoding RNAs (ncRNAs) that in animals regulate gene expression by inhibiting mRNA translation. Each miRNA is initially processed from a large (approximately 200 nucleotide (nt) to several thousand nt) RNA transcript, the 'primary miRNA' (pri-miRNA) to a smaller (approximately 58-137 nt) hairpin precursor miRNA (pre-miRNA) by a protein complex, the 'microprocessor', and then by DICER1 (alias Dicer) to the mature miRNA [6]. The mature miRNA joins with the RNA-induced silencing complex (RISC), and then binds the RISC to a partially complementary target region in an mRNA to accelerate mRNA degradation or inhibit translation. Some 474 RNA hairpins (pre-miRNAs) are known to be transcribed in humans, yielding 471 distinct, mature miRNAs, and there are in addition over 800 predicted human miRNAs. The associated control systems might regulate expression of thousands of human genes [7-9]. In particular, seminal experiments have shown that miRNAs regulate a variety of key biological functions, including cell proliferation and differentiation [10-15], insulin secretion [16], and apoptosis [17]. Emerging evidence suggests that miRNAs also regulate brain development [18,19], dendritic spine morphology [20], and neurite outgrowth [21], that is, certain processes that are hypothesized to be associated with schizophrenia neuropathology.
In addition to critical regulatory roles in development and cellular functions, miRNAs have now been implicated in several human diseases [22]. For example, the etiology of some cases of Tourette's syndrome, a disorder characterized by vocal and motor tics, has been shown to be related to either the absence of or a mutation in the miR-189 target site in the 3' untranslated region (UTR) of gene SLITRK1 [23]. Fragile X syndrome, one of the most common genetic disorders affecting brain function, is characterized by deficits that range from learning disabilities in individuals with normal intelligence to severe intellectual deficits and behavioral disturbances. The genetic basis is most commonly a CGG repeat expansion in the 5' UTR of FMRP causing transcriptional silencing [24]. FMRP might regulate the translation of mRNAs through association with RISCs and miRNAs, and, in particular, might regulate translation of mRNAs locally in the dendrites [24-26].
Given the critical role that miRNAs might play in regulating brain development early in life and mediating synaptic plasticity later in life, we have hypothesized that the etiopathology of schizophrenia might be associated with altered expression or function of miRNAs [27]; the association might be causative or part of compensatory reactions to some other causative agents. As a first step we compared the expression of human miRNAs from postmortem prefrontal cortex (PFC) of individuals with schizophrenia to that of unaffected individuals.
Results
General description of prefrontal cortical miRNA expression
From the 265 distinct, human miRNAs included on our array, 244 were detected (1.5-fold over background) in the PFC tissue of ≥60% of the study subjects. These included robust detection of miRNAs previously known to be expressed in the brain (for example, let-7a to let-7i) as well as brain-specific miRNAs (for example, miR-124a and miR-125b) (Additional data file 1) [11,28].
miRNA expression in schizophrenia versus unaffected comparison subjects
Assuming a false discovery rate (FDR) of 5%, 16 miRNAs were differentially expressed in PFC of schizophrenia subjects (n = 13) or schizoaffective disorder (n = 2) versus PFC of 21 psychiatrically unaffected individuals (Table 1). Of the 16 distinguished miRNAs, 15 were expressed at lower (fold change 0.63 to 0.89) and one at higher (fold change 1.77) levels than in the psychiatrically unaffected comparison subjects. A heat map based on cluster analysis illustrates the differentiated expression levels of these probes (Figure 1). Controlling on brain pH, postmortem interval (PMI), and hemisphere, and excluding the two subjects with schizoaffective disorder from the analyses did not substantially affect these results (Additional data file 2).
Table 1.
Differentially expressed miRNAs from the prefrontal cortex of subjects with schizophrenia compared to psychiatrically healthy subjects
| miRNA | Fold change | Chromosome location(s) |
| hsa-miR-26b | 0.63 | 2q35 |
| hsa-miR-30b | 0.68 | 8q24.22 |
| hsa-miR-29b | 0.69 | 1q32.2, 7q32.3 |
| hsa-miR-195 | 0.73 | 17p13.1 |
| hsa-miR-92 | 0.76 | 13q31.3, Xq26.2 |
| hsa-miR-30a-5p | 0.79 | 6q13 |
| hsa-miR-30d | 0.80 | 8q24.22 |
| hsa-miR-20b | 0.81 | Xq26.2 |
| hsa-miR-29c | 0.82 | 1q32.2 |
| hsa-miR-29a | 0.82 | 7q32.2 |
| hsa-miR-212 | 0.82 | 17p13.3 |
| hsa-miR-106b | 1.77 | 7q22.1 |
| hsa-miR-7 | 0.70 | 9q21.32, 15q26.1, 19p13.3 |
| hsa-miR-24 | 0.79 | 9q22.32, 19p13.12 |
| hsa-miR-30e | 0.89 | 1p34.2 |
| hsa-miR-9-3p | 0.77 | 1q22, 5q14.3, 15q26.1 |
Figure 1.

An miRNA expression map shows differentiated genes as determined by SAM analysis. Yellow indicates low expression and blue indicates high expression, relative to the median.
Quantitative RT-PCR verification of microarray results
The expression levels of 12 selected miRNAs were also determined by quantitative RT-PCR (qRT-PCR) in our lab (Additional data file 3). For the eight miRNAs distinguished by being expressed at lower microarray levels in schizophrenia samples versus comparison samples, seven were also expressed at lower levels with qRT-PCR (Figure 2). For four of the seven, the difference in expression was significant with p < 0.05, consistent with microarray findings for the same miRNAs. The eighth miRNA, miR-7, was found by qRT-PCR to have higher levels in schizophrenia than comparison subjects, but the difference in expression was not significantly different between groups (p = 0.23); we have not determined a cause for this one discrepancy of PCR versus microarray results. We also compared expression of four miRNAs that were not differentially expressed in the microarray results, and found none to be differentially expressed by qRT-PCR (p > 0.05).
Figure 2.

miRNA microarray fold changes can be compared with delta-delta C(t) functions of qRT-PCR data (see Materials and methods). The comparisons are over four samples from schizophrenia patients and four samples from psychiatrically unaffected comparison subjects. Seven of the eight comparisons are consistent.
Effect of haloperidol exposure on miRNA expression
Since all of the schizophrenia subjects were treated or had previously been treated with antipsychotics and none of the psychiatrically unaffected subjects were reported to have such a treatment history, we endeavored to evaluate the effect of antipsychotic treatment on miRNA expression. We compared expression of 179 rat miRNAs in haloperidol-treated and -untreated rats. With a FDR of 5% we found that three miRNAs were expressed at higher levels in the haloperidol-treated rats: miR-199a, miR-128a, and miR-128b. None of these miRNAs was differentially expressed in the PFC of schizophrenia patients (Additional data file 4).
miRNA and Affymetrix U133A probe relationships
We considered whether the observed pattern of lower expression of some miRNAs in schizophrenia subjects was related to lower pri-miRNA transcription. We adopted the previously published method of Thomson and colleagues [29], where the pri-miRNA expression was determined from existing archived mRNA microarray results from the PFC of the same study subjects. A total of 52 of the miRNAs included in this study could be mapped to a primary transcript that was present among the mRNA transcripts accessible with the Affymetrix U133A array. All but three of the miRNAs with corresponding U133A probes were from the introns of protein-coding genes (host genes). The mean expression of only two of the Affymetrix U133A probes was significantly different between groups (ELM2 hosting miR-330 with p = 0.03; MYH6 hosting miR-208 with p = 0.03). However, these differences were not significant after correction for multiple comparisons (p > 0.05).
We then focused on the five miRNAs expressed at significantly lower levels in schizophrenia that also had a U133A probe that included the pri-miRNA transcript (miR-26b, miR-9-3p (alias miR-9*), miR-24, miR-7, and miR-30e). The ratio of mature miRNA to primary miRNA transcripts was lower for schizophrenia versus controls for all 5 miRNAs, and the difference in ratios reached statistical significance for 3 of the 5 (miR-26b, p = 0.009; miR-9-3p, p = 0.002; and miR-24, p = 0.037). For the one miRNA that was expressed at a significantly higher level in schizophrenia subjects, miR-106b, the ratio was also significantly higher (p = 0.003 and p = 0.006 for the two associated Affymetrix pri-miRNA probes). In the remaining 46 miRNAs with a corresponding Affymetrix U133A probe for their pri-miRNA transcripts, the ratio of miRNA to host mRNA was significantly lower for two pri-miRNAs (primary transcripts for miR-218, p = 0.021, and miR-9, p = 0.006) and significantly higher for five (miR-482, p = 0.015; miR-190, p = 0.018; miR-105, p = 0.02; miR-148b, p = 0.027; miR-218, p = 0.02). Thus, we found that the miRNA:U133A probe ratios of the schizophrenia group were significantly different from those of the comparison group for 4 of the 6 differentiated miRNAs but only 7 of the 46 nondifferentiated miRNAs (p = 0.013, Fisher's exact test) (Additional data file 5).
Common motifs near the pre-miRNA:pri-miRNA junction
We hypothesized that the system regulating processing of the pri-miRNA to pre-miRNA might involve a motif within the pri-miRNA and upstream of the single-stranded RNA (ssRNA)-double-stranded RNA (dsRNA) junction that would lend selectivity to this process. Specifically, we hypothesized that an upstream motif of some kind is shared by the 15 miRNAs that were found to be downregulated in our tests of schizophrenia PFC samples. To seek bioinformatic indications, we focused on source pre-miRNAs that were isolated (no other pre-miRNAs within 1,000 bases), yielding 11 distinguished, isolated pre-miRNAs: miR-7-1, miR-7-2, miR-7-3, miR-9-1, miR-9-2, miR-9-3, miR-26b, miR-30a, miR-30b, miR-30d, miR-30e. Of these, miR-9-1 and miR-30a can yield two mature miRNAs; the others yield one. Furthermore, miR-7-2, miR-9-2, miR-9-3, miR-30b, and miR-30d are intergenic, and the others are intronic in coding genes.
Using a combination of approaches, we found the motif UGAGNCUU upstream of pre-miRNA sequences for miR-26b, miR-30a, miR-30b, and miR-7-1. We also found GUCNCUUC upstream of pre-miRNAs miR-9-1, miR-9-2, miR-9-3, miR-7-3, and miR-30e. Thus, both 8 nt motifs are found upstream of 9 of the 11 isolated, distinguished pre-miRNAs. Lastly, instances of UGUUNNAAGAUG were found upstream of pre-miRNAs for miR-30d and miR-7-2 at the same distance, 108 bases, and not within 500 bases upstream of any other human, isolated pre-miRNAs. For displays of the motifs and the bases between motifs and junctions, see Additional data file 6; clustering of the number of bases in each such interval is displayed in Figure 3. Bioinformatic searches by us have found neither shared motifs that are positioned at similarly clustered distances from the junctions nor strong general homology among the 11 upstream regions.
Figure 3.
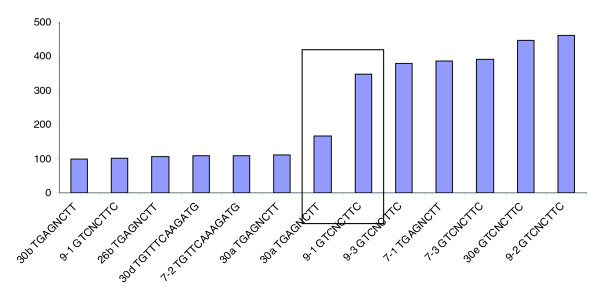
Regarding the 11 isolated miRNAs distinguished in schizophrenia, this figure shows the distances (numbers of bases) from shared 5' motifs we discovered (two 8 nt and two 12 nt motif sequences in the pri-miRNA) to the ssRNA-dsRNA junctions at starts of pre-miRNAs. Pre-miR-30a and -9-1 have double motif instances; second instances are in the rectangle. Ignoring the second instances as redundant leaves some motif distances in sharp clusters.
Importantly, the same 8 nt motifs UGAGNCUU and GUCNCUUC are absent from the 500-base 5' regions of most undistinguished pre-miRNAs. That is, the same motifs are also upstream of only 13 isolated, undistinguished pre-miRNAs among a total of 192 isolated, human pre-miRNAs, and some of the 13 are sequentially similar as mature miRNAs to the 11 distinguished ones. However, carefully designed and executed in vivo experiments would be needed to determine whether the above or any other motifs are actually functional; the above motifs are intriguing, but their bioinformatic properties are certainly not a proof of common regulation of coordinated pre-miRNA excision.
Discussion
miRNAs, with their key roles in regulating both synaptic plasticity and brain development, are candidate genetic contributors to the etiopathology of schizophrenia. miRNA expression for 16 miRNAs was significantly different in the PFC of schizophrenia versus comparison subjects, with all but one of the differentiated miRNAs decreased in the schizophrenia subjects. To our knowledge this study is the first to associate altered expression of miRNAs with schizophrenia. Possibly the association is etiologic, but it could also be part of a complex response to other factors.
A hypothesized role for altered miRNA biogenesis
Our follow-up analyses were designed to generate hypotheses about possible mechanisms that could explain the downregulation of miRNAs reported in this study. For miRNAs hosted in introns of coding genes, we found that the ratios of microarray expression levels of miRNA versus mRNA (of host gene) were significantly different for miRNA distinguished by schizophrenia. That is, 4 of the 6 hosted, distinguished miRNAs exhibited the difference, but only 7 of the 46 undistinguished miRNAs did so. This suggests a role for altered biogenesis of miRNAs rather than altered transcription of pri-miRNAs. In addition, our bioinformatic investigations found 2 common motifs located at approximately 100 or approximately 400 bases from the pri-miRNA:pre-miRNA junction in 9 of the 11 isolated, distinguished miRNAs; but the same motifs are absent in almost all of the undistinguished miRNAs. We speculate (see Figure 4) that these motifs might represent binding sites for factors like heteronuclear ribonuclear proteins (hnRNPs) [30], known to chaperone other RNA events.
Figure 4.
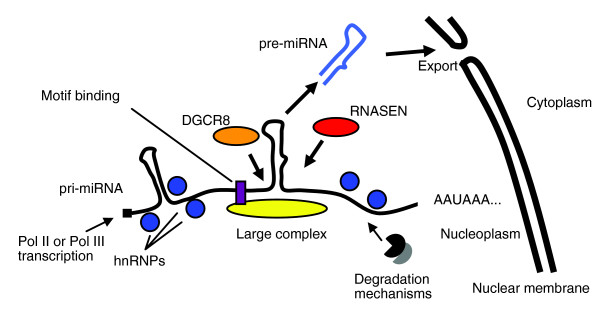
Transcription yields a continuous supply of some types of pri-miRNA transcripts, capped and polyadenylated. hnRNPs are hypothesized to shape the pri-miRNA into linear and hairpin sections. A signaling system somehow recruits and activates unknown factors that select particular pre-miRNA hairpins on a particular pri-miRNA for excision and processing in the miRNA pathway. We hypothesize that this system might include a binding motif. RNASEN and DGCR8 are products of genes 29102 and 54487.
The bioinformatic similarities involving motifs, though not yet investigated in vivo, are consistent with the hypothesis that the coordinated downregulation of 15 miRNAs reported in this study might be related to alternative processing during the pre-miRNA biogenesis process, rather than altered pri-miRNA transcription. There is evidence that, in some cases, miRNA biogenesis regulates mature miRNA levels. Thomson et al. [29] found that in mice, levels of mature miRNAs hsa-let-7g and hsa-let-7f-2/miR-98 increased over 4,000-fold in day 14.5 embryos from levels in embryonic stem cells. However, over the same developmental period the primary transcript pri-miRNA expression levels did not change, and pre-miRNA levels were essentially undetectable. Also, the same Thomson analysis indicates that the widespread downregulation of miRNAs observed in cancer [31,32] might be due to a failure in miRNA processing that is post-transcriptional (transcription of pri-miRNA). Discovery of parallel mechanisms of regulation of other sets of miRNAs, such as the 15 downregulated miRNAs in schizophrenia, would, therefore, be of considerable interest.
Further study is required to test the hypothesis that altered regulation of miRNA biogenesis might be involved in the etiopathology of schizophrenia, and whether the above motifs are involved in regulating miRNA processing from pri-miRNA to pre-miRNA.
As a final note, DiGeorge critical region 8 (DGCR8), involved in miRNA biogenesis as a component of the microprocessor, is located in a genomic region of chromosome 22q11 where microdeletions have been associated with a 30-fold increased risk of schizophrenia [33,34]. Microdeletions in 22q11 occur in approximately 1 in 3,000 live births but are present in 0.5% to 3% of individuals with schizophrenia [35,36]. Possibly, DGCR8 polymorphisms that alter expression or function through haploinsufficiency or other genetic variants might also contribute to the etiopathology of schizophrenia by impacting miRNA biogenesis and regulation of gene expression.
Common potential mRNA targets
Dysregulation of miRNA levels would be anticipated to affect the translation of multiple protein coding genes. Bioinformatic strategies are now developed to identify potential miRNA target sites in the 3' UTR of a protein coding gene, for example the program miRanda [7]. The potential targets of miRNAs often include hundreds of genes because the reverse complement of some 'seeds' (bases 2 through 8 of the mature miRNA) appears in multiple locations in many pre-mRNA 3' UTRs. However, only a few of these potential target sites have been verified as potent in vivo [37]. With the understanding that identification of mRNA targets is speculative, we explored whether there might be common mRNA targets for the 15 distinguished, downregulated miRNAs and whether these targeted genes are over-represented in any Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway through the KEGG website [38].
The differentially expressed miRNAs are currently annotated in the Memorial Sloan-Kettering Cancer Center Computational Biology Center web site. These 15 miRNAs are identified using miRanda to potentially target the 3' UTRs of over 4,600 genes, with 1,539 targeted by 2 or more of them [39]. Using the programs offered by the Database for Annotation, Visualization, and Integrated Discovery (DAVID) to identify over-represented pathways, we found that the genes that were commonly targeted by the miRNAs were significantly clustered in 12 KEGG pathways (Table 2) [40]. It is of interest that the most significantly differentiated pathways are involved in synaptic plasticity at the level of dendritic spines. For example, the MAPK and phosphatidylinositol signaling pathways are involved in the regulation of dendritic spine morphogenesis, size, and shape [41,42] and act through regulation of the actin cytoskeleton [43]. In addition, the focal adhesion pathways mediated through extracellular matrix receptor interactions have also been shown to control dendritic spine plasticity [44]. Translation of mRNA into proteins that are important to synaptic plasticity can occur locally in dendrites [45]. Thus, the miRNAs differentiated in this study might be involved in the regulation of synaptic plasticity, and in that manner associated with characteristics of synaptic plasticity in schizophrenia.
Table 2.
KEGG pathways of gene categories that are over-represented by targets of two or more miRNAs distinguished by schizophrenia
| KEGG Pathway Term | N | % | P value |
| HSA04810:REGULATION OF ACTIN CYTOSKELETON | 43 | 2.81 | 1.7E-07 |
| HSA04510:FOCAL ADHESION | 45 | 2.94 | 3.5E-07 |
| HSA04010:MAPK SIGNALING PATHWAY | 41 | 2.68 | 3.9E-05 |
| HSA04512:ECM-RECEPTOR INTERACTION | 17 | 1.11 | 0.0029 |
| HSA04070:PHOSPHATIDYLINOSITOL SIGNALING | 18 | 1.17 | 0.0076 |
| HSA04020:CALCIUM SIGNALING PATHWAY | 28 | 1.83 | 0.0093 |
| HSA00271:METHIONINE METABOLISM | 6 | 0.39 | 0.0099 |
| HSA04540:GAP JUNCTION | 16 | 1.04 | 0.0109 |
| HSA04530:TIGHT JUNCTION | 18 | 1.17 | 0.0173 |
| HSA04910:INSULIN SIGNALING PATHWAY | 21 | 1.37 | 0.0193 |
| HSA04630:JAK-STAT SIGNALING PATHWAY | 22 | 1.44 | 0.0326 |
| HSA04710:CIRCADIAN RHYTHM | 5 | 0.33 | 0.0370 |
N, number of potential target genes in pathway; %, percent of pathway genes that are targeted by differentiated miRNAs.
Conclusion
Although the functions of most human miRNAs have yet to be discovered, miRNAs have emerged as key regulators of gene expression. The findings of this study implicate a role for miRNAs in schizophrenia, and lead us to the hypothesis that there is altered processing of miRNAs during the miRNA biogenesis process in schizophrenia. This hypothesis is analogous to that for altered miRNA transcription in cancer by Thomson et al. [29].
Materials and methods
Postmortem tissue
This study was approved by the Institutional Review Board of the University of North Carolina School of Medicine. Postmortem human brain tissue was obtained from the Harvard Brain Tissue Resource Center [46]. Tissue consisted of frozen blocks (300-500 mg/block) from the PFC (Brodmann area nine from 15 individuals with schizophrenia and 21 unaffected comparison subjects (Table 3)). The tissue was group-matched for age, gender, PMI, and hemisphere. Postmortem neuropathological examinations were performed by an experienced neuropathologist, and all subjects included in the collection were free of neurodegenerative pathology. Postmortem neurotoxicological studies showed no evidence of illicit substance use at the time of death.
Table 3.
Demographics
| Subject | Age (years) | PDx | Sex | PMI | pH | Hemisphere |
| 1003 | 51-60 | Ctrl | F | 24 | 5.8 | Right |
| 1006 | 51-60 | Ctrl | M | 24.2 | 6.53 | Left |
| 1008 | 61-70 | Ctrl | F | 22.5 | 6.26 | Left |
| 1013 | 31-40 | Ctrl | M | 18.75 | 6.68 | Right |
| 1014 | 31-40 | Ctrl | M | 20 | 5.97 | Left |
| 1020 | 71-80 | Ctrl | M | 20.53 | 6.05 | Right |
| 1021 | 31-40 | Ctrl | M | 25.67 | 6.33 | Right |
| 1022 | 80+ | Ctrl | M | 7.42 | 6.39 | Right |
| 1024 | 71-80 | Ctrl | M | 20.92 | 6.74 | Left |
| 1025 | 71-80 | Ctrl | F | 23.91 | 6.67 | Right |
| 1026 | 31-40 | Ctrl | M | 28.83 | 6.53 | Left |
| 1028 | 61-70 | Ctrl | F | 24.25 | 6.4 | Right |
| 1029 | 61-70 | Ctrl | F | 7.42 | 6.03 | Right |
| 1030 | 41-50 | Ctrl | M | 18.33 | 6.78 | Left |
| 1032 | 41-50 | Ctrl | M | 24.13 | 6.01 | Left |
| 1033 | 80+ | Ctrl | M | 28.58 | 6.42 | Right |
| 1034 | 31-40 | Ctrl | M | 16.6 | 6.24 | Left |
| 1035 | 31-40 | Ctrl | M | 24.5 | 6.26 | Left |
| 1047 | 61-70 | Ctrl | M | 15.3 | 6.88 | Right |
| 1057 | 31-40 | Ctrl | M | 28 | 6.5 | Right |
| 1066 | 21-30 | Ctrl | M | 18.25 | 7.06 | Left |
| 1001 | 61-70 | SZ | M | 22.1 | 6.43 | Right |
| 1009 | 71-80 | SZ | F | 24 | 6.08 | Right |
| 1016 | 61-70 | SZ | M | 22.35 | 6.55 | Right |
| 1036 | 41-50 | SZ | M | 19 | 6.05 | Left |
| 1037 | 31-40 | SZ | M | 28 | 6.25 | Left |
| 1038 | 41-50 | SZ | M | 18.1 | 6.26 | Left |
| 1039 | 71-80 | SA | F | 13.4 | 6.81 | Left |
| 1040 | 41-50 | SZ | M | 18.5 | 6.31 | Left |
| 1042 | 21-30 | SZ | M | 16 | 6.75 | Right |
| 1043 | 41-50 | SZ | M | 27.1 | 6.64 | Right |
| 1044 | 41-50 | SZ | M | 19.25 | 6.57 | Right |
| 1052 | 80+ | SZ | F | 23.25 | 5.91 | Right |
| 1060 | 71-80 | SZ | F | 21.75 | 6.65 | Right |
| 1061 | 41-50 | SA | F | 33.78 | 6.63 | Left |
| 1065 | 41-50 | SZ | M | 19.08 | 6.6 | Left |
Ctrl, control; F, female; M, male; PDx, primary diagnosis; PMI, postmortem interval hours; SA, schizoaffective; SZ, schizophrenia.
Animals
Experimental protocols were approved by the UNC Institutional Animal Care and Use Committee. Singly housed, male Sprague-Dawley rats (150-200 g; Charles River, Raleigh, NC, USA) received daily intraperitoneal injections of haloperidol 1 mg/kg/d (n = 6) or saline 0.9% (n = 6) for 4 weeks. One hour after the final dose, rats were briefly anesthetized with ether and sacrificed; their brains were removed and hemisected. Right anterior medial frontal cortex was dissected out and frozen on dry ice. All tissue was kept frozen at -80°C until use.
miRNA microarray procedures
miRNA microarray expression analysis was performed as previously described [47]. Tissue disruption by Dounce homogenization was followed by total RNA isolation with TRIZOL™ reagent (Invitrogen, Carlsbad, California, USA). RNA (5 μg) was labeled with T4-RNA ligase and precipitated with 0.3 M sodium acetate, 2 volumes ethanol, and re-suspended in water.
Oligonucleotide probes were synthesized in duplicate for 264 human miRNAs antisense to the mature sequence reported in the Sanger miRNA registry [48]. Probes were spotted in duplicate on Corning (Corning, New York, USA) GAPS-2 coated slides using a robotic spotter and cross-linked by UV. Hybridization and washing were performed as described. All arrays were from the same batch, and the microarrays were run on the same day by the same two persons. Our prior research indicates that our in-house miRNA microarrays have excellent reliability and validity [49].
Microarray data analysis began with data extraction from the GPR files. Data points were eliminated if foreground was not 1.5 times local background and a probe was removed if >40% of the data points were missing. A total of 239 miRNA remained after this pre-processing. Data were background subtracted, log-transformed, and missing values were imputed using k-NN [50]. For comparisons across samples, data were normalized using rank invariant normalization [51]. The per-sample mean of the two rank invariant normalized probes was used for analyses. Univariate calculations of differential expression were estimated using Statistical Analysis of Microarrays (SAM; two-class, unpaired test; 500 permutations; FDR of 5%) [52]. All analysis procedures were done using R [53]. Cluster analysis was done with GeneCluster© [54] and displayed using TreeView© [55] (Figure 1).
mRNA microarray analysis procedures
Previous to our research, mRNA microarray profiling of PFC tissue from these same subjects (but different samples) was performed at the Harvard Brain Tissue Resource Center with Affymetrix U133A© arrays using standard methods and quality control procedures. The cel files and information on sample acquisition, preparation, and microarray analysis are publicly available and were downloaded from the Center's National Brain Databank.
The U133A microarrays were normalized using GC Robust Multi-Array (GCRMA), and analysis of probe expression levels was done with SAM. We used the March 2006 version of the UCSC Human (Homo sapiens) Genome Browser [56] to determine the U133A probes that corresponded to miRNA locations in host genes.
qRT-PCR procedures
Total RNA (5 μg) was DNase I (Promega, Madison, Wisconsin, USA) treated according to the manufacturer's instructions, phenol:chloroform extracted, ethanol precipitated, and dissolved in DEPC-treated dH2O (DEPC; diethylpyrocarbonate). RNA (5 μg) was polyadenylated using Poly(A) polymerase (Ambion, Austin, Texas, USA) according to the manufacturer's instructions, phenol:chloroform extracted, ethanol-precipitated, and dissolved in DEPC-treated dH2O. A modified cDNA was made as follows: 5 μg of polyadenylated RNA was reverse-transcribed using Superscript II reverse transcriptase (Invitrogen, Carlsbad, California, USA) with 2.5 μg of random hexamers and 500 ng of oligo(dT) adapter primer (5'-GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN-3') according to the manufacturer's instructions. The reaction was terminated by incubation at 70°C for 10 minutes and diluted into 2 ml of dH2O (5 ng/μl). Quantitative PCR was used to measure the mature miRNA transcript as follows: 5 μl of cDNA was mixed with 5 pmol of both the forward and reverse primers in a final volume of 12.5 μl and mixed with 12.5 μl of 2× SYBR Green PCR master mix (Applied Biosystems, Foster City, California, USA). Primer sequences are in Additional data file 7. All reactions were run in triplicate on a DNA Engine Opticon 2 (Bio-Rad Laboratories, Hercules, California, USA). The amplification protocol for mature miRNA PCR was performed according to the high-stringency protocol of Shi and Chiang [57] except the reverse primer Mir-qPCR-3-3' (5'-GCAGCA CAGAATTAATACGACTCAC-3') was used in conjunction with an exact sequence-specific primer to each miRNA. Mature miRNA expression used the reference gene U6 snRNA (U6-F, 5'-CGCTTC GGCAGCACATATAC-3'; U6-R, 5'-TTCACGAATTTGCGTGTCAT-3'). The expression was determined for eight subjects, four with schizophrenia and four healthy subjects (Additional data file 3). Expression was calculated using the delta-delta C(t) method: 2ΔCT healthy-ΔCT schizophrenia with ΔCT = (CT miRNA - CT reference RNA U6) [58].
Additional data files
The following additional data are available with the online version of this paper. Additional data file 1 is a table listing the tested miRNAs and their expression levels and fold changes. Additional data file 2 is a table showing data conditioning on PMI, pH, and hemisphere. Additional data file 3 is a table of qRT-PCR results. Additional data file 4 includes characterization and microarray results on rats treated with haloperidol. Additional data file 5 is a table of host mRNA data. Additional data file 6 lists putative motifs within regions upstream of some distinguished pre-miRNAs. Additional data file 7 is a table listing primer sequences.
Supplementary Material
Tested miRNAs and their expression levels and fold changes
Data conditioning on PMI, pH, and hemisphere
qRT-PCR results
Characterization and microarray results on rats treated with haloperidol
Host mRNA data
Putative motifs within regions upstream of some distinguished pre-miRNAs
Primer sequences
Acknowledgments
Acknowledgements
Many thanks are due to the referees for insightful and valuable comments that led to significant improvements. We are also very thankful to the Harvard Brain Tissue Resource Center, which is supported in part by PHS grant number R24 MH068855, for tissue. This project was supported in part by NIGMS Public Health Service grant GM070674 (SMH), NIH grant MH-01752 (LFJ), Elsa U Pardee Foundation and NIH Public Health Service 5-P20-RR020751-01-02 (CDJ), the Foundation of Hope (DOP), and the American Cancer Society (JMT). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official view of any granting agency.
Contributor Information
Diana O Perkins, Email: diana_perkins@med.unc.edu.
Clark D Jeffries, Email: clark_jeffries@med.unc.edu.
L Fredrik Jarskog, Email: lars_jarskog@med.unc.edu.
J Michael Thomson, Email: mike_thomson@med.unc.edu.
Keith Woods, Email: keith_woods@med.unc.edu.
Martin A Newman, Email: newman80@email.unc.edu.
Joel S Parker, Email: jparker@constellagroup.com.
Jianping Jin, Email: jjin@unc.edu.
Scott M Hammond, Email: hammond@med.unc.edu.
References
- Rossler W, Salize HJ, van Os J, Riecher-Rossler A. Size of burden of schizophrenia and psychotic disorders. Eur Neuropsychopharmacol. 2005;15:399–409. doi: 10.1016/j.euroneuro.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Sullivan PF. The genetics of schizophrenia. PLoS Med. 2005;2:e212. doi: 10.1371/journal.pmed.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. image 45. [DOI] [PubMed] [Google Scholar]
- Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, Wayland M, Freeman T, Dudbridge F, Lilley KS, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. doi: 10.1038/sj.mp.4001532. [DOI] [PubMed] [Google Scholar]
- Dracheva S, Elhakem SL, McGurk SR, Davis KL, Haroutunian V. GAD67 and GAD65 mRNA and protein expression in cerebrocortical regions of elderly patients with schizophrenia. J Neurosci Res. 2004;76:581–592. doi: 10.1002/jnr.20122. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Shcherbata HR, Hatfield S, Ward EJ, Reynolds S, Fischer KA, Ruohola-Baker H. The microRNA pathway plays a regulatory role in stem cell division. Cell Cycle. 2006;5:172–175. doi: 10.4161/cc.5.2.2343. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. Rna. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. MicroRNA therapeutics: a new niche for antisense nucleic acids. Trends Mol Med. 2006;12:99–101. doi: 10.1016/j.molmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DO, Jeffries C, Sullivan P. Expanding the 'central dogma': the regulatory role of nonprotein coding genes and implications for the genetic liability to schizophrenia. Mol Psychiatry. 2005;10:69–78. doi: 10.1038/sj.mp.4001577. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. Rna. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Debbane M, Glaser B, David MK, Feinstein C, Eliez S. Psychotic symptoms in children and adolescents with 22q11.2 deletion syndrome: Neuropsychological and behavioral implications. Schizophr Res. 2006;84:187–193. doi: 10.1016/j.schres.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Bassett AS, Chow EW, AbdelMalik P, Gheorghiu M, Husted J, Weksberg R. The schizophrenia phenotype in 22q11 deletion syndrome. Am J Psychiatry. 2003;160:1580–1586. doi: 10.1176/appi.ajp.160.9.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Gogos JA. The molecular genetics of the 22q11-associated schizophrenia. Brain Res Mol Brain Res. 2004;132:95–104. doi: 10.1016/j.molbrainres.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Horowitz A, Shifman S, Rivlin N, Pisante A, Darvasi A. A survey of the 22q11 microdeletion in a large cohort of schizophrenia patients. Schizophr Res. 2005;73:263–267. doi: 10.1016/j.schres.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. Rna. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEGG Genes Database http://www.genome.jp/kegg/genes.html
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- Nahorski SR, Young KW, John Challiss RA, Nash MS. Visualizing phosphoinositide signalling in single neurons gets a green light. Trends Neurosci. 2003;26:444–452. doi: 10.1016/S0166-2236(03)00178-4. [DOI] [PubMed] [Google Scholar]
- Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Shi Y, Ethell IM. Integrins control dendritic spine plasticity in hippocampal neurons through NMDA receptor and Ca2+/calmodulin-dependent protein kinase II-mediated actin reorganization. J Neurosci. 2006;26:1813–1822. doi: 10.1523/JNEUROSCI.4091-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Local translational control in dendrites and its role in long-term synaptic plasticity. J Neurobiol. 2005;64:116–131. doi: 10.1002/neu.20152. [DOI] [PubMed] [Google Scholar]
- Harvard Brain Tissue Resource Center http://www.brainbank.mclean.org/
- Igloi GL. Nonradioactive labeling of RNA. Anal Biochem. 1996;233:124–129. doi: 10.1006/abio.1996.0016. [DOI] [PubMed] [Google Scholar]
- miRBase http://microrna.sanger.ac.uk/
- Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Project http://www.r-project.org/
- GeneCluster http://www.broad.mit.edu/cancer/software/genecluster2/gc2.html
- Saldanha AJ. Java Treeview - extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. 10.1101/gr.229102. Article published online before print in May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tested miRNAs and their expression levels and fold changes
Data conditioning on PMI, pH, and hemisphere
qRT-PCR results
Characterization and microarray results on rats treated with haloperidol
Host mRNA data
Putative motifs within regions upstream of some distinguished pre-miRNAs
Primer sequences


