Abstract
Ribosome recruitment to the majority of eukaryotic mRNAs is facilitated by the interaction of the cap binding protein, eIF4E, with the mRNA 5′ cap structure. eIF4E stimulates translation through its interaction with a scaffolding protein, eIF4G, which helps to recruit the ribosome. Metazoans also contain a homolog of eIF4E, termed 4EHP, which binds the cap structure, but not eIF4G, and thus cannot stimulate translation, but it instead inhibits the translation of only one known, and possibly subset mRNAs. To understand why 4EHP does not inhibit general translation, we studied the binding affinity of 4EHP for cap analogs using two methods: fluorescence titration and stopped-flow measurements. We show that 4EHP binds cap analogs m7GpppG and m7GTP with 30 and 100 lower affinity than eIF4E. Thus, 4EHP cannot compete with eIF4E for binding to the cap structure of most mRNAs.
Keywords: 4EHP, eIF4E isoforms, mRNA 5′ cap, binding affinity, stopped-flow
INTRODUCTION
All nuclear transcribed eukaryotic mRNAs possess a common structure called a “cap” at their 5′ end, which consists of 7-methylguanosine bound by a 5′-5′-triphosphate bridge to the first transcribed nucleotide. The cap structure is important for stabilizing the mRNA (Furuichi and Shatkin 2000), facilitating the splicing of pre-mRNAs (Konarska et al. 1984), promoting mRNA transport to cytoplasm (Izaurralde et al. 1992), and facilitating the binding of ribosomes to the mRNA (Mathews et al. 2000; Pestova et al. 2007). Cap-dependent translation begins with recognition of the cap structure by eIF4E, which forms a heterotrimeric complex with eIF4A, which is thought to melt the mRNA 5′ secondary structure and the scaffolding protein eIF4G that binds other factors to recruit the ribosome (Mathews et al. 2000). The interaction of eIF4E with eIF4G is controlled by a group of proteins generally known as eIF4E inhibitory proteins, which share a common eIF4E-binding site with eIF4G. Whereas some 4EBPs repress translation of a large number of mRNAs by associating only with eIF4E (Raught et al. 2000), others, more recently discovered, such as Cup or Maskin, inhibit translation of specific mRNAs by binding simultaneously to eIF4E and additional proteins that interact with sequence elements in the mRNA 3′ UTR (Wilhelm et al. 2003; Cao and Richter 2002).
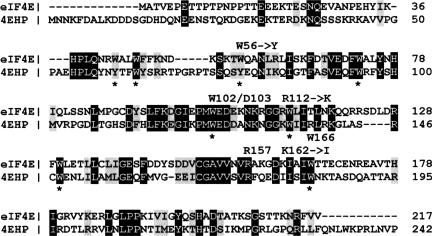
Crystallographic, NMR, and biophysical studies determined the amino acids in eIF4E that are of primary importance for cap structure recognition (Marcotrigiano et al. 1997; Matsuo et al. 1997; Niedzwiecka et al. 2002; Tomoo et al. 2003). The structural basis for the specificity of the eIF4E–cap interaction is the sandwiching of the 7-methylguanine base via π–π stacking interactions between tryptophan indol rings (Trp56 and Trp102). This interaction is stabilized by van der Waals contacts of the m7G with Trp166 and hydrogen bonds with Glu103. The phosphate chain of the cap structure forms direct or water-mediated hydrogen bonds with NH groups of Trp102 and Trp166 as well as with side chains of lysine and arginine residues (Arg112, Arg157, and Lys162).
eIF4E homologs have been identified in most organisms (except for Saccharomyces cervisiae). In mammals, there are three members of the eIF4E family: eIF4E (eIF4E-1), 4EHP (eIF4E-2), and eIF4E-3 (Rom et al. 1998; Joshi et al. 2004). Based on sequence alignments, h4EHP, together with nCBP from plants (Ruud et al. 1998), IF4E-4 from Caenorbiditis elegans and d4EHP (deIF4E-8) from Drosophila melanogaster are members of the eIF4E-2 class (Joshi et al. 2005). Members of this class possess mainly Tyr/Phe in the position corresponding to Trp43 and Tyr in the position of Trp56 of human eIF4E (Joshi et al. 2005). In h4EHP both of these positions are occupied by tyrosine (Fig. 1). Mouse and human 4EHP binds to m7GTP-Sepharose but does not interact with eIF4G (Rom et al. 1998; Joshi et al. 2004) and cannot rescue the lethality of eIF4E gene deletion in yeast (Joshi et al. 2004). Recently, the Drosophila 4EHP has been reported to be a translation repressor that is required for the asymmetric distribution of caudal protein in the egg, which is essential for the appropriate development of the embryo (Cho et al. 2005). d4EHP blocks translation by binding to the 5′ cap structure of Caudal mRNA and to the regulatory protein Bicoid, which interacts with 3′ UTR of the caudal mRNA (Cho et al. 2005).
FIGURE 1.
Amino acid alignment of human eIF4E (gi:4503535) with 4EHP (gi:3172339) performed with CLUSTALW (Thompson et al. 1994). Residues that are identical in proteins are shadowed in black and conserved substitutions in gray. The conserved residues that play a role in cap binding by eIF4E (Marcotrigiano et al. 1997), with changes in 4EHP, are labeled above. Stars below the lines indicate the positions of eight evolutionarily conserved tryptophan residues in eIF4E.
Here we present data on the binding affinities of cap analogs for human 4EHP versus human eIF4E using the fluorescence titration method and stopped-flow technique.
RESULTS AND DISCUSSION
Proteins preparation
All proteins were expressed in Escherichia coli in inclusion bodies, refolded by one-step dialysis, and purified using ion-exchange chromatography to avoid any contact with cap analogs (Niedzwiecka et al. 2002). Analysis of cell extracts and proteins after dialysis showed limited proteolysis of the N terminus. However, the application of ion-exchange column resulted in homogeneous full-length proteins (>95%; Fig. 2). The amount of proper refolded protein, obtained from fluorescence titrations by fitting concentration of the “active” protein as a free parameter of the equilibrium equation, was about 70%–85% (Niedzwiecka et al. 2002).
FIGURE 2.
Purified on ion-exchange column proteins visualized on a Comassie-stained 15% SDS-PAGE gel. Lane 1, protein marker weight standards (Sigma); lane 2, human eIF4E; lane 3, human eIF4EW56Y; and lane 4, human 4EHP.
Different binding affinities of 4EHP and eIF4E for cap analogs
The interaction between cap analogs and eIF4E results in quenching of intrinsic Trp fluorescence (Fig. 3A). 4EHP possesses six out of the eight conserved tryptophan residues of eIF4E and an additional tryptophan in the C terminus. Trp43 and Trp56 of eIF4E are replaced in 4EHP by tyrosines (Rom et al. 1998; Joshi et al. 2004). These differences result in slightly lower fluorescence intensities for equimolar protein solutions of 4EHP versus eIF4E. However, the emission spectra of both proteins are very similar (data not shown), suggesting no significant structural differences between the proteins as a result of the changes in the environment of Trp residues.
FIGURE 3.
(A) Fluorescence titration curves for binding m7GTP to human eIF4E (△), its eIF4EW56Y mutant (●), human 4EHP (■), and fitting residuals. Titrations were carried out in 50 mM HEPES/KOH (pH 7.2), 0.5 mM EDTA, and 1 mM DTT adjusting to I = 150 mM by KCl at 20°C. Protein fluorescence, presented as relative value, was excited at 280 nm and observed at 337 nm. The observed increasing fluorescence signal at a higher concentration of m7GTP originates from free-cap analog emission. (B) Graphical comparison of Gibbs free energy of binding (ΔGo) for association of cap analogs with human eIF4E, its mutant, and human 4EHP, calculated from obtained association constants (Kas).
To determine the accurate association constants (Kas) for the complexes of 4EHP with a series of mono- and dinucleotide cap analogs, we applied the time-synchronized fluorescence titration method (Niedzwiecka et al. 2002) and the stopped-flow technique with an emission detector (Dlugosz et al. 2002). The Kas values obtained from fluorescence titration are shown in Table 1, and the corresponding Gibbs free energy of binding (ΔG°) are graphically presented in Figure 3B. The kinetic parameters for the association reaction protein–cap analog are shown in Table 2. As a control, Kas for complexes of human eIF4E with all tested cap analogs were determined. It is striking that 4EHP binds all cap analogs significantly weaker than eIF4E. However, the differences among binding affinities depend on the nature of the cap analog. The association constant for 4EHP with m7GTP is about 100-fold lower compared to that for eIF4E (Fig. 3A), whereas the dinucleotide triphosphate cap analogs (m7GpppG, m7GpppA, and m7GpppC) are bound by 4EHP about 30-fold weaker. These data were also confirmed by stopped-flow measurements (Table 2). The results are in good agreement with the observations that human and Drosophila 4EHP bind m7GTP-Sepharose less efficiently than their eIF4E counterparts (Tee et al. 2004; Cho et al. 2005). However, the differences of two orders of magnitude in binding affinities for the cap structure between the two proteins are striking, particularly because 4EHP, compared to eIF4E, possesses only three substitutions in the cap-binding slot: two conservative and one nonconservative (Fig. 1).
TABLE 1.
Equilibrium association constants (Kas) for the complexes of cap analogs with the human eIF4E, eIF4EW56Y mutant, and human 4EHP obtained from analysis of steady-state fluorescence titrations at 20°C
TABLE 2.
Rate constants for association (k+1) and dissociation (k−1) of human eIF4E and 4EHP with cap analogs obtained from fitting one-step model to the kinetic traces registered during stopped-flow experiments at 20°C
Replacement of Trp56 by Tyr in eIF4E moderately stimulates cap binding affinity
A mutant of human eIF4E possessing a leucine substitution of Trp56 fails to bind m7GTP-Sepharose (Morino et al. 1996). However, substitution of the corresponding tryptophan by phenylalanine in yeast eIF4E reduces eIF4E's binding to the cap structure by 50% only (Altmann et al. 1988). To elucidate the contribution of the 4EHP tyrosine residue (corresponding to Trp56 in eIF4E) to the weak binding affinity of 4EHP for the cap, we generated a mutant of eIF4E possessing tyrosine instead of tryptophan in position 56. Surprisingly, this mutant exhibits a slightly enhanced (∼1.5-fold) binding to the cap structure (Table 1). The Kas for the complex of mutated eIF4E with m7GTP is 109.7 ± 5.0 μM−1, whereas for wild type eIF4E is 68.41 ± 5.09 μM−1. The increase of Kas for eIF4EW56Y is very similar for all tested cap analogs and does not depend on the number of phosphate groups. These results show that in eIF4E two tryptophans as well as tryptophan and tyrosine residues can sandwich 7-methylguanine through the π–π stacking interaction with similar efficiency. The recognition mode of the cap structure by the sandwich stacking of the m7G moiety via two aromatic rings of amino acids is exercised by other, eIF4E-unrelated, mRNA 5′ cap binding proteins. The vaccinia virus mRNA-cap-dependent 2′-O- methyltransferase, VP39, contains a tyrosine and a phenylalanine (Tyr22 and Phe180) (Hu et al. 1999) and the conserved nuclear cap binding protein, CBC20, contains two tyrosines (Tyr20 and Tyr43) (Mazza et al. 2002). The crystallographic structures of cap binding proteins supported by theoretical studies showed that the almost perfect alignment of the aromatic rings, an interplanar distance of 3.1–3.6 Å, a substantial area of overlap in stacking rings, and the positive charge of m7G all contribute to the strong interaction between the π-electrons of stacked rings (Quiocho et al. 2000; Fechter and Brownlee 2005). The VP39, which possesses phenylalanine in the stacking mode, had the lowest binding affinity (Kas ∼0.1 μM−1), and its replacement by tryptophan or tyrosine enhances binding. On the other hand, substitution of phenylalanine for Tyr22 cannot rescue the stacking interaction (Hu et al. 1999; Hsu et al. 2000). The CBC20, in which two tyrosines are involved in stacking, exhibits binding affinity for m7GTP similar to eIF4E: Kas = 29.7 ± 2.3 μM−1 for CBC (Worch et al. 2005) and Kas = 33.28 ± 1.04 μM−1 for mouse eIF4E in buffer containing 200 mM KCl (Zuberek et al. 2004). The ring system of tyrosine is more π-electron rich than that of phenylalanine due to the electron-donating character of phenolic oxygen, which engenders a stronger stacking interaction between tyrosine and the positively charged m7G base. Thus, all these data suggest that the presence of Tyr78 instead of tryptophan cannot be the major reason for the 100-fold weaker binding of cap analogs to 4EHP than to its eIF4E counterpart. In human eIF4E Trp56 is located in loop S1–S2 between two β sheets (Tomoo et al. 2003). This region in 4EHP contains five additional amino acids (Fig. 1), which can increase the flexibility of this loop. As a consequence, it may prevent the correct parallel alignment of the aromatic ring of Tyr78 and its substantial overlap with the m7G moiety essential for an efficient stacking interaction, which is adopted in the eIF4EW56Y mutant.
Recently, the cap binding affinities of four Leishmania eIF4E isoforms were determined (Yoffe et al. 2006). They all possess binding affinities for m7GTP similar to h4EHP. However, in LeishIF4E-1 two tryptophan residues and in LeishIF4E-4 one tyrosine and one tryptophan are involved in the stacking interaction. LeishIF4E-4 binds m7GTP even approximately fourfold stronger than LeishIF4E-1, which is in agreement with our results for the eIF4EW56Y mutant. These data suggest that not only is the type of aromatic amino acid (Tyr or Trp) engaged in the stacking interaction important for stabilizing the complex of the eIF4E-cap, but other structural elements also are important. However, homology modeling of either Leishmania isoforms (Yoffe et al. 2006) or h4EHP (Rom et al. 1998) did not predict significant differences in the three-dimensional structure compared to eIF4E.
Effect of the negative charge of cap analogs on their affinity for 4EHP
It has been shown previously that electrostatic interactions play a crucial role in the recognition of the cap structure by eIF4E (Niedzwiecka et al. 2002; Zuberek et al. 2004). The extension of the phosphate chain in cap analogs results in systematic, marked enhancement of the binding affinity for eIF4E. 4EHP carries only two substitutions, with reference to positive charged amino acid in the cap binding slot of eIF4E (Fig. 1). A hydrophobic isoleucine substitutes for Lys162, which forms a direct hydrogen bond with an oxygen atom of the β-phosphate group (Tomoo et al. 2003). In addition, a lysine replaces Arg112, which in eIF4E interacts with the α-phosphate group by a water-mediated hydrogen bond (Marcotrigiano et al. 1997; Tomoo et al. 2003). To establish the possible effect of substitutions on cap binding (especially the lack of a lysine residue), we analyzed the Gibbs free energy of binding in relation to the number of phosphate groups within the cap analogs (Fig. 3B). The addition of the β-phosphate group to the cap analog (m7GMP→m7GDP) gives a change in binding free energy of about −0.7 kcal/mol for 4EHP and about −1.8 kcal/mol for eIF4E. The difference between proteins of about 1.1 kcal/mol in the energetic cost (ΔΔGo) corresponds to one hydrogen bond. For both proteins the addition of a third (γ) phosphate group (m7GDP→m7GTP) causes a comparable change of binding free energy (4EHP of about −0.65 kcal/mol; eIF4E of about −0.8 kcal/mol). This suggests that the lack of the lysine residue in 4EHP is partially responsible for the ∼100-fold decrease in its binding affinity for m7GDP and m7GTP compared to eIF4E, whereas for m7GMP it is only 10-fold. The weaker binding of m7GMP by 4EHP presumably results from the replacement of arginine by lysine with a shorter side chain, thus preventing the formation of a water-mediated hydrogen bond. The twofold increase in the association constant for m7Gp4 (Kas = 1.20 ± 0.04 μM−1) in comparison with m7GTP (Kas = 0.70 ± 0.04 μM−1) suggests that the δ-phosphate of the cap analog in contrast to eIF4E (threefold increase of Kas) does not interact directly with the protein by forming hydrogen bonds.
CONCLUSIONS
Earlier studies suggested that h4EHP and d4EHP bind a cap structure weaker than eIF4E based on qualitative measurements using m7GTP-Sepharose (Tee et al. 2004; Cho et al. 2005). Here we used quantitative measurements to show that 4EHP binds to the cap structure with a very low affinity (30–100-fold lower than eIF4E). The weak binding of 4EHP to the cap prevents it from competing with eIF4E for association with the mRNA cap and to inhibit translation. Translational inhibition of caudal mRNA occurs only when 4EHP simultaneously binds the cap and Bicoid. The association of d4EHP with Bicoid might enhance its affinity for the cap. A similar mechanism of translational inhibition by h4EHP may occur by binding to as yet unidentified proteins.
MATERIALS AND METHODS
Synthesis of cap analogs
Syntheses of mono- and dinucleotide cap analogs were performed as described previously (Darzynkiewicz et al. 1985, 1990; Jemielity et al. 2003; Zuberek et al. 2004). The cap analog concentrations were determined spectrophotometrically (Cai et al. 1999).
Cloning and mutagenesis
The cDNA of human 4EHP, amplified from the pCDNA3-HA_4EHP vector by PCR, was subcloned into the expression vector pET30a (Novagen) in NdeI–BamHI sites.
The QuikChange PCR-based site-directed mutagenesis kit (Stratagene) was used to obtain the point-mutations replacement of Trp56 by tyrosine in human eIF4E. As an initial template the cDNA for eIF4E in a pET11d vector (Novagen) was used and site-directed mutagenesis was performed exactly according to the instructions provided by Stratagene. The presence of the mutation was confirmed by automatic DNA sequencing.
Protein expression and purification
For expression of the human eIF4E, an eIF4EW56Y mutant and 4EHP appropriate plasmids were transformed into E. coli BL21(DE3) cells. Bacteria were grown in LB medium to OD600 nm of 0.8, and induced for 3 h at 37°C by adding 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG). Cells were harvested, resuspended in lysis buffer (20 mM HEPES/KOH [pH 7.5], 100 mM KCl, 1 mM EDTA, 2 mM DTT, and 10% glycerol) and disrupted by sonication. After centrifugation of the lysate (30,000g for 30 min) the supernatant was removed and the pellet was washed three times with wash buffer (20 mM HEPES/KOH [pH 7.2], 1 M guanidine hydrochloride, 2 mM DTT, and 10% glycerol). The inclusion bodies were dissolved in 50 mM HEPES/KOH (pH 7.2), 6 M guanidine hydrochloride, 10% glycerol, and 2 mM DTT, and cell debris was removed by centrifugation (43,000g for 30 min). The protein (diluted to a concentration lower than 0.1 mg/mL) was refolded by one-step dialysis against 50 mM HEPES/KOH (pH 7.2), 100 mM KCl, 1.0 mM EDTA, and 2 mM DTT, and purified by ion exchange chromatography on a HiTrap MonoSP column (Amersham Bioscience). The purified proteins were analyzed by SDS-PAGE and their concentrations were determined by absorption, assuming the following: ε280 = 53,400 M−1cm−1 for eIF4E, ε280 = 49,200 M−1cm−1 for eIF4EW56Y, and ε280 = 47,600 M−1cm−1 for 4EHP (calculated from amino acid composition using an algorithm on the ExPASy Server).
Fluorescence binding titration
Fluorescence titration measurements were carried out on an LS-50B or LS-55 spectrofluorometer (Perkin-Elmer), in 50 mM HEPES/KOH (pH 7.2) and 0.5 mM EDTA 1 mM DTT, adjusting to an ionic strength of 150 mM by KCl at 20.0 ± 0.2°C. Aliquots of 1 μL increasing concentrations of cap analog solutions were added to 1.4 mL of 0.1 or 0.2 μM protein solutions. Fluorescence intensities (excitation at 280 nm or 295 nm with a 2.5-nm bandwidth and detection at 337 nm or 345 nm with a 4-nm bandwidth and a 290-nm cutoff filter) were corrected taking into account sample dilution and the inner filter effect. Equilibrium association constants (Kas) were determined by fitting the theoretical dependence of the fluorescence intensity on the total concentration of the cap analog to the experimental data points according to the equation described previously (Niedzwiecka et al. 2002). The concentration of protein was fitted as a free parameter of the equilibrium equation showing the amount of “active” protein. The final Kas was calculated as a weighted average of 3–10 independent titrations, with the weights taken as the reciprocals of the numerical standard deviations squared. Numerical nonlinear least-squares regression analysis was performed using ORGIN 6.0 (Microcal Software).
The Gibbs free energy of binding was calculated from the Kas value according to the standard equation ΔG° = −RTlnKas.
Stopped-flow measurements and analysis of kinetics transients
Kinetic measurements of interaction eIF4E and 4EHP proteins with m7GTP and m7GpppG were run on a SX.18MV stopped-flow reaction analyzer (Applied Photophysics) using fluorescence detection. The protein emission was excited at 290 nm (with a 0.5 mm slit) and its fluorescence was monitored after passage through a 320-nm cutoff filter. The path lengths in the stopped flow cell were 2 mm for absorption and 10 mm for emission. The reaction was initiated by mixing an equal volume of protein solution (1 μM) with the cap analog (0.5–8 μM for eIF4E and 0.5–20 μM for 4EHP). The measurements were performed in 50 mM HEPES/KOH (pH 7.2), 0.5 mM EDTA adjusted to an ionic strength of 150 mM by KCl at 20.0 ± 0.1°C. The fluorescence changes were monitored up to a 200-msec recording of 1000 data points using the oversampling option of the instrument. The kinetic traces for each concentration of the cap analog are an average of 14 independent runs.
The obtained kinetics traces were subjected to nonlinear least-squares regression assuming a one-step model for the protein–cap analog association:
 |
described by differential equations for rates of changes of concentration of molecular species
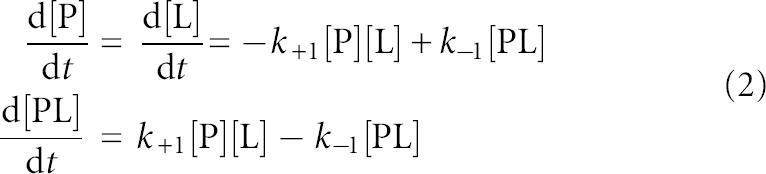 |
and its specific contributions to the monitored fluorescence signal
where k+1 and k−1 are association and dissociation rate constants, respectively; [P], [L], and [PL] are concentrations, and fP,fL, and fPL, are molar fluorescence of free protein, free ligand, and complex. Numerical analysis was performed using the DynaFit program (BioKin) created by Peter Kuzmic (Kuzmic 1996).
ACKNOWLEDGMENTS
We thank Dr. Jan M. Antosiewicz (Warsaw University) for making the SX.18MV stopped-flow reaction analyzer available to us. This work was supported by Howard Hughes Medical Institute Grant No. 55005604 to E.D., Polish Ministry of Science and Higher Education Grant No. 2P04A 006 28 to E.D., and the Canadian Institute of Health Research to N.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.453107.
REFERENCES
- Altmann, M., Edery, I., Trachsel, H., Sonenberg, N. Site-directed mutagenesis of the tryptophan residues in yeast eukaryotic initiation factor 4E. Effects on cap binding activity. J. Biol. Chem. 1988;263:17229–17232. [PubMed] [Google Scholar]
- Cai, A.L., Jankowska-Anyszka, M., Centers, A., Chlebicka, L., Stepinski, J., Stolarski, R., Darzynkiewicz, E., Rhoads, R.E. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry. 1999;38:8538–8547. doi: 10.1021/bi9830213. [DOI] [PubMed] [Google Scholar]
- Cao, Q., Richter, J.D. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, P.F., Poulin, F., Cho-Park, Y.A., Cho-Park, I.B., Chicoine, J.D., Lasko, P., Sonenberg, N. A new paradigm for translational control: Inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–423. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Ekiel, I., Tahara, S.M., Seliger, L.S., Shatkin, A.J. Chemical synthesis and characterization of 7-methylguanosine cap analogs. Biochemistry. 1985;24:1701–1707. [Google Scholar]
- Darzynkiewicz, E., Stepinski, J., Tahara, S.M., Stolarski, R., Ekiel, I., Haber, D., Neuvonen, K., Lehikoinen, P., Labadi, I., Lonnberg, H. Synthesis, conformation and hydrolytic stability of P1, P3-dinucleotide triphosphate related to mRNA 5′-cap, and comparative kinetic studies on their nucleoside and nucleotide monophosphate analogs. Nucleosides Nucleotides. 1990;9:599–618. [Google Scholar]
- Dlugosz, M., Bojarska, E., Antosiewicz, J.M. A procedure for analysis of stopped-flow transients for protein–ligand association. J. Biochem. Biophys. Methods. 2002;51:179–193. doi: 10.1016/s0165-022x(02)00015-5. [DOI] [PubMed] [Google Scholar]
- Fechter, P., Brownlee, G.G. Recognition of mRNA cap structure by viral and cellular proteins. J. Gen. Virol. 2005;86:1239–1249. doi: 10.1099/vir.0.80755-0. [DOI] [PubMed] [Google Scholar]
- Furuichi, Y., Shatkin, A.J. Viral and cellular mRNA capping: Past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P.C., Hodel, M.R., Thomas, J.W., Taylor, L.J., Hagedorn, C.H., Hodel, A.E. Structural requirements for the specific recognition of an m7G mRNA cap. Biochemistry. 2000;39:13730–13736. doi: 10.1021/bi000623p. [DOI] [PubMed] [Google Scholar]
- Hu, G., Gershon, P.D., Hodel, A.E., Quiocho, F.A. mRNA cap recognition: Dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc. Natl. Acad. Sci. 1999;96:7149–7154. doi: 10.1073/pnas.96.13.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde, E., Stepinski, J., Darzynkiewicz, E., Mattaj, I.W. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol. 1992;118:1287–1295. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity, J., Fowler, T., Zuberek, J., Stepinski, J., Lewdorowicz, M., Niedzwiecka, A., Stolarski, R., Darzynkiewicz, E., Rhoads, R.E. Novel “anti-reverse” cap analogs with superior transnational properties. RNA. 2003;9:1108–1122. doi: 10.1261/rna.5430403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, B., Cameron, A., Jagus, R. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 2004;271:2189–2203. doi: 10.1111/j.1432-1033.2004.04149.x. [DOI] [PubMed] [Google Scholar]
- Joshi, B., Lee, K., Maeder, D.L., Jagus, R. Phylogenetic analysis of eIF4E-family members. BMC Evol. Biol. 2005;5:48. doi: 10.1186/1471-2148-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska, M.M., Padgett, R.A., Sharp, P.A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Kuzmic, P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano, J., Gingras, A.C., Sonenberg, N., Burley, S.K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- Mathews, M.B., Sonenberg, N., Hershey, J.W.B. Origins and principles of translation control. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 1–32. [Google Scholar]
- Matsuo, H., Li, H., McGuire, A.M., Fletcher, C.M., Gingras, A.C., Sonenberg, N., Wagner, G. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- Mazza, C., Segref, A., Mattaj, I.W., Cusack, S. Large-scale induced fit recognition of an m7GpppG cap analog by the human nuclear cap binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino, S., Hazama, H., Ozaki, M., Teraoma, Y., Shibata, S., Doi, M., Ueda, H., Ishida, T., Uesugi, S. Analysis of the mRNA cap-binding ability of human eukaryotic initiation factor-4E by use of recombinant wild-type and mutant forms. Eur. J. Biochem. 1996;23:597–601. doi: 10.1111/j.1432-1033.1996.0597u.x. [DOI] [PubMed] [Google Scholar]
- Niedzwiecka, A., Marcotrigiano, J., Stepinski, J., Jankowska-Anyszka, M., Wyslouch-Cieszynska, A., Dadlez, M., Gingras, A.C., Mak, P., Darzynkiewicz, E., Sonenberg, N., et al. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP proteins. J. Mol. Biol. 2002;319:615–635. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Lorch, J.R., Hellen, C.U.T. The mechanism of translation initiation in eukaryotes. In: Mathews M.B., et al., editors. Translation control in biology and medicine. Cold Spring Laboratory Press; New York, NY: 2007. pp. 87–128. [Google Scholar]
- Quiocho, F.A., Hu, G., Gershon, P.D. Structural basis of mRNA cap recognition by proteins. Curr. Opin. Struct. Biol. 2000;10:78–86. doi: 10.1016/s0959-440x(99)00053-6. [DOI] [PubMed] [Google Scholar]
- Raught, B., Gingras, A.C., Sonenberg, N. Regulation of ribosomal recruitment in eukaryotes. In: Sonenberg N., et al., editors. Translation control of gene expression. Cold Spring Laboratory Press; New York, NY: 2000. pp. 245–293. [Google Scholar]
- Rom, E., Kim, H.C., Gingras, A.C., Marcotrigiano, J., Favre, D., Olsen, H., Burley, S.K., Sonenberg, N. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J. Biol. Chem. 1998;273:13104–13109. doi: 10.1074/jbc.273.21.13104. [DOI] [PubMed] [Google Scholar]
- Ruud, K.A., Kuhlow, C., Goss, D.J., Browning, K.S. Identification and characterization of a novel cap-binding protein from Arabidopsis thaliana. J. Biol. Chem. 1998;273:10325–10330. doi: 10.1074/jbc.273.17.10325. [DOI] [PubMed] [Google Scholar]
- Tee, A.R., Tee, J.A., Blenis, J. Characterizing the interaction of the mammalian eIF4E-related protein 4EHP with 4E-BP1. FEBS Lett. 2004;564:58–62. doi: 10.1016/S0014-5793(04)00313-8. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoo, K., Shen, X., Okabe, K., Nozoe, Y., Fukuhara, S., Morino, S., Sasaki, M., Taniguchi, T., Miyagawa, H., Kitamura, K., et al. Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J. Mol. Biol. 2003;328:365–383. doi: 10.1016/s0022-2836(03)00314-0. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J.E., Hilton, M., Amos, Q., Henzel, W.J. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J. Cell Biol. 2003;163:1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worch, R., Niedzwiecka, A., Stepinski, J., Mazza, C., Jankowska-Anyszka, M., Darzynkiewicz, E., Cusack, S., Stolarski, R. Specificity of recognition of mRNA 5′ cap by human nuclear cap-binding complex. RNA. 2005;11:1355–1363. doi: 10.1261/rna.2850705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoffe, Y., Zuberek, J., Lerer, A., Lewdorowicz, M., Stepinski, J., Altmann, M., Darzynkiewicz, E., Shapira, M. Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania. Eukaryot. Cell. 2006;5:1969–1979. doi: 10.1128/EC.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberek, J., Jemielity, J., Jablonowska, A., Stepinski, J., Dadlez, M., Stolarski, R., Darzynkiewicz, E. Influence of electric charge variation at residues 209 and 159 on the interaction of eIF4E with the mRNA 5′ terminus. Biochemistry. 2004;43:5370–5379. doi: 10.1021/bi030266t. [DOI] [PubMed] [Google Scholar]