Abstract
Morphological, cytogenetic, and biological evidence supports a relationship between congenital (infantile) fibrosarcoma (CFS) and congenital mesoblastic nephroma (CMN). These tumors have a very similar histological appearance, and they are both associated with polysomies for chromosomes 8, 11, 17, and 20. Recently, CFS was shown to contain a novel t(12;15)(p13;q25) translocation resulting in ETV6-NTRK3 gene fusion. The aims of this study were to determine whether congenital mesoblastic nephroma contains the t(12;15)(p13;q25) translocation and ETV6-NTRK3 gene fusion and whether ETV6-NTRK3 fusions, in CMN and CFS, antedate acquisition of nonrandom chromosome polysomies. To address these aims, we evaluated 1) ETV6-NTRK3 fusion transcripts by reverse transcriptase polymerase chain reaction and sequence analysis, 2) genomic ETV6-region chromosomal rearrangement by fluorescence in situ hybridization, and 3) chromosomal polysomies by karyotyping and fluorescence in situ hybridization. We report ETV6-NTRK3 fusion transcripts and/or ETV6-region rearrangement in five of six CMNs and in five of five CFSs. The ETV6-NTRK3 fusion transcripts and/or ETV-region chromosome rearrangements were demonstrated in two CMNs and one CFS that lacked chromosome polysomies. These findings demonstrate that t(12;15) translocation, and the associated ETV6-NTRK3 fusion, can antedate acquisition of chromosome polysomies in CMN and CFS. CMN and CFS are pathogenetically related, and it is likely that they represent a single neoplastic entity, arising in either renal or soft tissue locations.
Congenital mesoblastic nephromas (CMNs) are uncommon renal tumors diagnosed generally within the first 3 months of life. 1 CMNs are characterized by a variably cellular proliferation of bland spindle cells arranged in interlacing bundles, and their clinical behavior is generally benign. 2 The CMNs described originally by Bolande in 1967 2 were characterized by low cellularity, but it was subsequently appreciated that, more commonly, CMNs are cellular and mitotically active and may even have necrosis. 3-5 These more cellular CMNs have polygonal cells and can adopt a storiform or diffuse growth pattern as well. The hypocellular examples are referred to as classic histology CMNs, whereas those with the more common sarcoma-like appearance are termed cellular. CMNs with admixtures of the two patterns are designated as having mixed histology. The clinical outcome in all morphological forms is excellent, particularly after complete resection with negative margins. 6-8 However, local recurrences and even metastases can occur after subtotal resection. 9,10
Congenital (infantile) fibrosarcomas (CFSs) are uncommon soft tissue tumors, principally arising in the extremities, which are also diagnosed generally in the first year of life. 11,12 CFSs have broad histological overlap with CMNs, and their clinical course is relatively benign, especially in comparison with the aggressive clinical behavior of histologically similar fibrosarcomas in adult patients. 13
Cytogenetic studies have demonstrated a strikingly similar profile, consisting of multiple polysomies, in CMN 14,15 and CFS. 16-23 The more cellular tumors, whether CMN or CFS, often have clonal polysomies of chromosomes 8, 11, 17, and/or 20. On the other hand, these polysomies have not been demonstrated in less cellular CMNs and CFSs, and acquisition of polysomies is associated with progression from classic to cellular histology in mixed histology CMN. 15,24 Hence, it is likely that the chromosomal polysomies are secondary oncogenic events, responsible in part for histological progression within these tumors. Presumably, other genomic aberrations are responsible for initial transformation of CMN nonneoplastic progenitor cells. Inasmuch as CMNs and CFSs share histological, clinical, and cytogenetic features, it is reasonable to hypothesize a common pathogenesis in these tumors. 15 Recently, CFSs were shown to contain a novel t(12;15)(p13;q25) translocation, resulting in ETV6-NTRK3 gene fusion. 25 To date, the t(12;15) translocation has not been reported in CMN. However, this translocation could have been overlooked when evaluated by conventional chromosome banding methods. This is because the regions exchanged between chromosomes 12 and 15 are similar in size and banding characteristics. Given the above mentioned evidence for common pathogenetic pathways in CMN and CFS, we evaluated whether the t(12;15) translocation and ETV6-NTRK3 fusion are present in CMN. Furthermore, we evaluated whether the translocation is present, and therefore a potential initial transforming event, in CMNs and CFSs lacking chromosomal trisomies.
Materials and Methods
The study group consisted of six CMNs and five CFSs that were excisionally removed and/or biopsied at Children’s Hospital, Boston, MA (Table 1) ▶ . Histological material was reviewed in all cases. The CMNs included one case with classic histology, two cases with cellular histology, and three cases with mixed histology. Fresh material was available from four CMNs and five CFSs for cell culture and cytogenetic analysis; the remaining two CMNs were available only as paraffin blocks. Frozen tumor material for RNA isolation and reverse transcription polymerase chain reaction (RT-PCR) was available from the same four CMNs that were karyotyped and from three CFSs. One CFS was analyzed both at time of original biopsy (CFS 5a) and at definitive resection 1 month later (CFS 5b).
Table 1.
Clinicopathological Features, Karyotypes, ETV6-NTRK3 Fusion, and FISH Findings in Congenital Mesoblastic Nephromas and Congenital Fibrosarcomas
| Case | Age | Sex | Site/histology | Karyotype | ETV6-NTRK3 fusion | FISH | ||
|---|---|---|---|---|---|---|---|---|
| ETV6 region | Chr 8 | Chr 11 | ||||||
| CMN 1 | 1D | M | Kidney/Cellular | 47,XY,+11,t(12;15)(p13;q26) | Yes | Rearranged | Di | Tri |
| CMN 2 | 1D | F | Kidney/Mixed | 46,XX | Yes | Rearranged | Di | Di |
| CMN 3 | 6D | M | Kidney/Cellular | 47,XY,+11,t(12;15)(p13;q26) | Yes | Rearranged | Di | Tri |
| CMN 4 | 3D | M | Kidney/Classic | 46,XY | No | Nonrearranged | Di | Di |
| CMN 5 | 16D | F | Kidney/Mixed | ND | ND | Rearranged | Di | Di |
| CMN 6 | 1D | M | Kidney/Mixed | ND | ND | Rearranged | Tri | Di |
| CFS 1 | 23D | M | Back | 48,XY,+11,add(15)(q26),+20 | Yes | Rearranged | Di | Tri |
| CFS 2 | 3D | F | Back | 51,XX,add(5)(p15),+8,del(10)(p11.2), | Yes | Rearranged | Tri | Tri |
| +11,t(12;15)(p13;q26),+15,+17,+20 | ||||||||
| CFS 3 | 6M | M | Hand | 50,XY,+8,+8,+11,+11, | Yes | Rearranged | Tet | Tet |
| t(12;15)(p13;q26) | ||||||||
| CFS 4 | 11D | F | Neck | 49,XX,+11,t(12;15)(p13;q26), | ND | Rearranged | Di | Di |
| +del(17)(p12),+20 | ||||||||
| CFS 5a | 7D | F | Forearm | 46,XX | ND | Rearranged | Di | Di |
| CFS 5b | 1M | F | Forearm | 49,XX,+8,+11,+20 | ND | Rearranged | Tri | Tri |
CMN, congenital mesoblastic nephroma; M, male; F, female; CFS, congenital fibrosarcoma; D, day; M, month; ND, not determined; Chr, chromosome; Di, disomic; Tri, trisomic; Tet, tetrasomic.
Cytogenetics
CMN and CFS specimens were processed for cytogenetic analysis immediately after biopsy. A 2- to 3-mm 3 portion of each specimen was minced with scalpels, disaggregated with collagenase, and cultured as described previously. 26 Metaphase harvesting, fixation in 3:1 methanol:acetic acid, slide making, and trypsin-Giemsa staining were also performed as described previously. 26 Metaphase cells were harvested within 3 to 7 days after establishing the primary cultures.
FISH
Four-micron-thick, paraffin-embedded sections were prepared on silane-coated slides and baked overnight at 65°C. Tissue section pretreatment and proteinase K digestion were accomplished using the Oncor Tissue Kit (Oncor, Gaithersburg, MD), according to the manufacturer’s recommendations. Cytogenetic preparations were dehydrated and denatured according to standard protocols. 27 Hybridization and washing steps, for both tissue sections and cytogenetic preparations, were also performed according to standard protocols. 27 Rearrangements of the ETV6 region were evaluated by dual-color fluorescence in situ hybridization (FISH) using flanking yeast artificial chromosome (YAC) clones 788_g_5 (telomeric) and 916_d_8 (centromeric). YACs 788_g_5 and 916_d_8 were digoxigenin and biotin labeled, respectively. Numerical aberrations of chromosomes 8 and 11 were evaluated using D8Z2 and D11Z1 pericentromeric α-satellite probes, which were biotin and digoxigenin labeled, respectively. Chromosomes 17 and 20 were not evaluated by FISH, although these chromosomes are also involved frequently in CMN and CFS polysomies; very few CMNs or CFSs have chromosome 17 or 20 polysomies in the absence of chromosome 8 or 11 polysomies. FISH probes were detected using avidin-Texas Red (Vector, Burlingame, CA) and FITC anti-digoxigenin (Boehringer, Indianopolis, IN), respectively, and all slides were counterstained with 0.1 to 1.0 mg/ml 4,6-diamidino-2-phenylindole-dihydrochloride (DAPI). One hundred nonoverlapping interphase nuclei were scored for each paraffin section, and images were captured using a charge-coupled device camera (Photometrics, Tucson, AZ). The criteria used in scoring FISH signals were as recommend by Hopman et al. 28 Tumors were classified as trisomic or tetrasomic if more than 5% of nuclei had three or four pericentromeric α-satellite signals, respectively. Tumors were classified as ETV6-region rearranged if more than 20% of nuclei contained wide splits between the centromeric and telomeric ETV6-region YAC clones.
RT-PCR and DNA Sequencing
Total RNA was extracted from 20 to 30 mg of frozen tissue using Trizol (Gibco, Gaithersburg, MD), according to the manufacturer’s protocol. The resultant RNA pellets were dissolved in 25 μl of dH2O, and 1 μl of the RNA solution was reverse transcribed using random primers (GeneAmp Kit, Perkin Elmer, Norwalk, CT). Semi-nested PCR was performed using two ETV6 forward primers (F/ETV6/541, 5′-CCTCCCACCATTGAACTGTT-3′ 29 and F/ETV6/701, 5′-AGAACAACCACCAGGAGTCC-3′) 29 and a NTRK3 reverse primer (R/NTRK3/1838, 5′-CCGCACACTCCATAGAACTTGAC-3′). 25 First-round PCR was with F/ETV6/541 and R/NTRK3/1838 at 95°C for 15 seconds and 60°C for 2 minutes for 30 cycles. Second-round PCR was with F/ETV6/701 and R/NTRK3/1838 at 94°C for 2 minutes, then 94°C for 30 seconds, 60 to 55°C (touchdown) for 30 seconds, and 72°C for 1 minute for 10 cycles, and then 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute for 25 cycles. Five microliters of the second-round PCR product was electrophoresed on a 0.9% agarose gel containing ethidium bromide, and DNA fragments were purified using the Qiaquick gel extraction kit (Qiagen, Valencia, CA). All fragments were sequenced in forward and reverse directions using the F/ETV6/701 and R/NTRK3/1838 primers, respectively, by cycle sequencing with ABI BigDye terminators. Sequences were analyzed using an ABI Prism 377 sequencer.
Results
Clinical Information and Pathological Findings
The clinicopathological data are summarized in Table 1 ▶ . One CMN exhibited the classic histological pattern with thick interlacing bundles of elongate eosinophilic spindle cells with delicate cytoplasm (Figure 1A) ▶ . The neoplastic cells entrapped normal renal structures, and mitoses were rare. The cellular variants were composed of more polygonal or short spindle cells (Figure 1C) ▶ . They were diffusely cellular, contained focal necrosis, had numerous mitoses, and had pushing borders. The mixed variants consisted of an admixture of discrete areas characteristic of both the classic and cellular variants (Figure 1B) ▶ . The CFSs were composed of a monomorphic population of densely packed polygonal or short spindle cells with minimal pleomorphism and a fascicular growth pattern (Figure 1D) ▶ . There were numerous mitoses, and some cases contained focal areas of necrosis. The CFSs bore a striking resemblance to the cellular variants of CMN (Figure 1, C and D) ▶ .
Figure 1.
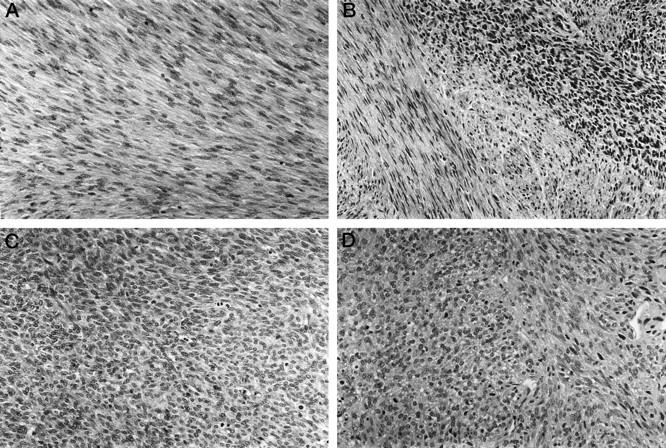
H&E-stained sections of classic CMN (A), mixed CMN (B), and cellular CMN (C) and CFS (D) are shown. The classic CMN consists of a moderately cellular proliferation of interlacing bundles of spindle cells whereas the cellular CMN exhibits a more densely cellular histology with increased mitotic activity. The mixed CMN contains a mixture of the two patterns. The CFS is very similar in appearance to the cellular CMN.
Cytogenetics
The t(12;15) translocation was subtle cytogenetically (Figure 2, A and B) ▶ and was manifested primarily by loss of the normal terminal dark band from the long arm of chromosome 15. This translocation was overlooked at the time of original cytogenetic analysis in most cases. However, review of all karyotypes, prompted by description of CFS t(12;15) translocations by Knezevich et al, 25 suggested similar t(12;15) translocations in two CMNs and three CFSs (Table 1) ▶ . Another case, CFS 2, had a rearrangement of the NTRK3 region (chromosome band 15q26) that could not be ascribed to t(12;15) based on the banding study. Additional chromosome aberrations were found in most tumors. Two cellular CMNs contained trisomy 11, whereas two mixed histology CMNs lacked apparent chromosome aberrations (Table 1) ▶ . Each of five CFSs contained polysomies of chromosomes 8, 11, 17, and/or 20 (Table 1) ▶ .
Figure 2.
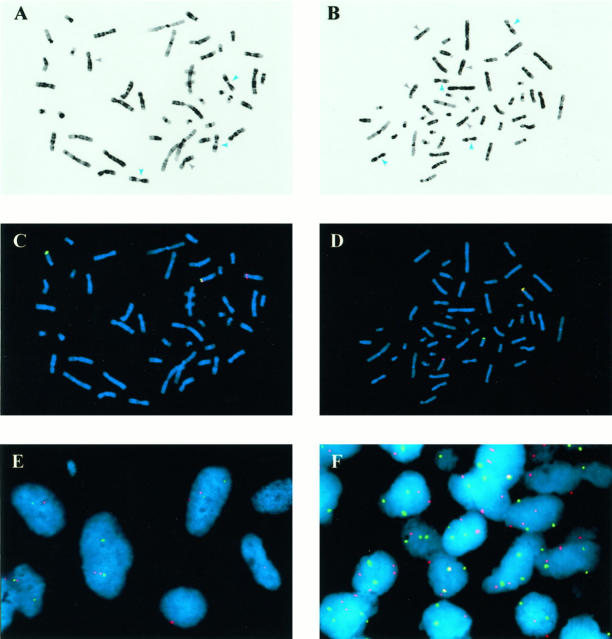
Molecular cytogenetic analyses of CMN and CFS. A and B: Giemsa emulations are derived from DAPI-stained metaphase cells of CMN 3 (A) and CFS 3 (B). Gray and blue arrows indicate chromosome 8 and 11 homologs, respectively. CMN 3 has disomy 8 and trisomy 11; CFS 3 has tetrasomy 8 and tetrasomy 11. C and D: ETV6-region FISH for the same metaphase cells shown in A and B. The t(12;15) translocations are revealed by splitting of the centromeric (rhodamine detection is red) and telomeric (FITC detection is green) ETV6-region FISH probes. E: ETV6-region FISH in 4-μm section from CMN 3. Several nuclei in center of field show wide splitting of the centromeric (red) and telomeric (green) components of the FISH probe. F: Chromosome 8 (rhodamine is red) and 11 (FITC is green) FISH in 4-μm paraffin section from CMN 3. Several nuclei show two copies of chromosome 8 and 3 copies (trisomy) of chromosome 11.
FISH
FISH analyses revealed rearrangement of the ETV6 gene region in five of six CMNs and five of five CFSs (Table 1 ▶ ; Figure 2, C–F ▶ ). Correlations with chromosome 8 and 11 polysomies were determined both by Giemsa emulation in DAPI-stained FISH metaphase cell preparations (Figure 2, A and B) ▶ and by sequential FISH analyses using chromosome 8 and 11 pericentromeric α-satellite probes (Figure 2F) ▶ . These analyses demonstrated that all tumors containing chromosome 8 and 11 polysomies also contained ETV6-region rearrangements. By contrast, ETV6 rearrangements were demonstrable in three specimens, CMN 2, CMN 5, and CFS 5a, which lacked chromosome polysomies. CFS 5a was a diagnostic needle biopsy in a 7-day-old girl, whereas CFS 5b, containing trisomies 8 and 11 along with the ETV6-region rearrangement, was the subsequent resection performed 3 weeks later. The CFS 5 data are consistent with intratumor cytogenetic heterogeneity resulting from acquisition of chromosomal trisomies in an ETV6-rearranged tumor population.
The rationale in undertaking the FISH chromosome 8 and 11 studies was to determine, particularly in mixed histology CMN, whether chromosome polysomies might be restricted to more cellular regions whereas ETV6-region rearrangements might be found in both less cellular and more cellular regions. However, because of the complex admixture of less cellular and more cellular areas in a given histological section, combined with a relative loss of histological detail after proteinase K treatment, it was difficult to ascertain whether a particular nucleus was in a more cellular or less cellular region within an individual case of mixed histology CMN.
RT-PCR and DNA Sequencing
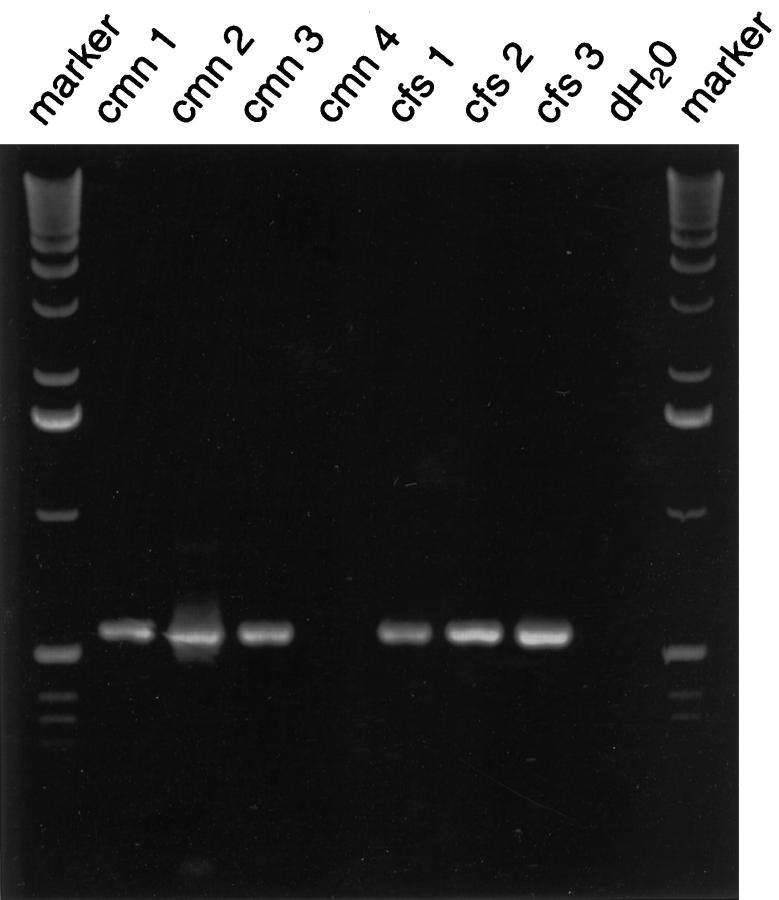
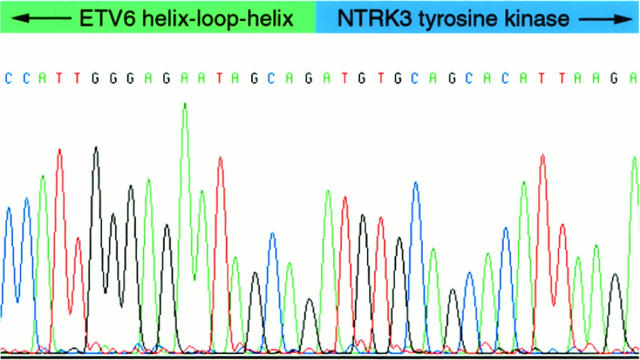
Nested RT-PCR, using ETV6 forward primers and an NTRK3 reverse primer, revealed ∼550-bp fragments in three of four CMNs and three of three CFSs (Figure 3) ▶ . Forward and reverse sequencing for each of these fragments demonstrated ETV6-NTRK3 fusion transcripts (Figure 4) ▶ , identical to those reported previously in CFS (GenBank accession number AF041811). No sequence variations were detected in any of these six fusion transcripts. The corresponding full-length ETV6-NTRK3 fusion transcripts, as demonstrated by Knezevich et al, 25 encode ETV6 helix-loop-helix and NTRK3 tyrosine kinase domains.
Figure 3.
ETV6-NTRK3 RT-PCR. Fusion transcripts are seen in three of four CMNs and in each of three CFSs. dH2O is control RT-PCR with no RNA template.
Figure 4.
Sequence analysis of ETV6-NTRK3 fusion cDNA from CMN 2. Identical fusion sequences were identified in three CMNs and three CFSs.
Discussion
Significant strides have been made in the histological classification of solid tumors, and both cytogenetic and molecular markers have assisted in establishing the distinctive identities and pathogenesis of certain tumors. However, molecular classification schema are less well developed for solid tumors than for hematological malignancies. One group of solid tumors that has been intensively and productively characterized, by cytogenetic and molecular approaches, are the mesenchymal neoplasms. Diagnostic chromosome translocations, often affecting genes that encode DNA-binding proteins, have been identified in soft-tissue neoplasms at both the benign and malignant ends of the spectrum. These diagnostic translocations have been useful in establishing pathogenetic relationships between neoplasms that have been regarded as different entities. A notable example is the role of the t(11;22) translocation, associated with EWS-FLI1 gene fusion, in supporting a common pathogenesis and a common cell lineage of origin in Ewing’s sarcoma and peripheral primitive neuroectodermal tumors. 30 Another example is the genetic characterization of dermatofibrosarcoma protuberans and giant-cell fibroblastoma, which were shown to contain the same t(17;22) translocation, resulting in deregulated expression of platelet-derived growth factor B. This evidence of a common pathogenesis is notable because giant-cell fibroblastomas have been regarded as juvenile (pediatric) forms of dermatofibrosarcoma protuberans. 31
Demonstration of a molecular relationship, in and of itself, does not establish a more general relationship between different clinicopathological tumor entities. It is well known, for example, that a wide variety of neoplasms acquire inactivating mutations of the same tumor suppressor genes. However, specific chromosome translocations are typically found only in tumors of related, or identical, histogenesis and pathogenesis. This is because nonrandom chromosome translocations involve juxtaposition, and often fusion, of genes from each of the participating chromosomes. These gene rearrangements are functional only if one of the genes is transcriptionally active in the nonneoplastic progenitor cell and if one or both of the genes, when overexpressed or rearranged, can serve an oncogenic role in that cell. Observations to date suggest that a specific translocation is unlikely to play a transforming role, in vivo, in widely divergent cell lineages. 32
Our studies reveal identical chromosome translocations, associated with ETV6-NTRK3 fusion transcripts, in CFS and CMN. CFS and CMN arise in the soft tissues and kidney, respectively, and share many clinicopathological features. CFSs are histologically similar, but clinically distinct, from fibrosarcomas in older children and adults. 11-13,16,33-36 Many CFSs follow a benign clinical course despite worrisome histological features, whereas adult fibrosarcomas are often lethal. Likewise, CMNs, which are very similar histologically to CFSs, are generally cured by complete resection or nephrectomy. 2,3 Karyotypic and molecular cytogenetic studies also support a pathogenetic relationship between CFS and CMN. Both tumors, particularly in cases with greater degrees of cellularity, are associated with gains of chromosomes 8, 11, 17, and 20. 14-23 In the present study, we demonstrate ETV6-NTRK3 fusion transcripts and/or ETV6-region chromosomal rearrangement in five of six CMNs and in five of five CFSs. Notably, Knezevich et al demonstrated that the CFS-associated t(12;15)(p13;q25) translocation is not found in adult fibrosarcomas. 25 Therefore, the accumulated evidence indicates that CFSs and CMNs are closely related neoplasms, which are distinct, clinically and pathogenetically, from adult fibrosarcomas. It remains unclear, however, whether CMN and CFS are the same entity, differing only in site of origin. We favor this viewpoint, given that the histological, clinical, cytogenetic, and molecular evidence support a common histogenesis and pathogenesis.
Cytogenetic t(12;15) translocations were not identified in several cases in this series (CMN4, CFS1, and CFS5), although RT-PCR and FISH analyses revealed ETV6-region rearrangements in those same cases (Table 1) ▶ . The chromosome banding was of average quality in these cases, but cytogenetic recognition of the t(12;15) translocation requires superior banding quality. Therefore, we view these cytogenetic analyses as uninformative, rather than negative, for the translocation. Given this experience, we would be reluctant to exclude a t(12;15) translocation, in CMN or CFS, based solely on chromosome banding findings.
Acquisition of the above mentioned chromosome polysomies is associated with histological progression in CMN. 15,24 The polysomies are often acquired only as CMNs become more cellular, and they are unlikely to be the oncogenetic events responsible for initial neoplastic transformation of the nonmalignant progenitor cells. Our present findings suggest that ETV6-NTRK3 fusion might represent the initial transforming event. This possibility is supported by demonstration of ETV6-region rearrangement, and/or ETV6-NTRK3 fusion, in two CMNs (cases CMN 2 and CMN 5) and one CFS (case CFS 5a) that lacked detectable chromosome polysomies (Table 1) ▶ . It is also notable that the original diagnostic needle biopsy, CFS 5a, lacked chromosome polysomies but had ETV6-region rearrangement, whereas the subsequent resection, CFS 5b, contained trisomy 8, 11, and 20, in addition to the ETV6-region rearrangement.
ETV6 (also known as TEL) was originally characterized as an oncogene in several types of leukemias and myeloproliferative syndromes. 37,38 ETV6 translocations, in these hematopoietic neoplasms, involve a variety of partner genes. Several of the ETV6 translocation partners, including PDGFRβ, ABL, and JAK2, 29,37-39 are tyrosine kinase genes; the transcripts associated with these translocations consist of the ETV6 5′ end fused to the 3′ end of the tyrosine kinase gene. The corresponding oncoproteins include the ETV6 helix-loop-helix (HLH) domain (amino-terminal end) and a tyrosine kinase domain (carboxyl-terminal end). The oncogenic mechanism, in the well characterized ETV6-ABL and ETV6-PDGFRβ fusion oncoproteins, involves ETV6 HLH-mediated dimerization, resulting in constitutive tyrosine kinase catalytic activity. 29,40,41 Similarly, HLH-mediated ETV6-NTRK3 homodimerization might engender ligand-independent activation of the NTRK3 tyrosine kinase, leading to autophosphorylation of specific tyrosine residues and activation of p21ras-related signal transduction cascades. 42 NTRK3 expression has been observed primarily in neuronal cells 43,44 and in neuroectodermal tumors, and Knezevich et al did not detect NTRK3 expression in fibroblasts. 25 These observations suggest that unscheduled NTRK3 tyrosine kinase domain expression, mediated by the ETV6 promotor, is important in CMN and CFS oncogenic transformation. However, as discussed above, it is also likely that ETV6 performs an oncogenic role above and beyond driving NTRK3 transcription. The critical role of the ETV6 HLH domain is evidenced by the invariant sequence of the ETV6-NTRK3 fusion transcripts in the six CFSs and CMNs reported herein and in the three CFSs reported by Knezevich et al. 25
Only one of six CMNs (case CMN 4) in this study lacked the t(12;15) translocation or ETV6-region rearrangement (Table 1) ▶ . It is reasonable to question the histological diagnosis in this case, because low-grade fibrous lesions of infancy represent a difficult area in pathological diagnosis. The differential diagnosis of CMN is complex, including such entities as fibromatosis, clear-cell sarcoma, stroma-predominant Wilms’ tumor, and low-grade malignant peripheral nerve sheath tumor. However, even upon additional review, this tumor was believed to be a CMN. It is possible that neither ETV6 nor NTRK3 was oncogenically activated in this case, but it is also possible that NTRK3 was activated by a point mutation undetected by the cytogenetic and RT-PCR assays. There is ample precedence for activating point mutations in other receptor tyrosine kinase (RTK) oncogenes. Oncogene mutations in RTK extracellular, transmembrane, or juxtamembrane domains can affect tyrosine kinase activity by promoting dimerization. 45 Other activating mutations modulate tyrosine kinase activity through direct involvement of the catalytic domain. 45
In summary, we have established that CMNs contain the same t(12;15)(p13;q25) translocation described recently in CFS. This translocation is associated with an ETV6-NTRK3 fusion gene, in which the ETV6 HLH domain is coupled with the NTRK3 tyrosine kinase domain. ETV6-NTRK3 fusion appears to be an early event in the oncogenesis of CMN and CFS, antedating the acquisition of several characteristic chromosome polysomies. Clinical behavior, histological features, cytogenetics, and molecular data all suggest a close relationship between CMN and CFS. In fact, the evidence is strong that these are one and the same neoplasm, albeit presenting in different anatomic sites.
Footnotes
Address reprint requests to Dr. Jonathan A. Fletcher, Department of Pathology, Brigham and Women’s Hospital, 75 Francis Street, Boston, MA 02115. E-mail: fletcher@bustoff.bwh.harvard.edu.
References
- 1.Marsden HB, Lawler W: Primary renal tumours in the first year of life: a population based review. Virchows Arch A 1983, 399:1-9 [DOI] [PubMed] [Google Scholar]
- 2.Bolande RP, Brough AJ, Izant RJ: Congenital mesoblastic nephroma of infancy: a report of eight cases and the relationship to Wilms’ tumor. Pediatrics 1967, 40:272-278 [PubMed] [Google Scholar]
- 3.Bolande RP: Congenital mesoblastic nephroma of infancy. Perspect Pediatr Pathol 1973, 1:227-250 [PubMed] [Google Scholar]
- 4.Richmond H, Dougall AJ: Neonatal renal tumors. J Pediatr Surg 1970, 5:413-418 [DOI] [PubMed] [Google Scholar]
- 5.Wigger HJ: Fetal hamartoma of kidney: a benign, symptomatic, congenital tumor, not a form of Wilms’ tumor. Am J Clin Pathol 1969, 51:323-337 [DOI] [PubMed] [Google Scholar]
- 6.Howell CH, Othersen HB, Kiviat NE, Norkool P, Beckwith JB, D’Angio GJ: Therapy and outcome in 51 children with mesoblastic nephroma: a report of the National Wilms’ Tumor Study. J Pediatr Surg 1982, 17:826-831 [DOI] [PubMed] [Google Scholar]
- 7.Chan HS, Cheng MY, Mancer K, Payton D, Weitzman SS, Kotecha P, Daneman A: Congenital mesoblastic nephroma: a clinicoradiologic study of 17 cases representing the pathologic spectrum of the disease. J Pediatr 1987, 111:64-70 [DOI] [PubMed] [Google Scholar]
- 8.Sandstedt B, Delemarre JFM, Krul EJ, Tournade MF: Mesoblastic nephromas: a study of 29 tumors from the SIOP nephroblastoma file. Histopathology 1985, 9:741-750 [DOI] [PubMed] [Google Scholar]
- 9.Joshi VV, Kasznica J, Walters TR: Atypical mesoblastic nephroma. Arch Pathol Lab Med 1986, 110:100-106 [PubMed] [Google Scholar]
- 10.Gonzalez-Crussi F, Sotelo-Avila C, Kidd JM: Malignant mesenchymal nephroma of infancy: report of a case with pulmonary metastases. Am J Surg Pathol 1980, 4:185-190 [DOI] [PubMed] [Google Scholar]
- 11.Stout AP: Fibrosarcomas in infants and children. Cancer 1962, 15:1028-1040 [DOI] [PubMed] [Google Scholar]
- 12.Coffin CM, Jaszcz W, O’Shea PA, Dehner LP: So-called congenital-infantile fibrosarcoma: does it exist and what is it? Pediatr Pathol 1994, 14:133-150 [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki H, Enjoji M: Infantile and adult fibrosarcomas of the soft tissues. Acta Pathol Jpn 1979, 29:377. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein R, Zeltzer PM, Lin F, Carpenter PM: Trisomy 11 and other nonrandom trisomies in congenital fibrosarcoma. Cancer Genet Cytogenet 1994, 78:82-86 [DOI] [PubMed] [Google Scholar]
- 15.Schofield DE, Yunis EJ, Fletcher JA: Chromosome aberrations in mesoblastic nephroma. Am J Pathol 1993, 143:714-724 [PMC free article] [PubMed] [Google Scholar]
- 16.Schofield DE, Fletcher JA, Grier HE, Yunis EJ: Fibrosarcoma in infants and children: application of new techniques. Am J Surg Pathol 1994, 18:14-24 [DOI] [PubMed] [Google Scholar]
- 17.Adam LR, Davison EV, Malcolm AJ, Pearson AD, Craft AW: Cytogenetic analysis of a congenital fibrosarcoma. Cancer Genet Cytogenet 1991, 52:37-41 [DOI] [PubMed] [Google Scholar]
- 18.Argyle JC, Tomlinson GE, Stewart D, Schneider NR: Ultrastructural, immunocytochemical, and cytogenetic characterization of a large congenital fibrosarcoma. Arch Pathol Lab Med 1992, 116:972-975 [PubMed] [Google Scholar]
- 19.Dal Cin P, Brock P, Casteels-Van Daele M, De Wever I, Van Damme B, Van den Berghe H: Cytogenetic characterization of congenital or infantile fibrosarcoma. Eur J Pediatr 1991, 150:579-581 [DOI] [PubMed] [Google Scholar]
- 20.Gorman PA, Malone M, Pritchard J, Sheer D: Deletion of part of the short arm of chromosome 17 in a congenital fibrosarcoma. Cancer Genet Cytogenet 1990, 48:193-198 [DOI] [PubMed] [Google Scholar]
- 21.Mandahl N, Heim S, Rydholm A, Willen H, Mitelman F: Nonrandom numerical chromosome aberrations (+8, +11, +17, +20) in infantile fibrosarcoma. Cancer Genet Cytogenet 1989, 40:137-139 [DOI] [PubMed] [Google Scholar]
- 22.Sankary S, Dickman PS, Wiener E, Robichaux W, Swaney WP, Malone PS, Gollin SM: Consistent numerical chromosome aberrations in congenital fibrosarcoma. Cancer Genet Cytogenet 1993, 65:152-156 [DOI] [PubMed] [Google Scholar]
- 23.Speleman F, Dal Cin P, De Potter K, Laureys G, Roels HJ, Leroy J, Van Den Berghe H: Cytogenetic investigation of a case of congenital fibrosarcoma. Cancer Genet Cytogenet 1989, 39:21-24 [DOI] [PubMed] [Google Scholar]
- 24.Mascarello JT, Cajulis TR, Krous HF, Carpenter PM: Presence or absence of trisomy 11 is correlated with histologic subtype in congenital mesoblastic nephroma. Cancer Genet Cytogenet 1994, 77:50-54 [DOI] [PubMed] [Google Scholar]
- 25.Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH: A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nature Genet 1998, 18:184-187 [DOI] [PubMed] [Google Scholar]
- 26.Fletcher JA, Kozakewich HP, Hoffer FA, Lage JM, Weidner N, Tepper R, Pinkus GS, Morton CC, Corson JM: Diagnostic relevance of clonal cytogenetic aberrations in malignant soft-tissue tumors. N Engl J Med 1991, 324:436-442 [DOI] [PubMed] [Google Scholar]
- 27.Xiao S, Renshaw AA, Cibas ES, Hudson TJ, Fletcher JA: Novel fluorescence in situ hybridization approaches in solid tumors: characterization of frozen specimens, touch preparations, and cytological preparations. Am J Pathol 1995, 147:896-904 [PMC free article] [PubMed] [Google Scholar]
- 28.Hopman AH, Ramaekers FC, Raap AK, Beck JL, Devilee P, van der Ploeg M, Voojis GP: In situ hybridization as a tool to study numerical chromosome aberrations in solid bladder tumors. Histochemistry 1988, 89:307-316 [DOI] [PubMed] [Google Scholar]
- 29.Carroll M, Tomasson MH, Barker GF, Golub TR, Gilliland DG: The TEL/platelet-derived growth factor β receptor (PDGFβR) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGFβR kinase-dependent signaling pathways. Proc Natl Acad Sci USA 1996, 93:14845-14850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delattre O, Zucman J, Melot T, Garau XS, Zucker JM, Lenoir GM, Ambros PF, Sheer D, Turc-Carel C, Triche TJ, Aurias A, Thomas G: The Ewing family of tumors: a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med 1994, 331:294-299 [DOI] [PubMed] [Google Scholar]
- 31.Simon MP, Pedeutour F, Sirvent N, Grosgeorge J, Minloletti F, Coindre JM, Terrier-Lacomb MJ, Mandahl N, Craver RD, Blin N, Sozzi G, Turc-Carel C, O’Brien KP, Kedra D, Fransson I, Guilbaud C, Dumanski JP: Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nature Genet 1997, 15:95-98 [DOI] [PubMed] [Google Scholar]
- 32.Rabbitts TH: Chromosomal translocations in human cancer. Nature 1994, 372:143-149 [DOI] [PubMed] [Google Scholar]
- 33.Balsaver AM, Butler JJ, Martin RG: Congenital fibrosarcoma. Cancer 1967, 20:1607-1616 [DOI] [PubMed] [Google Scholar]
- 34.Chung EB, Enzinger FM: Infantile fibrosarcoma. Cancer 1976, 38:729-739 [DOI] [PubMed] [Google Scholar]
- 35.Dahl I, Save-Soderbergh J, Angervall L: Fibrosarcoma in early infancy. Pathol Eur 1973, 8:193-209 [PubMed] [Google Scholar]
- 36.Hays DM, Mirabal VQ, Karlan MS, Patel HR, Landing BH: Fibrosarcomas in infants and children. J Pediatr Surg 1970, 5:176-183 [DOI] [PubMed] [Google Scholar]
- 37.Papadopoulos P, Ridge SA, Boucher CA, Stocking C, Wiedemann LM: The novel activation of ABL by fusion to an ets-related gene, TEL. Cancer Res 1995, 55:34-38 [PubMed] [Google Scholar]
- 38.Golub TR, Barker GF, Lovett M, Gilliland DG: Fusion of PDGF receptor β to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 1994, 77:307-316 [DOI] [PubMed] [Google Scholar]
- 39.Peeters P, Raynaud SD, Cols J, Wlodarska I, Grosgeorge J, Philip P, Monpoux F, Van Rompaey L, Baens M, Van den Berghe H, Marynen P: Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood 1997, 90:2535-2540 [PubMed] [Google Scholar]
- 40.Okuda K, Golub TR, Gilliland DG, Griffin JD: p210BCR/ABL, p190BCR/ABL, and TEL/ABL activate similar signal transduction pathways in hematopoietic cell lines. Oncogene 1996, 13:1147-1152 [PubMed] [Google Scholar]
- 41.Golub TR, Goga A, Barker GF, Afar DE, McLaughlin J, Bohlander SK, Rowley JD, Witte ON, Gilliland DG: Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol Cell Biol 1996, 16:4107-4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephens RM, Loeb DM, Copeland TD, Pawson T, Greene LA, Kaplan DR: Trk receptors use redundant signal transduction pathways involving SHC and PLC-gamma 1 to mediate NGF responses. Neuron 1994, 12:691-705 [DOI] [PubMed] [Google Scholar]
- 43.Lamballe F, Klein R, Barbacid M: trkC, a new member of the trk family of tyrosine protein kinases, is a receptor for neurotrophin-3. Cell 1991, 66:967-979 [DOI] [PubMed] [Google Scholar]
- 44.Shelton DL, Sutherland J, Gripp J, Camerato T, Armanini MP, Phillips HS, Carroll K, Spencer SD, Levinson AD: Human trks: molecular cloning, tissue distribution, and expression of extracellular domain immunoadhesins. J Neurosci 1995, 15:477-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM, Bergsagel PL: Frequent translocation t(4;14)(p16.3;q32.2) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nature Genet 1997, 16:260-264 [DOI] [PMC free article] [PubMed] [Google Scholar]