Abstract
There is an immense collection of underpowered Affymetrix gene array experiments. Although a majority of these experiments generated biologically feasible results, the considerable fraction of assays failed to identify expected transcriptional changes. There is an unused potential of Affymetrix probe-set redundancy for common exonic and UTR regions. We hypothesized that group analysis of multiple probe-sets which hybridize to the same exon or UTR will increase array discriminating power of transcriptional changes. To test this hypothesis, we analyzed Affymetrix mouse probe-sets that share the same exon using blocking feature of the Significance Analysis of Microarrays (SAM). Two-thousand two-hundred one exon-sharing probe-sets targeting 1,011 transcripts were identified by mapping 36701 MG-U74v2 probe-sets to genomic alignments of 3,971,086 known mouse transcripts. Using the blocking feature of SAM with an underpowered (two microarrays per experimental condition) mouse hypoxia-induced pulmonary hypertension model, we identified 24 genes that were significantly (FDR<5%) affected by hypoxia but were not detected by regular SAM. The relevance of the four newly identified genes (Mig6, F3, Bmp6, and Ndrg1) to known hypoxia-associated responses was confirmed by PubMatrix; and hypoxia-induced up-regulation of Mig6 expression was validated by real-time RT-PCR. We demonstrated that analysis of exon-sharing probe-sets allowed discovery of additional hypoxia-affected genes in an underpowered array experiment. This method will facilitate re-evaluation of existing underpowered Affymetrix gene expression profiles.
Keywords: gene expression profiling, microarray, oligonucleotide probe, mouse model, hypoxia, pulmonary hypertension
1. Introduction
Global gene expression profiling is a robust technique for the detection of differentially-regulated genes, which can facilitate the identification of novel markers and potential therapeutic targets in a large variety of human diseases. The Affymetrix microarray (GeneChip) platform [1] offers identification of all up-to-date transcripts using uniquely designed probe-sets (http://www.affymetrix.com/technology/design/index.affx). However, due to the considerable resource requirement of GeneChip technology, a large fraction of experiments are conducted using a low number of array replicates [2] resulting in a considerable fraction of assays fail to identify expected transcriptional changes. We hypothesized that the low discriminating power of transcriptional changes of genes that are represented by multiple probe-sets can be increased by simultaneous analysis of such probe-sets. Although the redundancy of Affymetrix probe-sets for a single transcript was previously applied for the improvement of gene expression analysis [3-5], the more stringent group analysis of probe-sets hybridizing to the same exon or UTR has not been employed.
Our lab and the others have demonstrated that expression analysis of underpowered array experiments complemented with compatible expression information from similar experimental settings produced biologically feasible results [6-8]. In current studies, we further utilized complementary information obtained from multiple probe-sets that target the same exon or UTR . Based on these exon-sharing probe-sets identified by mapping probe-set sequences to genomic alignments of known mouse transcripts were analyzed and additional candidate genes identified, The detection of transcriptional changes was evaluated using an underpowered mouse hypoxia experiment (two GeneChips per condition) and validated by real time RT-PCR.
2. Materials and Methods
2.1. Mouse genomic alignments
The genomic alignments of mouse transcripts were retrieved from the GoldenPath [9] database of the University of California Santa Cruz (UCSC) Build 33 assembly of the mouse genome (mm5, May 2004). These alignments were generated by the basic local aligning tool (BLAT) [10] against mouse genome draft using expressed sequence tags (EST) (all_est.txt.gz, 22-Nov-2004), mRNA sequences (all_mrna.txt.gz, 22-Nov-2004), and known genes (knownGene.txt.gz, 16-Jul-2004) and stored at http://hgdownload.cse.ucsc.edu/goldenPath/mm5/database. All genomic alignments have been filtered for common repeats using the RepeatMasker software designed by Kent et al (http://genome-archive.cse.ucsc.edu/goldenPath/algo.html). In addition, alignments were passed through the “near best in genome filter,” which discarded alignments that had 1% or greater divergence from the best among the multiple alignments. The detailed flowchart and description of filtering procedure is provided in Supplemental File 1.
2.2. Affymetrix MG_U74v2 probe-set genomic alignment
Given that genomic alignment of Affymetrix probe-sets (affyU74.txt.gz, 2-Aug-2004) were provided for target consensus sequences rather than actual probe-set sequences, we generated alignments of the exact (starting with the first base of probe #1 and ending with the 25th base of probe #11) MG_U74v2 probe-set sequences using standalone BLAT v.27 application [10] (http://www.soe.ucsc.edu/∼kent/exe). Alignments that covered more than 80% of probe-set sequence with cumulative unaligned stretch less than 75 bases (combined length of 3 individual probes) were selected for mapping to mouse genomic alignments.
2.3. Identification of exon-sharing probe-sets
For identification of exon-sharing probe-sets, we utilized a clustering algorithm developed by our group based on exon-exon overlap [11]. Briefly, for computational efficiency, clusters were formed in two steps. Each genomic alignment was first considered as a continuous line from the start to the end point of the transcript on the genome, then the start and the end points of exonic regions were identified. The alignments that share at least one exon were clustered. The Affymetrix probe-sets were matched against generated clusters and 12,676 probe-sets that were mapped to the same cluster (transcript) were named ‘target-sharing’. The 2,256 target-sharing probe-sets that mapped to the same exon were termed ‘exon-sharing probe-sets’. The detailed clustering procedure is described in Supplemental File 1. The exon- and target-sharing probe-sets are listed in Supplemental Files 2 and 3, respectively.
2.4. Mouse hypoxia model
All procedures were approved by the Animal Care and Use Committee of the Johns Hopkins University School of Medicine. C57BL/6J 8-week old mice were placed in a hypoxic chamber for 10 hours. The chamber was continuously flushed with a mixture of room air and N2 (10 ± 0.5 % O2) to maintain low CO2 concentrations (<0.5 %). Chamber O2 concentration was continuously monitored (PRO-OX, RCI Hudson, Anaheim, CA). Normoxic control animals were kept in room air next to the hypoxic chamber. At the end of exposure, animals were anesthetized with sodium pentobarbital (130 mg/kg i.p.); lung tissue was collected, snap-frozen and stored at −80°C.
2.5. Gene expression profiling
The sample description and GeneChip Cell files (CEL) for 0h and 10h hypoxia were uploaded from HOPGENE (http://www.hopkins-genomics.org/pulmHyper/pulmHyper005/index.html) and the Public Expression Profiling Resource (PEPR) [12] http://pepr.cnmcresearch.org/browse.do?action=list_prj_exp&projectId=124, respectively. The Bioconductor affy package [13] was used to extract the probe level data from CEL files and was converted into gene expression values by background correction, across array normalization, and summarization. The extracted data was analyzed by the Robust MultiChip Average (RMA) module of the affy package [14]. The resulting expression values of target-sharing probe-sets were formatted for the blocking Significance Microarray Analysis (SAM) [6]. Three data subsets were generated based on the numbers of probe-sets that recognized the same transcript: 2 probe-sets per transcript (6,092 probe-sets), 3 probe-sets per transcript (3,882 probe-sets) and 4 probe-sets per transcript (2,256 probe-sets). The regular SAM was also applied to unblocked 12,676 probe-sets (Supplemental File 7. Changes in expression that were greater than 20% with false discovery rate less than 5% (based on 1000 permutations) were considered significant.
2.6. PubMatrix analysis
The relevance of newly identified candidate genes to hypoxia was evaluated using PubMatrix (http://pubmatrix.grc.nia.nih.gov), the automated biomedical literature search engine. The PubMatrix-selected citations of journal articles that referenced “mouse”, “lung”, “hypoxia”, and “pulmonary hypertension” terms in the same context with a given candidate gene were manually evaluated.
2.7. Real-time RT-PCR
Transcript levels of Riken cDNA 1300002F13 (mouse mitogen-induced-gene-6, Mig6) in control mouse lungs and lungs subjected to 10% O2 for 10 hrs. were measured (n=3 per condition) as previously described [8]. Briefly, the 96-well microtiter plate setting of an ABI Prism 7700 Sequence Detector Systems (Perkin-Elmer/Applied Biosystems) was employed. TaqMan® 18S rRNA Control Reagent was used as an internal control for normalization. Primers and probes designed against third and fourth exons were purchased from Applied Biosystems Inc. (Cat.# Mm00505292_m1). All experimental protocols were based on manufactures' recommendation using the TaqMan Gold RT-PCR Core Reagents Kit (Perkin-Elmer/Applied Biosystems, P/N 402876). Experimental parameters were 48°C for 30 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. A relative quantitative method was used to calculate Mig6 transcript levels in the hypoxic sample relative to an untreated control sample and expressed as fold difference. The significance of the obtained difference was assessed using t-test where p < 0.05 was considered significant.
3. Results
3.1. Clusters of target- and exon-sharing Affymetrix probe-sets
The MG_U74v2 array set (A, B, and C GeneChips) comprises 36,701 probe-sets (http://www.affymetrix.com/support/downloads/package_inserts/mu74v2_insert.pdf). The readily available genomic probe-set alignment information provided by the Affymetrix NetAffx tool (http://www.affymetrix.com/analysis/index.affx) proved to be inadequate. NetAffx generates alignments for consensus sequences which are much longer than their corresponding probe-sets. Furthermore, only one alignment per consensus sequence is reported, while alignments to other highly homologous genomic regions are ignored. Our procedure utilized exact sequences of the MG_U74v2 probe-sets for alignment to the mouse genome draft and retained high quality multiple genomic alignments as described in Section 2.2 of Materials and Methods. Twenty-six thousand seventy-six probe-set sequences were mapped to the mouse genome and produced 31,073 alignments. The matching of these alignments to the clusters of known mouse transcripts identified 5,527 clusters that contained multiple Affymetrix probe-sets (target-sharing). Further classification of these target-sharing probe-sets by their location on the target identified 1,011 clusters, where all probe-sets were mapped to the same exon (exon-sharing).
3.2. Gene expression analysis of target-sharing probe-sets
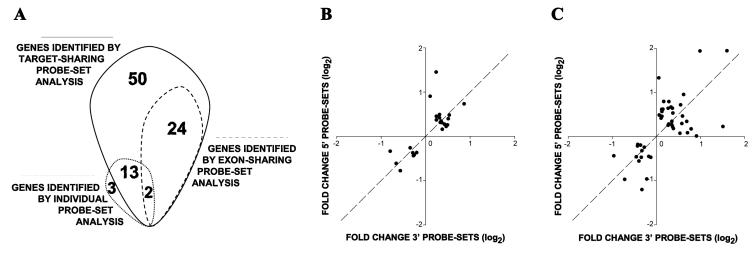
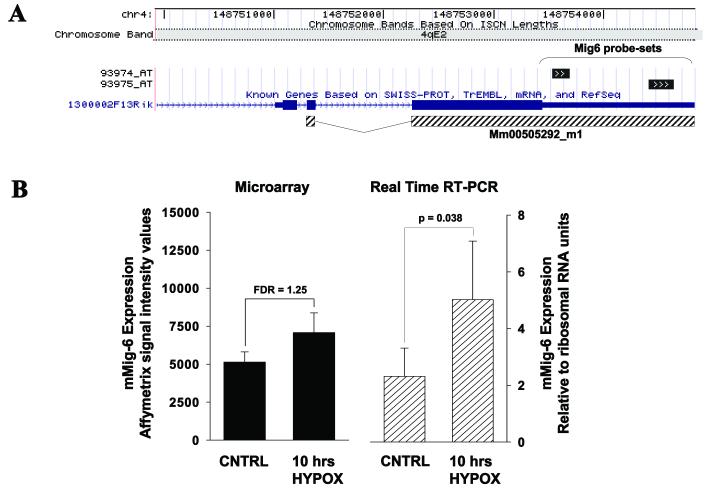
An underpowered array experiment (two arrays per experimental condition) of a mouse hypoxia-induced pulmonary hypertension model was selected for evaluation of target-sharing probe-set analysis. While the regular SAM identified only 18 hypoxia-affected genes in this experiment, the blocking SAM of target-sharing probe-sets identified 89 hypoxia-affected genes, including 26 genes identified by the exon-sharing fraction (Figure 1A). Transcriptional changes detected by 26 exon-sharing probe-sets (Figure 1B) were compared to those detected by the rest of the target-sharing probe-sets (63 probe-sets, Figure 1C) and demonstrated higher correlation of transcriptional changes detected by the exon-sharing probe-sets. The PubMatrix automated biomedical literature search of 24 recovered (Figure 1A) candidate genes identified several well-known hypoxia or pulmonary hypertension-related genes (Table 1), including Mig6, F3, Bmp6, and Ndrg1 [15-19]. The mouse Mig6 gene was selected for further validation. The aligning pattern of probe-sets targeting Mig6, is shown in Figure 2A, where the MG_U74v2 probe-sets 93974_at and 93975_at hybridize to different regions of the last exon of the Mig6 transcript. The regular SAM analysis of these probe-sets as stand alone entities was unable to classify transcriptional changes of the Mig6 gene as significant (Table 1). However, the simultaneous analysis of both probe-sets classified hypoxia-induced transcriptional changes of Mig6 as significant (Table 1). The subsequent real-time RT-PCR analysis of the Mig6 region, which is targeted by both probe-sets, confirmed this observation (Figure 2B).
Figure 1.
Classification and properties of target-sharing probe-sets.
Panel A. Overlaps between genes identified by SAM of individual, exon- and target-sharing probe-sets. The sum of numbers inside of each paddle represents the total number of genes identified using corresponding class of probe-sets. Panel B. Transcriptional changes of 24 genes identified by two exon-sharing probe-sets. Plotted results represent expression fold change (X axis - values detected by probe-sets that located at the 3′ end of the exon; Y axis - values identified by probe-sets that located at the 5′ end of the exon). Panel C. Transcriptional changes of 45 genes identified by two target-sharing probe-sets. Plotted results represent expression fold change (X axis - values detected by probe-sets that align to the 3′ exon; Y axis - values identified by probe-sets that align to the 5′ exon). The dashed line depicts the position of the exact agreement between transcriptional changes identified by 3′ and 5′ prone probe-sets.
Table 1.
Identification of the transcriptional changes induced by 10 hour hypoxia using individual and exon-sharing probe-set analyses.
| Gene | Probe-set ID | Individual |
Exon-sharing |
PubMatrix terms |
Citations† | |||
|---|---|---|---|---|---|---|---|---|
| FC | FDR* | FC | FDR | HPX | PAH | |||
| Mig6 | 93974_at | 1.31 | 36.88 | 1.37 | 1.30 | 3 | 1 | Saarikoski et al [17] |
| 93975_at | 1.43 | 14.70 | ||||||
| F3 | 163380_f_at | 1.67 | 24.38 | 1.74 | 1.30 | 31 | 13 | Collados et al [15] |
| 97689_at | 1.82 | 5.79 | ||||||
| Bmp6 | 161028_at | −1.54 | 54.62 | −1.55 | 1.30 | 0 | 1 | Yu et al [18] |
| 92372_at | −1.56 | 54.62 | ||||||
| Ndrg1 | 160464_s_at | 1.26 | 25.42 | 1.27 | 1.30 | 25 | 0 | Salnikow et al [19] |
| 96596_at | 1.28 | 57.19 | ||||||
Mig6-mitogen inducible gene 6; Bmp6-bone morphogenetic protein 6; F3-coagulation factor III (tissue factor); Ndrg1-N-myc downstream regulated gene 1.
HPX-hypoxia, PAH-pulmonary arterial hypertension
FC-fold change; FDR-false discovery rate (%)
FDR< 5% was considered significant.
most representative citation.
Figure 2.
Genome alignment and hybridization pattern of M ig6 probe-sets and expression levels of M ig6 mRNA in lung tissues detected by GeneChip array and real time RT-PCR.
The snap shot of GoldenPath browser (mouse genome May 24 Build) of chromosomal alignments of Mig6 gene and its corresponding probe-sets (Panel A). Each exon has shown as horizontal solid rectangular where narrow region of the last exon represents 3′ UTR. The intronic regions represented by solid lines with arrows showing the direction of transcription. The hatched horizontal bars represent region of A&B real time RT-PCR assay. The relative Mig6 message abundance was detected by hybridization of total mouse lung RNA to the MG-U74Av2 GeneChip (solid bars) or by real time RT-PCR (hatched bars) and shown in Panel B. The SAM calculated fold-change and false discovery rate values are reported in Table 1. Error bars represent standard deviation (SD). CNTRL-control, HYPOX-hypoxia.
4. Discussion and conclusions
In current studies, we have demonstrated that identification and group analysis of exon-sharing probe-sets of the MG_U74v2 GeneChips can be successfully employed for in silico recovery of transcriptional changes in underpowered array experiments. This approach identified several hypoxia-related genes that were missed by regular SAM analysis, including the mouse Mig6 gene (Table 1). Involvement of Mig6 in response to experimental hypoxia [17] is well documented, however, the standard SAM analysis of an underpowered hypoxia experiment failed to identify a significant transcriptional response of this gene to hypoxia (Table 1). We speculated that this was mainly due to a limited number of expression profiles (2 control and 2 hypoxia arrays) and hypothesized that the discriminating power of transcriptional changes can be improved by the group analysis of probe-sets that hybridize to the same transcript. This hypothesis was successfully tested by simultaneous analysis of two Mig6-sharing probe-sets that aligned to the last exon of the Mig6 transcript (Figure 2A). As anticipated, the transcriptional changes detected by these neighboring probe-sets were unidirectional (Table 1) and their simultaneous analysis detected significant transcriptional changes in Mig6 expression.
To identify all exon-sharing probe-sets that exist on the MG_U74v2 array, an automated probe-set mapping tool was needed. This approach required knowledge of the exon boundaries on the target transcript. Therefore, previously described techniques that used Affymetrix probe-set mapping to cDNA clones [3] or matching to the NCBI RefSeq database [4] without consideration of splicing events were not applicable. The direct clustering of probe-sets based on their description information was not considered for the same reason. In addition, it has been reported that description of approximately 19% of Affymetrix probe-sets do not correspond to their appropriate mRNA reference [5]. Therefore, we designed an exon-based clustering algorithm using our previously reported genomic alignment method [11]. We built a mouse exon dataset using the GoldenPath database of mouse genomic alignments. This database represents a collection of high quality genomic alignments of known mouse transcripts, including multiple alignments of identical sequences, which represents segmental repeats of homologous sequences common to mammalian genomes [20-23]. This is different from the NetAffx approach, which retains only the best matches for a given sequence, ignoring other homologous genomic regions which can be recognized by certain probe-sets. The conceptual simplicity of this exon-based mapping and the ease of computation make this method the procedure of choice for identification of exon-sharing probe-sets.
This mapping procedure identified 12,676 target-sharing probe-sets on the MG_U74v2 GeneChips that recognize 5,527 mouse transcripts. The large fraction (more than 80%) of these probe-sets aligned to multiple exons of their corresponding targets, thus introducing an extra alternative splicing-related variable. Predictably, the probe-sets that hybridize to the same exon demonstrated higher concordance in detection of transcriptional changes (Figure 1B) than probe-sets that hybridize to different exons (Figure 1C). This observation confirmed our selection of exon-sharing probe-sets for in silico amplification of the discriminating power of transcriptional changes. The group analysis of exon-sharing probe-sets was successfully tested on an underpowered array experiment of hypoxia-induced pulmonary hypertension mouse model. The exon-sharing approach recovered 24 hypoxia-affected genes which went undetected by regular SAM (Table 1). The biological relevance of 4 newly identified genes to hypoxia and pulmonary hypertension was suggested by an automated biomedical literature search. The subsequent manual evaluation of PubMatrix citations confirmed involvement of Mig6, F3, Bmp6, and Ndrg1 genes in hypoxia-triggered response [15-19]. Up-regulation of Mig6 expression was validated by real-time RT-PCR.
In the present studies, we have demonstrated that genome-based identification of exon-sharing probe-sets combined with blocking SAM can be successfully applied to gene expression analysis of underpowered array experiments. The panel of more than 1,000 genes that are targeted by exon-sharing probe-sets was successfully employed for the re-evaluation of underpowered hypoxia MG-U74v2 array experiment. We are confident that this new analytical approach will facilitate microarray studies and provide an efficient tool for the recovery of transcriptional changes in previously uninformative experiments.
Supplementary Material
Acknowledgements
We thank Eric Hoffman for generating, organizing and publishing pertinent to this manuscript array data on his PEPR website (http://pepr.cnmcresearch.org), and Karen Maresso for helpful suggestions on the manuscript compilation. This work was supported by the NHLBI – sponsored HopGene Program in Genomics Application (HL-69340), SCCOR (HL-073994), and individual (DNG) NRSA grant (F32 HL74590-01A1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21(1 Suppl):20–4. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 2.Jain N, Thatte J, Braciale T, Ley K, O'Connell M, Lee JK. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics. 2003;19(15):1945–51. doi: 10.1093/bioinformatics/btg264. [DOI] [PubMed] [Google Scholar]
- 3.Carter SL, Eklund AC, Mecham BH, Kohane IS, Szallasi Z. Redefinition of Affymetrix probe sets by sequence overlap with cDNA microarray probes reduces cross-platform inconsistencies in cancer-associated gene expression measurements. BMC Bioinformatics. 2005;6(1):107. doi: 10.1186/1471-2105-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gautier L, Moller M, Friis-Hansen L, Knudsen S. Alternative mapping of probes to genes for Affymetrix chips. BMC Bioinformatics. 2004;5:111. doi: 10.1186/1471-2105-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mecham BH, Wetmore DZ, Szallasi Z, Sadovsky Y, Kohane I, Mariani TJ. Increased measurement accuracy for sequence-verified microarray probes. Physiol Genomics. 2004;18(3):308–15. doi: 10.1152/physiolgenomics.00066.2004. [DOI] [PubMed] [Google Scholar]
- 6.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grigoryev DN, Lee B, Ma SF, Johns RA, Garcia JGN. Paralogous analysis of global changes in gene expression profiles during onset of pulmonary hypertension. Proceedings of the American Thoracic Society. 2005;2:A527. [Google Scholar]
- 8.Ma SF, Grigoryev DN, Taylor AD, Nonas S, Sammani S, Ye SQ, Garcia JG. Bioinformatic identification of novel early stress response genes in rodent models of lung injury. Am J Physiol Lung Cell Mol Physiol. 2005;289(3):L468–77. doi: 10.1152/ajplung.00109.2005. [DOI] [PubMed] [Google Scholar]
- 9.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maitra R, Grigoryev DN, Bera TK, Pastan IH, Lee B. Cloning, molecular characterization, and expression analysis of Copine 8. Biochem Biophys Res Commun. 2003;303(3):842–7. doi: 10.1016/s0006-291x(03)00445-5. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Zhao P, Massaro D, Clerch LB, Almon RR, DuBois DC, Jusko WJ, Hoffman EP. The PEPR GeneChip data warehouse, and implementation of a dynamic time series query tool (SGQT) with graphical interface. Nucleic Acids Res. 2004;32(Database issue):D578–81. doi: 10.1093/nar/gkh003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irizarry R, Gautier L, Cope L. The Analysis of Gene Expression Data: Methods and Software. In: Parmigiani G, Garrett ES, Irizarry RA, Zeger SL, editors. An R package for analyses of Affymetrix oligonucleotide arrays. Springer; New York: 2003. [Google Scholar]
- 14.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collados MT, Velazquez B, Borbolla JR, Sandoval J, Masso F, Montano LF, Guarner V. Endothelin-1 and functional tissue factor: a possible relationship with severity in primary pulmonary hypertension. Heart Vessels. 2003;18(1):12–7. doi: 10.1007/s003800300002. [DOI] [PubMed] [Google Scholar]
- 16.Solovey A, Kollander R, Shet A, Milbauer LC, Choong S, Panoskaltsis-Mortari A, Blazar BR, Kelm RJ, Jr., Hebbel RP. Endothelial cell expression of tissue factor in sickle mice is augmented by hypoxia/reoxygenation and inhibited by lovastatin. Blood. 2004;104(3):840–6. doi: 10.1182/blood-2003-10-3719. [DOI] [PubMed] [Google Scholar]
- 17.Saarikoski ST, Rivera SP, Hankinson O. Mitogen-inducible gene 6 (MIG-6), adipophilin and tuftelin are inducible by hypoxia. FEBS Lett. 2002;530(13):186–90. doi: 10.1016/s0014-5793(02)03475-0. [DOI] [PubMed] [Google Scholar]
- 18.Yu PB, Beppu H, Kawai N, Li E, Bloch KD. Bone morphogenetic protein (BMP) type II receptor deletion reveals BMP ligand-specific gain of signaling in pulmonary artery smooth muscle cells. J Biol Chem. 2005;280(26):24443–50. doi: 10.1074/jbc.M502825200. [DOI] [PubMed] [Google Scholar]
- 19.Salnikow K, Kluz T, Costa M, Piquemal D, Demidenko ZN, Xie K, Blagosklonny MV. The regulation of hypoxic genes by calcium involves c-Jun/AP-1, which cooperates with hypoxia-inducible factor 1 in response to hypoxia. Mol Cell Biol. 2002;22(6):1734–41. doi: 10.1128/MCB.22.6.1734-1741.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Complete sequence and gene map of a human major histocompatibility complex.The MHC sequencing consortium. Nature. 1999;401(6756):921–3. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 21.Irwin DM. Ancient duplications of the human proglucagon gene. Genomics. 2002;79(5):741–6. doi: 10.1006/geno.2002.6762. [DOI] [PubMed] [Google Scholar]
- 22.Bailey JA, Yavor AM, Viggiano L, Misceo D, Horvath JE, Archidiacono N, Schwartz S, Rocchi M, Eichler EE. Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22. Am J Hum Genet. 2002;70(1):83–100. doi: 10.1086/338458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichler EE. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17(11):661–9. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.