Abstract
The c-ski protooncogene encodes a transcription factor that binds DNA only in association with other proteins. To identify co-binding proteins, we performed a yeast two-hybrid screen. The results of the screen and subsequent co-immunoprecipitation studies identified Smad2 and Smad3, two transcriptional activators that mediate the type β transforming growth factor (TGF-β) response, as Ski-interacting proteins. In Ski-transformed cells, all of the Ski protein was found in Smad3-containing complexes that accumulated in the nucleus in the absence of added TGF-β. DNA binding assays showed that Ski, Smad2, Smad3, and Smad4 form a complex with the Smad/Ski binding element GTCTAGAC (SBE). Ski repressed TGF-β-induced expression of 3TP-Lux, the natural plasminogen activator inhibitor 1 promoter and of reporter genes driven by the SBE and the related CAGA element. In addition, Ski repressed a TGF-β-inducible promoter containing AP-1 (TRE) elements activated by a combination of Smads, Fos, and/or Jun proteins. Ski also repressed synergistic activation of promoters by combinations of Smad proteins but failed to repress in the absence of Smad4. Thus, Ski acts in opposition to TGF-β-induced transcriptional activation by functioning as a Smad-dependent co-repressor. The biological relevance of this transcriptional repression was established by showing that overexpression of Ski abolished TGF-β-mediated growth inhibition in a prostate-derived epithelial cell line.
Keywords: ski oncogene, transcription, melanoma
The products of the cellular and retroviral ski genes are nuclear proteins that either activate or repress transcription, depending on the cellular and promoter context (1–6). The v-Ski oncoprotein of the SKV retrovirus (7) is truncated by 312 amino acids relative to its cellular homolog, c-Ski (8), but this truncation per se is not oncogenic. Rather, overexpression of c-ski or v-ski similarly induces morphological transformation and anchorage-independent growth in avian fibroblasts (9). Ski overexpression also induces muscle differentiation in embryo fibroblasts and causes postnatal hypertrophy of type II fast muscle fibers in transgenic mice (2, 3, 10). Moreover, germline inactivation in mice demonstrates that loss of ski function results in decreased myofiber development in addition to other abnormalities (11).
c-ski is highly expressed in several human tumor cell lines, including those derived from neuroblastomas and carcinomas of the stomach, vulva, and prostate (12). During mouse development, ski is expressed at relatively high levels in neural crest cells, and human ski is expressed at elevated levels in many human melanomas (13), which are tumors of neural crest-derived cells. This finding is of interest in light of early studies showing that v-ski induces transformation of pigmented primary melanocytes (14).
c-Ski and v-Ski do not bind DNA on their own, but we have shown that they participate in a multiprotein complex that binds specifically to the consensus element, GTCTAGAC (5). Binding to this element mediates transcriptional repression by Ski, and these activities are tightly linked to Ski's ability to transform cells and induce muscle differentiation (6). Transcriptional repression is an intrinsic property of an N-terminal domain of Ski, which is capable of repressing transcription when fused to a heterologous DNA binding domain (5, 6). Ski has also been shown to act as a co-repressor of the retinoic acid receptor, and that activity is linked to its ability to transform hematopoietic cells (15). In this context, repression by Ski may be accomplished through interactions with N-CoR and mSin3A (16).
The GTCTAGAC sequence that we had found to be bound by Ski-containing protein complexes was subsequently identified as a Smad binding element (SBE) that is bound directly by Smad3 and Smad4 (17). These proteins and the related Smad2 are transcription factors that activate gene expression in response to type β transforming growth factor (TGF-β). Smad2 and Smad3 are direct substrates of activated TGF-β receptors and signal directly to the nucleus (18, 19). On TGF-β receptor activation, Smad2 and Smad3 become phosphorylated and form heteromeric complexes with Smad4 (18). These complexes translocate to the nucleus, where they activate the transcription of target genes. Smad2 and Smad3 proteins synergize with Smad4 in stimulating TGF-β-induced transcription (20, 21).
TGF-β is a member of a family of proteins that signal a wide variety of biological responses through transcriptional regulation of genes encoding critical determinants of cell fate, cell-cycle progression, differentiation, extracellular matrix association, motility, and death (22). TGF-β signaling has been implicated in the transformation of normal melanocytes to melanoma and in the progression of several other human tumors (23, 24). Inactivation of this pathway, by mutation of the TGF-β receptor or Smad proteins, has been observed in a variety of human cancers (reviewed in ref. 22).
The present studies were prompted by our interest in defining the role of Ski in human cancers expressing high levels of this protein. Because Ski modulates transcription through protein-protein interactions, we sought to identify protein(s) that interact with Ski. Here we provide evidence that Smad2 and Smad3 are two Ski-interacting proteins that form a complex with the SBE. We show that Ski acts as a Smad-dependent co-repressor of TGF-β-responsive promoters via the SBE. Repression of TGF-β responsive genes by Ski is opposite of the usual activation mediated by the Smad proteins, suggesting the Ski might counteract biological responses to this cytokine. Indeed, we show that overexpression of Ski abrogates the TGF-β-induced block of proliferation in prostate epithelial cells.
Materials and Methods
Cell Culture.
The human melanoma cell lines UCD-Mel-N and a derivative line overexpressing the human ski gene, UCD-Mel-N(Ski+), were cultured as described for other melanoma cell lines (25). UCD-Mel-N (Ski+) cells were generated by infection of the parental lines with an amphotropic recombinant retrovirus (pBABEpuro-H-ski) (26). Mink lung epithelial cells CCL-64 (Mv1Lu) were obtained from the American Type Culture Collection (ATCC) and were maintained in penicillin, streptomycin, and nonessential amino acids. The HCT116 human colon carcinoma cell line and its Smad4-deficient derivative (27) (generous gifts of B. Vogelstein, Johns Hopkins University) were maintained in DMEM with 10% FBS and gentamycin. Chicken embryo fibroblasts (CEFs) and CEFs transformed by infection with a c-ski retroviral vector (cSki-CEFs) were obtained and cultured as described (9).
The rat prostate cell line DP-153 was infected with the amphotropic retroviruses pBABEpuro-H-ski and pBABE-puro. Infected cells were plated in 12-well Falcon dishes at 8 × 104 cells/well with 2 ml of DMEM/F12 (vol/vol, 1:1) supplemented with 15 mM Hepes, 1% calf serum, and 0.1 μM dexamethasone. After overnight culture for attachment, wells received either 10 ng/ml of recombinant human TGF-β1 or vehicle (2 μl of 4 mM HCl, 1 mg/ml BSA). Cells were trypsinized off dishes into single cell suspensions and were counted daily with a hemocytometer. The values represent the average of triplicate cultures ± standard deviation.
Construction of the Yeast Two-Hybrid cDNA Library.
Poly(A)+ mRNA (5 μg) from IIB-Mel-J cells was used to prepare cDNAs with an EcoRI site at their 5′ends and a XhoI site at their 3′ ends by standard procedures. The cDNA fragments of 0.5–3.0 kilobase pairs were cloned into the HybriZAP yielding 3 × 106 independent plaques, more than 90% of which contains a cDNA insert. A phagemid cDNA library with the cDNAs fused to the Gal4 activation domain was isolated from the lambda HybriZAP library by in vivo excision with Exassist helper phages according to the manufacturer's recommendation (Stratagene).
Plasmids.
A cDNA encoding the full-length human Ski (pSHH-ski) was cloned in frame with the Gal4 DNA binding domain in vector pAS2–1 (CLONTECH), yielding pAS2–1-ski. The mammalian ski expression vector pcDNA3.1-ski was constructed by cloning the cDNA of h-ski into pcDNA3.1(+) (Invitrogen). The SBE2 × 2-tkluc reporter contains the herpes simplex virus thymidine kinase promoter with four copies of the SBE from GTCT2 × 2-tkCAT (5) upstream of the luciferase gene in pGL3-Basic (Promega).
Yeast Two-Hybrid System.
The yeast strain PJ-X, which carries HIS3, ADE2, and Lac Z reporter genes, was used for library screening. We confirmed expression of the bait fusion protein (Gal4 BD-ski) by Western blotting using anti Gal4-DBD monoclonal antibody (CLONTECH. These yeast cells transformed with the two-hybrid cDNA library (28) were plated on the selective synthetic dextrose medium lacking adenine, tryptophan, and leucine (SD-ATL). Colonies that grew in the presence of 3-aminotriazole (5 mM) and in the absence of adenine and histidine were further analyzed for β-galactosidase activity by a liquid assay (29). cDNA inserts derived from triple positive (Ade+, His+, Lac Z+) yeast colonies were tested for bait specificity by retransformation with different Gal4-DBD fusion proteins and pAS2–1 expressing the Gal4-DBD only.
Immunoprecipitation.
UCD-Mel-N(Ski+) melanoma cells, treated with 20 ng/ml TGF-β1 for 0, 0.5, and 24 h, were scraped, were solubilized in 0.1% Nonidet P-40, 50 mM Tris⋅HCl (pH 7.5), and 100 mM NaCl containing protease inhibitors, and were centrifuged at 10,000 × g for 15 min at 4°C. Supernatants were precleared by centrifugation after 1 h of incubation at 4°C with 50 μl of immobilized protein-G Sepharose beads (Pierce). The resulting supernatants were incubated with either goat anti-Smad2, goat anti-Smad3 serum (Santa Cruz Biotechnology), or normal goat serum for 2 h at 4°C. Immunocomplexes were collected by centrifugation after incubation with protein-G Sepharose beads and were analyzed by SDS/PAGE and immunoblotting with G8 anti-Ski monoclonal antibody (mAb) and the ECL chemiluminescent detection system (Amersham Pharmacia).
Nuclear extracts (80 μg protein) of CEFs or cSki-CEFs were prepared as described (5), were suspended in 50 mM Tris⋅Cl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 1 mM PmSF, and 1 mM Pefabloc, and were incubated on ice for 1 h with 3 μg of either G8 mAb, Smad2/3 polyclonal antibody (N-19) (Santa Cruz Biotechnology), or normal goat or rabbit serum (Sigma). Immunocomplexes adsorbed to protein G-Agarose beads (Boehringer Mannheim) were collected by centrifugation and were analyzed by SDS/PAGE and immunoblotting with either G8 mAb or Smad3(H-2) (Santa Cruz Biotechnology). Chemiluminescent detection was accomplished by using CDP-Star (Tropix, Bedford, MA) after incubation with a goat anti-mouse κ chain AP conjugate (Southern Biotechnology Associates) at a 1:1,000 dilution. Western blot analysis of Smad2 and Smad4 was attempted, but none of the available antibodies detected the chicken homologs of these proteins.
Electrophoretic Mobility-Shift Assay (EMSA) and Antibody Supershift Analysis.
EMSAs and supershifts were performed as described (5) except that 15 μg of nuclear extracts were used. An oligonucleotide probe containing two tandem copies of the SBE element and was 32P-labeled according to the method of Mertz and Rashtchian (30). For supershifts, anti-Ski mAb M6 and 68 (1 μg of protein A-purified mAb), goat anti-Smad2/Smad3 (Santa Cruz Biotechnology, N-19, 3 μl), or rabbit anti-Smad4 (Santa Cruz Biotechnology, H-552, 3 μl) were used. Oligonucleotide competitors containing two copies of the SBE or two copies of a mutated sequence (CTGTACAG) were used at 100-fold molar excess of the probe.
Transfection and Luciferase Assay.
Mv1Lu, R1B/L17, or HCT116 and HCT116 Smad4−/− cells were transfected by using the FuGENE 6 reagent, as specified by the manufacturer (Boehringer Mannheim). Transfections used 0.5 μg of the p3TP-Lux (20, 21), SBE2 × 2-tkluc, (CAGA)12MLP (31), p800Luc (32), or TRE-luc (33, 34) reporter plasmids, and combinations of 1 μg each of Smad2, Smad3, Smad4, and Ski expression plasmids, together with a β-galactosidase expression plasmid for normalization of transfection efficiencies. Luciferase and β-galactosidase activities were measured 22 h after transfection by using the appropriate reporter assay kits according to manufacturer's instructions (Promega). For TGF-β stimulation, cells were transfected and incubated overnight. The cells then received medium containing 0.2% FBS with or without 1 ng/ml TGF-β1 (Sigma). Luciferase and β-galactosidase activities were measured 20 h later, as described above.
Results
Identification of Smad2 and Smad3 as Ski Binding Proteins.
To identify proteins that interact with Ski, we screened a yeast two-hybrid IIB-Mel-J human melanoma cDNA library using the full-length coding region of human ski as bait. A screen of approximately 1.5 × 106 yeast transformants yielded 16 colonies that grew in selective medium and expressed β-galactosidase. A GenBank search revealed that three of the insert sequences are identical to Smad2 and two identical to Smad3.
Re-transformation assays showed that the Smad2 and Smad3 fusion proteins interact specifically with the Ski bait because no interaction was detected with either the Gal4-DBD alone or Gal4-DBD-p53 (data not shown). The latter construct produces a p53 fusion protein that showed strong interaction using SV40 large T as bait. The Smad2 and Smad3 clones obtained in the two-hybrid screen do not encode full-length proteins. Rather, the clones contain a segment of the linker region and the C-terminal transcriptional activation domains (MH2) of both Smad2 and Smad3 (Fig. 1A). The linker regions of these proteins are quite dissimilar, and the Smad2 clones contain only nine residues from the linker region. Thus, it is likely that Ski binds the MH2 domains Smad2 and Smad3, which are virtually identical.
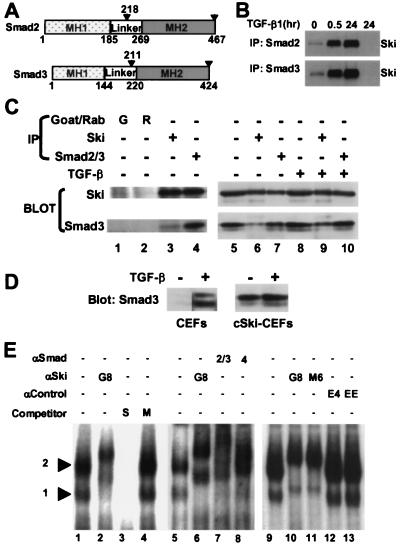
Figure 1.
Smad2 and Smad3 associate with c-Ski in vivo and bind the SBE. (A) The diagram shows major domains (MH1 and MH2) of Smad2 and Smad3 relative to the Ski-interaction regions identified in the two-hybrid screen, which extend from the residues numbered above the arrowheads to the C termini. (B) Lysates of UCD(Ski+) cells treated with TGF-β for the indicated times were analyzed by immunoprecipitation (IP) with anti-Smad2, anti-Smad3, or normal goat serum followed by Western blotting using the G8 anti-Ski monoclonal antibody, as described in Materials and Methods. (C) The influence of TGF-β on the in vivo association of Ski with Smad3 in cSki-CEFs. Nuclear extracts of untreated and TGF-β-treated (30 min) cSki-CEFs were either analyzed directly (lanes 5 and 8) or were subjected to immunoprecipitation (IP) with Smad2/Smad3 antiserum (lanes 4, 7, and 10), G8 monoclonal anti-Ski (lanes 3, 6, and 9), or normal sera from goat (lane 1) and rabbit (lane 2) followed by Western blotting using the G8 mAb or anti-Smad3 mAb as described in Materials and Methods. (D) Comparison of TGF-β dependence of Smad3 nuclear accumulation in CEFs and cSki-CEFs. Nuclear extracts of untreated and TGF-β-treated (30 min) CEFs and cSki-CEFs were analyzed by Western blotting using anti-Smad3 mAb as described in Materials and Methods. (E) EMSAs of nuclear extracts of cSki-CEFs were performed by using a probe containing two SBE sequences, as described in Materials and Methods. Reactions were incubated with no antibody (lanes 1, 3, 4, 5, and 9), with M6 anti-Ski mAb (lanes 2, 6, and 11), G8 anti-Ski mAb (lane 10), goat Smad2/3 antiserum (lane 7), or rabbit Smad4 antiserum (lane 8) or two irrelevant antibodies (lanes 12 and 13). Binding assays performed in the presence of 100-fold excess of specific (S) and mutant (M) oligonucleotide competitors are shown in lanes 3 and 4, respectively.
We next examined the intracellular association of endogenous Smad proteins with Ski by co-immunoprecipitation. Lysates of control and TGF-β-treated UCD(Ski+) melanoma cells, which stably overexpress human Ski, were immunoprecipitated with anti-Smad antibodies and were immunoblotted for Ski. As shown in Fig. 1B, a small amount of Ski co-precipitated with either Smad2 or Smad3 before TGF-β addition, but the amount increased dramatically after 30 min and 24 h of TGF-β treatment. No Ski was precipitated from the 24-h TGF-β lysate by a control antiserum. Thus, the association of these Smad proteins with Ski depends on their translocation to the nucleus as a result of receptor activation. In other experiments involving transient overexpression of FLAG-tagged Smad proteins in these cells, we found that, even in the absence of TGF-β, Smad2 and Smad3 were largely nuclear and co-precipitated a substantial amount of Ski protein (data not shown). Nuclear localization of exogenously overexpressed Smads has been reported for other cell types (35) and is likely responsible for their ligand-independent transcriptional activity.
Transformation of embryo fibroblasts by ski occurs in the absence of added TGF-β and requires only overexpression of the Ski protein. We therefore asked whether endogenous Smad proteins associate with overexpressed Ski in these cells (cSki-CEFs) and whether the association is influenced by addition of TGF-β. As shown in Fig. 1C, neither the amount of Ski in the nucleus nor its co-precipitation with Smad3 was influenced by TGF-β treatment (compare lanes 5–7 and 8–10). Moreover, Ski was quantitatively co-precipitated with Smad3, suggesting that all of the Ski in these cells is in a complex with Smad3 (Fig. 1C, compare lanes 6 and 7 with 9 and 10). Consistent with these observations, the amount of Smad3 in the nuclear extracts increased by only about two-fold on TGF-β treatment (Fig. 1C, compare lanes 5 and 8 with 7 and 10). About half of this Smad3 was co-precipitated with Ski (Fig. 1C, compare lanes 6 and 7 and 9 and 10).
The relatively small effect of TGF-β on the amount of nuclear Smad3 in cSki-CEFs was unexpected and contrasted with the results obtained with UCD(Ski+) melanoma cells (Fig. 1B). To determine whether this was caused by overexpression of Ski in CEFs, we compared the effect of TGF-β on the amount of nuclear Smad3 in CEFs and cSki-CEFs. As shown in Fig. 1D, TGF-β treatment resulted in a dramatic increase in nuclear Smad3 in CEFs compared with the small effect in cSki-CEFS. Thus, overexpression of Ski in CEFs results in apparent ligand-independent nuclear translocation of Smad3. Overexpressed Ski protein might bind and escort Smad3 into the nucleus, or it might stimulate an autocrine loop of receptor activation.
Ski, Smad2/Smad3, and Smad4 Form a Complex with the Smad/Ski Binding Element (SBE) GTCTAGAC.
Ski-containing protein complexes in nuclear extracts of cSki-CEFs have been shown to bind the DNA sequence GTCTAGAC that has also been identified as the Smad binding element (SBE) bound by Smad3 and Smad4 (5, 17). Although the cSki-CEFs used in those studies were not treated with TGF-β, the results shown in Fig. 1 C and D suggest that the Ski-containing complexes that bind this element also contain Smad proteins. To test this possibility, we performed EMSAs using antibodies to these proteins and nuclear extracts from cSki-CEFs (Fig. 1D). Both complexes formed with the SBE probe were supershifted by anti-Ski monoclonal antibodies (Fig. 1E, lanes 1 and 2, 5 and 6, 9 and 10, and 11). These complexes were also supershifted by polyclonal antibodies against Smad2 and Smad3 (Fig. 1E, lane 6) or Smad4 (lane 7) but not by either of two irrelevant antibodies (lanes 12 and 13). Competition EMSAs employing a specific SBE competitor (Fig. 1E, S, lane 7) or mutated competitor (M, lane 8) demonstrated that the two Ski/Smad-containing complexes represent sequence-specific binding to the SBE. The results presented in Fig. 1 show that, intracellularly, Ski associates with a multi-Smad complex that specifically binds to the SBE.
Ski Represses the Activation of Reporter Genes by TGF-β and by Combinations of Smad Proteins.
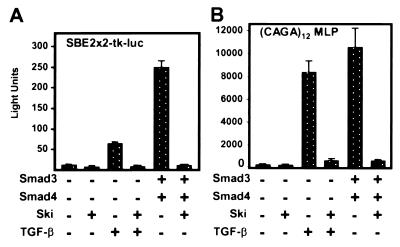
Ski has been shown to repress reporters containing upstream copies of SBE whereas TGF-β was shown to elicit Smad-dependent activation of SBE reporters and of reporters with multiple copies of the SBE-related CAGA element (5, 17, 31). Because Ski binds the SBE in association with Smad proteins, we tested whether TGF-β induced expression of these reporters would be counteracted by Ski. As shown in Fig. 2 A and B, TGF-β stimulated expression of SBE2 × 2-tkluc and to an even greater extent that of (CAGA)12MLP in mink lung cells. For both reporters, TGF-β activation was efficiently blocked by co-expression of Ski. In addition to the herpes simplex virus thymidine kinase (SBE2 × 2-tkluc) and adenovirus major late (CAGA)12MLP promoters used here, we have also found that Ski represses TGF-β-activated expression of an SBE reporter (SBE4-luc) driven by a minimal SV40 promoter (data not shown). Thus, both activation by TGF-β and repression by Ski are independent of the promoter.
Figure 2.
Repression by Ski of TGF-β and Smad-activated expression of SBE reporters. Transient expression assays in Mv1Lu cells in the presence or absence of TGF-β and with the indicated co-expressed Ski and Smad proteins were performed as described in Materials and Methods. (A) Assays with the SBE2 × 2-tkluc reporter, which contains two sets of tandem repeats (four copies) of the SBE upstream of a herpes simplex virus thymidine kinase promoter. (B) Assays with the (CAGA)12MLP reporter, which contains 12 copies of CAGA, upstream of the adenovirus major late promoter.
Previous work has shown that overexpressed Smad proteins activate expression of TGF-β responsive promoters, and that activation by combinations of Smad proteins is synergistic (20, 36). We therefore asked whether Smad activation of SBE reporters was repressed by Ski. The data in Fig. 2 A and B show that co-expression of Smad3 and Smad4 produced strong activation of both the SBE reporter and the CAGA reporter and that this activation was blocked by co-expression of Ski.
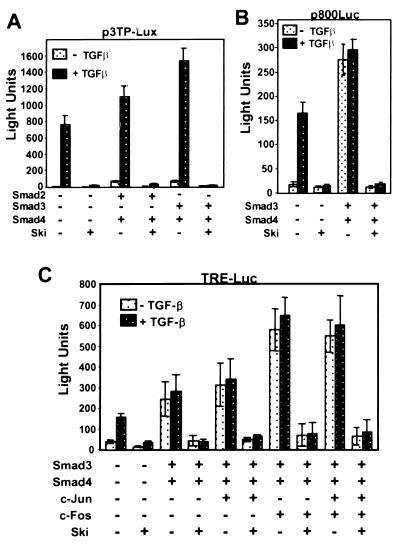
Ski Represses Chimeric and Natural TGF-β Responsive Promoter-Reporters.
Transcriptional activation by TGF-β and combinations of Smad proteins has been extensively studied using 3TP-Lux (20, 21, 37, 38), which is a chimeric reporter containing upstream regions from both the human plasminogen activator inhibitor 1 (PAI-1) gene (32) and the human collagenase gene (39). Because these upstream regions contain more complex TGF-β response elements than the isolated SBE elements used in the studies shown in Fig. 2, we sought to determine whether Ski would repress transcription from this reporter. The results in Fig. 3A agree with published data (38) by showing that 3TP-Lux was responsive to TGF-β and that this activity was enhanced to a greater extent by Smad3/4 than by Smad2/4. The results also demonstrate that reporter activation in response to TGF-β alone, or in combination with Smad2 plus Smad4 or Smad3 plus Smad4, was completely blocked by co-expression of Ski.
Figure 3.
Ski represses TGF-β and Smad activity in chimeric and natural PAI-1 promoter-reporters. (A) 3TP-Lux expression in TGF-β-responsive Mv1Lu cells. (B) PAI-1 promoter reporter p800Luc expression in M1vLu cells. (C) TRE-Luc expression in Mv1Lu cells. Cells were transfected with p3TP-Lux, p800Luc, or TRE-Luc reporter plus or minus combinations of Smad2, Smad3, Smad4, Jun, Fos, and c-Ski expression plasmids, as indicated. Luciferase activity, normalized to β-galactosidase activity, was determined as described in Materials and Methods. Where indicated, cells were transferred to medium containing 0.2% FBS plus or minus 1 ng/ml TGF-β, 20 h after transfection. Luciferase activity and β-galactosidase activity were measured 22 h later. The results shown are representative of at least three independent experiments, and data are the average of triplicate measurements plus and minus the standard error.
The PAI-1-derived sequence in the 3TP-Lux reporter is only about 100 base pairs of an 800-base pair upstream region that mediates responsiveness to TGF-β (32). We were therefore interested in determining whether Ski also represses the expression of a reporter driven by the natural PAI-1 promoter (p800Luc) (32). The results in Fig. 3B show that activation of PAI-1 by Smad3/Smad4 or TGF-β is dramatically suppressed by Ski. These results demonstrate that Ski functions as a co-repressor of Smad proteins bound to TGF-β responsive elements in a natural gene regulatory region.
In addition to SBE-related elements, 3TP-Lux contains AP-1 binding sites (TREs) that have been shown to function as TGF-β responsive elements in this and in other promoters (33, 40). With TREs, DNA binding is accomplished by AP-1, and Smad proteins function as co-activators through association with Fos and/or Jun. To determine whether Ski could also function as a repressor in this context, we performed transient expression assays with a TRE reporter (34). As expected from previous results, the TRE reporter was activated by TGF-β and by the combination of Smad3 and Smad4 (Fig. 3C). Further stimulation was observed on addition of Jun or Fos or both. In every case, activation of the TRE reporter was blocked by expression of Ski. Combined with the data in Fig. 2, these results show that Ski counteracts Smad activation of 3TP-Lux and of the two major TGF-β responsive elements in this chimeric reporter.
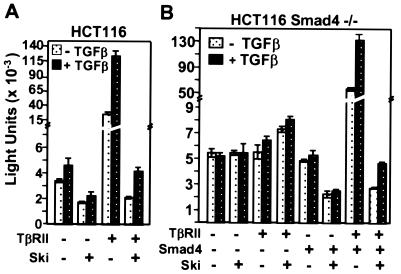
Ski Fails To Repress an SBE Reporter in Cells Lacking Smad4.
We have shown that Ski co-binds the SBE with Smad proteins and represses their activation of TGF-β-responsive reporters. To determine whether repression of SBE reporters by Ski requires interaction with functional Smad protein complexes, we asked whether Ski could repress an SBE reporter in a line of HCT116-derived cells that lack the obligatory common Smad subunit, Smad4 (27). The parental HCT116 cell line lacks the TGF-β RII receptor and as shown in Fig. 4A, transfection of the receptor greatly stimulated expression of SBE2 × 2-tkluc and restored its response to TGF-β. In these cells, Ski repressed expression of the reporter in both the absence and presence of the receptor.
Figure 4.
Repression of SBE2 × 2-tkluc reporter expression by Ski requires expression of Smad4. Shown is SBE2 × 2-tkluc activity in the TGF-β RII-deficient parental cell line HCT116 (A) and in HCT116 cells lacking Smad4 (B). Cells were transfected with the reporter plus or minus combinations of TGF-β RII, Smad4, and c-Ski as indicated. Cells were transferred to medium containing 0.2% FBS plus or minus 1 ng/ml TGF-β, 20 h after transfection. Luciferase activity was measured 22 h later as described in Materials and Methods. The results shown are averages of two independent experiments performed in duplicate.
In Smad4-deficient HCT116 cells (Fig. 4B), expression of TGF-β RII or Smad4 alone had no effect on reporter activity. However, expression of Smad4 and TGF-β RII in combination dramatically increased reporter activity and restored responsiveness to TGF-β. In these Smad4-deficient cells, Ski did not repress reporter expression, either in the vector-transfected controls or in the TGF-β RII-transfected cells. However, expression of Smad4 alone or in combination with TGF-β RII restored repression by Ski to that observed in HTC116 cells. Smad4 is the obligatory common member of active Smad protein complexes. Thus, the dependence of Ski's repression activity on Smad4 does not mean that these two proteins directly interact. Rather, it indicates that interaction with functional Smad complexes is required for Ski's binding to the SBE and repressing transcription.
Ski Antagonizes TGF-β-Mediated Growth Inhibition in Prostate Epithelial Cells.
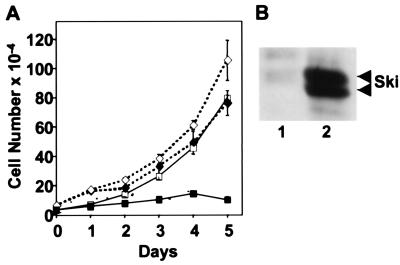
We next sought to assess the biological significance of Ski's transcriptional opposition to Smad activation. In most epithelial cells, TGF-β functions as an inhibitor of cellular proliferation. Therefore, we tested whether overexpression of Ski could counteract the potent growth inhibitory effect of TGF-β on a rat prostate epithelial cell line (DP-153). As shown in Fig. 5A, TGF-β treatment drastically reduced the proliferation of control DP-153 cells whereas it had only a modest effect on the cells expressing ski (DP-153-ski). These dramatic results may actually underestimate the ability of Ski to block the growth-inhibitory activity TGF-β. That is because although Western analysis showed that this polyclonal population of infected cells expresses Ski at high levels (Fig. 5B), examination of individual cells by immunofluorescence revealed a significant fraction (>10%) that did not express detectable levels of Ski (data not shown).
Figure 5.
Overexpression of Ski blocks the TGF-β-mediated growth inhibition in a prostate-derived, epithelial cell line. Shown is the rat prostate epithelial cell line DP-153 infected with the amphotropic retroviruses pBABEpuro or pBABEpuro-H-ski as described in Materials and Methods. Puromycin-resistant clones were pooled and tested for Ski expression by immunoblotting and for sensitivity to growth inhibition by TGF-β. (A) The number of DP-153 cells (□, ■) and DP-153-Ski+ cells (♦, ⋄) per well in triplicate cultures (average +/− standard error) was determined at the indicated days in the absence (□, ⋄) or presence (■, ♦) of TGF-β1 (10 ng/ml). (B) Immunoblot showing high levels of the Ski protein in the polyclonal populations of control DP-153 cells (lane 1) and DP-153-Ski (lane 2) cells.
Discussion
Our results show that Ski associates with Smad2 and Smad3 and counteracts their activation of gene expression in response to TGF-β. The Ski-interacting domains of Smad2 and Smad3 reside within the MH2 regions, which also contain their transcriptional activation domains. This suggests that Ski could block Smad activity by binding to the activation domains and preventing interactions with co-binding transcription factors, co-activators (reviewed in ref. 41) or the basal transcription machinery. However, Ski contains an active repression domain that functions when tethered to DNA via a heterologous DNA binding domain (5, 6). Thus, Ski's action may not be to merely block activation by Smad proteins. Rather, Ski may function to actively repress the transcription of genes that are targeted by TGF-β activation of Smads. Ski has been shown to associate with both NcoR and mSin3, so it is tempting to speculate that its repression mechanism in association with Smad proteins involves histone deacetylation. However, preliminary results in transient expression assays indicate that repression of SBE reporters by Ski is insensitive to the deacetylase inhibitor, trichostatin A.
Regardless of its mechanism of repression, our results provide a model for the action of Ski as an oncogene by showing that it counteracts the TGF-β pathway, which usually functions to suppress cellular proliferation, and transformation. In human tumors, high levels of Ski expression could produce a disruption of TGF-β signaling similar to that resulting from mutation of genes encoding TGF-β receptors or Smad proteins. This might be relevant to certain melanoma cells and other tumor cells that are unresponsive to this cytokine, even though most melanoma cell lines tested have adequate and active TGF-β receptors and Smad proteins (24, 42). Dominant disruption of Ski function in a subset of these tumors that overexpress high levels of Ski might confirm this suggestion by restoring gene activation and growth inhibition by TGF-β.
Although a plausible model for Ski's possible role in tumorigenesis, the above proposal does not account for its ability to transform CEFs in vitro, which occurs in the absence of TGF-β. On the other hand, we found that overexpressed Ski increases the nuclear accumulation of Smad3 so that all of the Ski protein in transformed CEFs is associated with Smads. Thus, it is likely that dominant repression of TGF-β-inducible genes that are negative regulators of cell cycle progression plays a major role in Ski's transforming activity. This hypothesis should be testable by determining whether TGF-β signaling and/or association with Smad proteins are essential for transformation of CEFs by Ski.
Acknowledgments
We are grateful to R. Derynck for C-terminal Flag-Smad2 and Smad3 plasmids, Joan Massagué for p3TP-lux, N-terminal Flag-Smad2 and Smad4 plasmids and R1B/L17 cells, Sandy Markowitz for the TGF-β RII plasmid, J-M Gauthier for the (CAGA)12 MLP plasmid, and Michael Karin for TRE-Luc plasmid. This work was supported by National Institutes of Health Grants AG-3663 (E.E.M.), AG-09990 (J.C.), and CA-43600 (E.S.), and in part by Cancer Center Development Grant P30CA43703 (D.D.). M.M.H. was supported by a Fellowship from the Dermatology Foundation.
Abbreviations
- SBE
Smad binding element
- TGF-β
type β transforming growth factor
- CEF
chicken embryo fibroblast
- EMSA
electrophoretic mobility-shift assay
- PAI-1
plasminogen activator inhibitor 1
Note
While this manuscript was under review, three other groups (43–45) reported that Ski acts as a co-repressor and regulator of TGF-β signaling by direct interaction with Smad proteins.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090097797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090097797
References
- 1.Barkas A E, Brodeur D, Stavnezer E. Virology. 1986;151:131–138. doi: 10.1016/0042-6822(86)90111-x. [DOI] [PubMed] [Google Scholar]
- 2.Sutrave P, Kelly A M, Hughes S H. Genes Dev. 1990;4:1462–1472. doi: 10.1101/gad.4.9.1462. [DOI] [PubMed] [Google Scholar]
- 3.Sutrave P, Copeland T D, Showalter S D, Hughes S H. Mol Cell Biol. 1990;10:3137–3144. doi: 10.1128/mcb.10.6.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engert J C, Servaes S, Sutrave P, Hughes S H, Rosenthal N. Nucleic Acids Res. 1995;23:2988–2994. doi: 10.1093/nar/23.15.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicol R, Stavnezer E. J Biol Chem. 1998;273:3588–3597. doi: 10.1074/jbc.273.6.3588. [DOI] [PubMed] [Google Scholar]
- 6.Nicol R, Zheng G, Sutrave P, Foster D, Stavnezer E. Cell Growth Differ. 1999;10:243–254. [PubMed] [Google Scholar]
- 7.Li Y, Turck C M, Teumer J K, Stavnezer E. J Virol. 1986;57:1065–1072. doi: 10.1128/jvi.57.3.1065-1072.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stavnezer E, Brodeur D, Brennan L A. Mol Cell Biol. 1989;9:4038–4045. doi: 10.1128/mcb.9.9.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colmenares C, Sutrave P, Hughes S H, Stavnezer E. J Virol. 1991;65:4929–4935. doi: 10.1128/jvi.65.9.4929-4935.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colmenares C, Stavnezer E. Cell. 1989;59:293–303. doi: 10.1016/0092-8674(89)90291-2. [DOI] [PubMed] [Google Scholar]
- 11.Berk M, Desai S Y, Heyman H C, Colmenares C. Genes Dev. 1997;11:2029–2039. doi: 10.1101/gad.11.16.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nomura N, Sasamoto S, Ishii S, Date T, Matsui M, Ishizaki R. Nucleic Acids Res. 1989;17:5489–5500. doi: 10.1093/nar/17.14.5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fumagalli S, Doneda L, Nomura N, Larizza L. Melanoma Res. 1993;3:23–27. doi: 10.1097/00008390-199304000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Barkas A. Ph. D. dissertation. New York: New York University; 1986. [Google Scholar]
- 15.Dahl R, Kieslinger M, Beug H, Hayman M J. Proc Natl Acad Sci USA. 1998;95:11187–11192. doi: 10.1073/pnas.95.19.11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nomura T, Khan M M, Kaul S C, Dong H D, Wadhwa R, Colmenares C, Kohno I, Ishii S. Genes Dev. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zawel L, Dai J L, Buckhaults P, Zhou S, Kinzler K W, Vogelstein B, Kern S E. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 18.Massague J. Cell. 1996;85:947–950. doi: 10.1016/s0092-8674(00)81296-9. [DOI] [PubMed] [Google Scholar]
- 19.Derynck R, Zhang Y. Curr Biol. 1996;6:1226–1229. doi: 10.1016/s0960-9822(96)00702-6. [DOI] [PubMed] [Google Scholar]
- 20.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Feng X, We R, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 22.Massague J. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 23.Krasagakis K, Garbe C, Zouboulis C C, Orfanos C E. Recent Results Cancer Res. 1995;139:169–182. doi: 10.1007/978-3-642-78771-3_12. [DOI] [PubMed] [Google Scholar]
- 24.Rodeck U, Bossler A, Graeven U, Fox F E, Nowell P C, Knabbe C, Kari C. Cancer Res. 1994;54:575–581. [PubMed] [Google Scholar]
- 25.Guerra L, Mordoh J, Slavutsky I, Larripa I, Medrano E E. Pigm Cell Res. 1989;2:504–509. doi: 10.1111/j.1600-0749.1989.tb00246.x. [DOI] [PubMed] [Google Scholar]
- 26.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou S, Zawel L, Lengauer C, Kinzler K W, Vogelstein B. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]
- 28.Gietz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breeden L, Nasmyth K. Cold Spring Harb Symp Quant Biol. 1995;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 30.Mertz L M, Rashtchian A. Anal Biochem. 1994;221:160–165. doi: 10.1006/abio.1994.1392. [DOI] [PubMed] [Google Scholar]
- 31.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J M. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keeton M R, Curriden S A, van Zonneveld A J, Loskutoff D J. J Biol Chem. 1991;266:23048–23052. [PubMed] [Google Scholar]
- 33.Zhang Y, Feng X H, Derynck R. Nature (London) 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 34.Renshaw M W, McWhirter J R, Wang J Y. Mol Cell Biol. 1995;15:1286–1293. doi: 10.1128/mcb.15.3.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Lebrun J J, Vale W. Proc Natl Acad Sci USA. 1997;93:12992–12997. doi: 10.1073/pnas.93.23.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Musci T, Derynck R. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 37.Wrana J L, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang X F, Massague J. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 38.Yingling J M, Datto M B, Wong C, Frederick J P, Liberati N T, Wang X F. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Groot R P, Kruijer W. Biochem Biophys Res Commun. 1990;168:1074–1081. doi: 10.1016/0006-291x(90)91139-j. [DOI] [PubMed] [Google Scholar]
- 40.Jin G, Howe P H. J Biol Chem. 1997;272:26620–26626. doi: 10.1074/jbc.272.42.26620. [DOI] [PubMed] [Google Scholar]
- 41.Derynck R, Zhang Y, Feng Z H. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 42.Krasagakis A V, Garbe C, Orfanos C E. Melanoma Res. 1993;3:425–433. [PubMed] [Google Scholar]
- 43.Luo K, Stroschein S L, Wang W, Chen D, Martens E, Zhou S, Zhou Q. Genes Dev. 1999;13:2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Y, Liu X, Eaton E N, Lane W S, Lodish H F, Weinberg R A. Mol Cell. 1999;4:499–509. doi: 10.1016/s1097-2765(00)80201-4. [DOI] [PubMed] [Google Scholar]
- 45.Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. J Biol Chem. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]