Abstract
Background
Claudin-4, a tight junction (TJ) protein and receptor for the C-terminal fragment of Clostridium perfringens enterotoxin (C-CPE), is overexpressed in epithelial ovarian cancer (EOC). Previous research suggests DNA methylation is a mechanism for claudin-4 overexpression in cancer and that C-CPE acts as an absorption-enhancing agent in claudin-4-expressing cells. We sought to correlate claudin-4 overexpression in EOC with clinical outcomes and TJ barrier function, investigate DNA methylation as a mechanism for overexpression, and evaluate the effect of C-CPE on the TJ.
Methods
Claudin-4 expression in EOC was quantified and correlated with clinical outcomes. Claudin-4 methylation status was determined, and claudin-4-negative cell lines were treated with a demethylating agent. Electric cell-substrate impedance sensing was used to calculate junctional (paracellular) resistance (Rb) in EOC cells after claudin-4 silencing and after C-CPE treatment.
Results
Claudin-4 overexpression in EOC does not correlate with survival or other clinical endpoints and is associated with hypomethylation. Claudin-4 overexpression correlates with Rb and C-CPE treatment of EOC cells significantly decreased Rb in a dose- and claudin-4-dependent noncytotoxic manner.
Conclusions
C-CPE treatment of EOC cells leads to altered TJ function. Further research is needed to determine the potential clinical applications of C-CPE in EOC drug delivery strategies.
Keywords: Ovarian cancer, claudin-4, gene expression, therapy, methylation
Introduction
Gene expression profiling has led to the discovery of novel genes and pathways thought to be important in ovarian cancer tumorigenesis [1]. One such gene identified by our group [2] and others [3–7] is claudin-4 (CLDN4). The claudins are a group of over 20 proteins that are expressed in a tissue-specific manner and form the backbone of tight junction (TJ) strands [8,9]. Tight junctions play an important role in the maintenance of cell polarity and the regulation of paracellular transport. Through their homophilic and heterophilic interactions, claudins help to define the ability of TJs to affect the strength and selectivity of paracellular resistance [10]. Furthermore, recent evidence suggests that claudins play a role in a wide variety of cellular signaling processes through their cytosolic carboxyterminal interaction with PDZ-containing proteins such as ZO-1 [8,9]. As such, evidence is mounting to suggest that cell adhesion molecules in general [11], and TJs and claudins specifically, influence a wide variety of processes including cell signaling, proliferation, differentiation, trafficking, and polarity, and may thus play an important role in the coordinated molecular biochemistry of the tumor microenvironment [9].
Recent studies have implicated alterations of a number of claudin isotypes in malignancy [12]. Claudin-7 was shown to be underexpressed in ductal carcinomas of the breast [13], whereas claudin-3 is overexpressed in prostate, breast, and ovarian cancers [6,14,15]. These changes have been noted in preinvasive carcinomas [13,16], and in some cases, can be correlated with tumor grade and subtype [13]. In addition to ovarian cancer, claudin-4 has been found to be elevated in prostate, pancreatic, gastric, and breast cancers [14,15,17,18]. Interestingly, the role of claudin-4 in each of these tissue types appears to be variable. In pancreatic cancer, claudin-4 overexpression leads to decreased invasiveness and metastatic potential while having no effect on proliferation, cell cycle, or matrix metalloproteinase activity [17]. In contrast, claudin-3 and claudin-4 expression have been associated with increased invasiveness and metallo-proteinase-2 activity in ovarian cancer [19]. Although the mechanism of claudin-4 overexpression has yet to be fully elucidated [20], recent studies have shown that the methylation status of claudins appears to be important. Decreased expression of claudin-7 in breast cancer has been associated with promoter hypermethylation, whereas CLDN4 hypomethylation has been associated with overexpression in pancreatic cancer [21].
Several investigators have sought to exploit the therapeutic potential of claudin-4 overexpression in a variety of cancers. Both in vitro and animal studies in breast, prostate, and ovarian cancers have shown that Clostridium perfringens enterotoxin (CPE), which binds to claudin-4, may have an important therapeutic benefit, as it is rapidly cytotoxic in tissues overexpressing claudin-4 [7,14,15,17]. The recent report by Santin et al. [7] showing the successful treatment of epithelial ovarian cancer (EOC) with intraperitoneal (IP) CPE in SCID mice is promising; however, claudin-4 is expressed in a number of normal tissues [15], and some authors have suggested that IP CPE may have potentially significant toxic side effects [14,22]. However, the noncytotoxic receptor-binding C-terminus of CPE (C-CPE) can disrupt TJs and has been shown to be an absorption-enhancing agent in epithelia [23,24]. The use of IP “chemosensitizers” that can decrease tumor density and improve drug penetration (by surface diffusion) [25] may be an attractive method of improving cytotoxic drug delivery, given the important role of adjuvant IP chemotherapy in advanced EOC [26], Alternatively, tumor-specific drug delivery may be improved by using C-CPE as a claudin-4 targeting molecule by fusing it to cytotoxic drugs directly [27].
Our previously described oligonucleotide array [2] suggested that CLDN4 was one of the most highly overexpressed genes in ovarian cancer. We sought to validate the overexpression of claudin-4 in EOC and to correlate expression with several clinical outcomes. We also sought to investigate the possibility that the methylation status of CLDN4 contributes to its overexpression in ovarian cancer. Lastly, we explored the relationship between claudin-4 overexpression and junctional (paracellular) resistance, a measure of TJ “barrier function,” and examined the ability of C-CPE to act as a nontoxic modulator of junctional resistance in ovarian cancer cell lines.
Materials and Methods
Tissue Samples
Paraffin-embedded tissue was collected and archived from patients undergoing surgery at Brigham and Women's Hospital (Boston, MA). All patient-derived specimens were collected under protocols approved by the institutional review board. Clinical stage, as defined by the International Federation of Gynecology and Obstetrics system, was determined by chart review, and histologic subtype and grade were assigned by pathologists according to World Health Organization guidelines.
Cell Lines
A total of 20 ovarian cancer cell lines were used, and included cell lines derived from serous (ALST, CAOV3, DOV13, OVCAR3, OVCA420, OVCA429, OVCA432, OVCA433, OVCA633, PEO4, REST, SKOV3, 2008), mucinous (MCAS, RMUG-L, RMUG-S), clear cell (ES2, TOV21G, RMG1), and endometrioid adenocarcinomas (TOV112D). Cell lines SKOV3, RMG1, ES2, OVCAR3, MCAS, RMUG-L, RMUG-S, and TOV112D were purchased from American Type Culture Collection (Rockville, MD) or from the Japanese Collection of Research Bioresources (Tokyo, Japan), and 2008 was obtained from The University of Texas Southwestern Medical Center (Dallas, TX). The remaining cell lines were established in our laboratory. Normal human ovarian surface epithelium (HOSE) cells were obtained from fresh ovarian scrapings at the time of surgery for benign nonovarian conditions. Immortalized HOSE cells were obtained by HPVE6E7 immortalization of normal HOSE cells [28]. Cells were maintained at 37°C in a 5% carbon dioxide incubator and were cultured in a filter-sterilized medium consisting of equal parts M199 (Sigma, St. Louis, MO) and MCDB105 (Sigma), supplemented with 10% fetal bovine serum and l-glutamine-penicillin-streptomycin (10 ml/L, Sigma). Cell culture medium was changed every 1 to 2 days, depending on the rate of cell growth, and cells were subcultured at 80% confluence using 1 x trypsin solution.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted, purified, and quantified from laser microdissected tissue samples or cell lines and then converted to cDNA as described previously [2,29]. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed by use of the 7300 Real-Time PCR System. TaqMan Gene Expression Assay probes for cyclophilin A (an endogenous control) and claudin-4 (assay ID Hs00533616_s1, assay location 1484, reference sequence NM_001305.3) were purchased from Applied Biosystems (Foster City, CA). For each qRT-PCR reaction, claudin-4 and cyclophilin A were multiplexed as follows: 2 µl of cDNA was added to 10 µl of TaqMan Universal PCR Master Mix (Applied Biosystems), 1 ml of claudin-4 TaqMan primer/probe, 1 µl of cyclophilin A TaqMan primer/probe, and 6 µl of autoclaved distilled water. Reactions started with a 10-minute hold at 95°C, followed by 40 cycles of denaturation at 95°C for 125 seconds, followed by annealing/extending at 60°C for 1 minute. All experiments were run in duplicate (values averaged) and repeated at least twice. The 7300 Real-Time PCR System software monitored the amplification process and determined the threshold cycle (Ct) for each reaction. Quantification (fold-change calculation) was relative to a HOSE sample and was determined by the ΔΔCt method as described previously [30].
Immunohistochemistry
Paraffin-embedded tissue samples were cut to 7-µm sections and mounted on Superfrost Plus microscope slides (Fisher Scientific, Pittsburgh, PA). Deparaffinization and antigen retrieval were performed by using BORG Decloaker (Biocare Medical, Walnut Creek, CA) solution and the Decloaking Chamber 2002 (Biocare Medical) electric pressure cooker. Slides were sequentially washed with 1x Hot Rinse (Biocare Medical), deionized water, and 1x TBS, then incubated with mouse anti-claudin-4 antibody (Zymed Laboratories Inc., South San Francisco, CA) at 1:50 dilution (in 1x TBS) for 60 minutes in a moist chamber at room temperature. Immunohistochemistry (IHC) was performed by using the EnVision-AP system (DakoCytomation, Carpinteria, CA) according to the manufacturer's recommendations. Slides were washed with deionized water, counterstained with hematoxylin and 5% ammonium hydroxide, and mounted in Accergel (Accurate Chemical and Scientific Corp., Westbury, NY). Control slides (incubated in 1x TBS without anti-claudin-4 antibody) were similarly prepared.
Western Blot
For the isolation of protein lysates, cells were first washed twice with 1x PBS, then lysed with a solution of RIPA Buffer-2 (Boston BioProducts, Worcester, MA), PMSF (Sigma), and Protease Inhibitor Cocktail (Sigma). After briefly vortexing and centrifuging at 4°C, the Micro BCA Protein Assay Kit (Pierce Chemical Co., Rockford, IL) was used for protein quantification of the cell lysates. Total cell lysates were resolved using a 1-D 10% SDS-PAGE gel (25 mg per sample). The resolved proteins were transferred to a (polyvinylidene fluoride) membrane (Perkin Elmer, Boston, MA) using the SEMI-DRY Transfer Cell (Bio-Rad Laboratories, Hercules, CA). After transfer, the PVDF membrane was washed and blocked with 5% nonfat dry milk at 4°C overnight. The membrane was subsequently incubated in anti-claudin-4 (Zymed) antibody in blocking solution at 1:2500 dilution for 1 hour, after which it was washed and incubated with antimouse IgG secondary antibody (1:2500) for 1 hour. For signal detection, the membrane was incubated with SuperSignal West Pico Chemiluminescent Substrate (Pierce) for 5 minutes, after which the membrane was exposed to film and the film was developed. To normalize for protein loading and transfer, β-actin was used as a positive control (1:50,000, Sigma). Western blot was performed on 13 ovarian cancer and 3 HOSE cell lines and blot intensity was quantified by using computer imaging software (Bio-Rad). Blot intensity was normalized to β-actin expression and correlated to claudin-4 expression as determined by qRT-PCR.
Methylation-Specific PCR and Bisulfite Sequencing
CpG Island Searcher software (http://ccnt.hsc.usc.edu/cpgislands/) was used to identify a CLDN4 CpG island using the following criteria: %GC = 55, ObsCpG/ExpCpG = 0.65, length of CpG island, ≥500 bp, distance between islands, ≥100 bp. Methylation-specific PCR (MSPCR) was performed using the CpGenome DNA Modification Kit (Chemicon International, Temecula, CA) [31]. Genomic DNA was extracted and purified from cell lines and tissue samples as described previously [32]. Bisulfite modification was performed according to the manufacturer's instructions. PCR was then performed using primer sequences for methylated reactions [5′-CTA CCG ATA AAA ACC GTC ACG-3′ (forward), 5′-GTG TAT TTT GCG AAC GTT AAG TTC-3′ (reverse)] and unmethylated reactions [5′-AAT ATT ACT ACC AAT AAA AAC CAT CAC AC-3′ (forward), 5′-TGT ATT TTG TGA ATG TTA AGT TTG T-3′ (reverse)] (Sigma-Genosys, The Woodlands, TX). The PCR mixture contained 1x PCR Gold Buffer, 5 mmol/l MgCl2, 2 µl dNTP 10 mmol/l, 1 µl each of forward and reverse primers, 0.125 µl AmpliTaq Gold, and 2 µl of DNA and brought to a total volume of 25 µl by addition of autoclaved deionized water (all reagents from Applied Biosystems). PCR reactions were hot started at 94°C for 12 minutes, followed by 35 cycles (30 seconds at 94°C, 30 seconds at the annealing temperature, 30 seconds at 72°C) and a final 7-minute hold at 72°C. Different annealing temperatures (57°C, 54°C) were used to account for the different melting temperatures of the methylated and unmethylated primers, respectively. Each PCR product was loaded into a 1% agarose or 10% acrylamide gel, stained with ethidium bromide, and visualized under UV illumination. Nine ovarian cancer tissue samples, 11 ovarian cancer cell lines, and 3 HOSE cell lines underwent MSPCR, and the CLDN4 methylation status was compared to its expression. On average, MSPCR was repeated three to four times for each cell line. MSPCR was also performed on unmodified genomic DNA, as a negative control, and on methylated control DNA (Universal Methylated DNA, Chemicon).
Bisulfite-modified DNA was prepared for bisulfite sequencing to assess for the completeness of the bisulfite treatment and the accuracy of the MSPCR results. The OVCA429 ovarian cancer cell line, which demonstrated a hypomethylated CLDN4 allele only, was used for this purpose. PCR amplification was performed on 100 ng of bisulfite-modified genomic DNA and the product was used for sequencing. The PCR primers used were 5′-TTG GAA GGA ATT GGT TTG TTT ATA T-3′ (forward) and 5′-CAT AAA CCC TCC CAA ATA ATC TAC-3′ (reverse); the annealing temperature was 57°C. The amplification product was cloned and sequenced; 21 CpG dinucleotides sites were located within the PCR product.
To assess the effect of DNA methylation, cell lines DOV13 and TOV112D (neither of which expressed claudin-4) were treated with 1 µmol/l 5-AZA-2′-deoxycytidine (5-AZA, Sigma), an inhibitor of DNA methylation. Treatment of cell cultures started at 30% to 40% confluence for a total of four days. The medium and drug were changed every 24 hours and the cell cultures were confluent for at least 48 hours before collection. MSPCR and qRT-PCR was then performed, in duplicate, as described above.
Preparation and Cell Culture with C-CPE
The C-terminal fragment of CPE (C-CPE) was attained using a 3′ fragment of the cpe gene cloned into the pTrcHis B expression vector, as described previously [33]. Sonic lysates from Escherichia coli transformants expressing C-CPE were filtered with a MILLEXHV 0.45-µm PVDF Filter (Millipore, Billerica, MA) and then applied to a HiTrap metal chelating column (Amersham Biosciences, Piscataway, NJ) preequilibrated with HiTrap Binding Buffer (20 mmol/l NaH2PO4, 500 mmol/l NaCl, 60 mmol/l imidazole, pH 7.4). C-CPE was eluted from the column with an imidazole gradient, and C-CPE containing fractions were pooled and dialyzed against distilled water (pH 7.0). The amount of C-CPE recovered from the purifications was estimated by the Lowry method, and the final enriched C-CPE product (90–95% purity) was stored frozen at -80°C until use.
C-CPE was thawed at 4°C and serially diluted in cell culture medium, after which it was immediately used in experiments. The Micro BCA Protein Assay Kit (Pierce) was used to determine protein concentrations. The effect of C-CPE on cell viability was assessed by the CellTiter-Blue Cell Viability Assay (Promega Corp., Madison, WI). The OVCA429 ovarian cancer cell line, one of the highest overexpressers of claudin-4, was subcultured on 96-well black-walled/clear-bottomed plates (BD Biosciences, Franklin Lakes, NJ) at 1 x 104 cells per well and incubated overnight at 37°C before treatment. Subsequently, the cell culture medium was changed and medium containing C-CPE was added, after which the cells were incubated for 44 to 48 hours at 37°C. Cell morphology was visually inspected, after which the Spectra Max Gemini (Molecular Devices Co., Sunnyvale, CA) microplate spectrofluorometer was used for cell viability measurements. Each set of experiments contained wells to control for background fluorescence.
Electric Cell-Substrate Impedance Sensing Measurements
The electric cell-substrate impedance sensing (ECIS) system was used to measure transepithelial impedance in ovarian cancer cell lines alone and after exposure to C-CPE. Transepithelial impedance measurements were used to calculate the junctional resistance Rb, a measure of TJ barrier function and paracellular resistance, by data fitting and model analysis, as briefly described below [34,35]. First, transepithelial impedance was measured in nine ovarian cancer cell lines during 5 days of exposure to cell culture medium alone. These cell lines all overexpressed claudin-4, albeit at varying levels: low (TOV112D, RMUG-L, ES2, SKOV3), higher (OVCAR3, RMUG-S, CAOV3), and highest (OVCA429, RMG1). The parameter Rb was calculated by using ECIS, after which it was statistically correlated to the level of claudin-4 overexpression (as assessed by qRT-PCR). To further demonstrate the relationship between claudin-4 expression and Rb, transepithelial impedance was measured during 7 days of cell culture and Rb was calculated and compared in the wild-type and small interfering RNA (siRNA) claudin-4 silenced SKOV3 cell lines. ECIS measurements were also obtained in ovarian cell lines OVCA429 and SKOV3 during drug treatment with increasing doses of C-CPE. C-CPE was added to the apical aspect of cells in culture and dose-response plots were generated based on the dynamic response of the parameter Rb 60 hours after exposure to C-CPE.
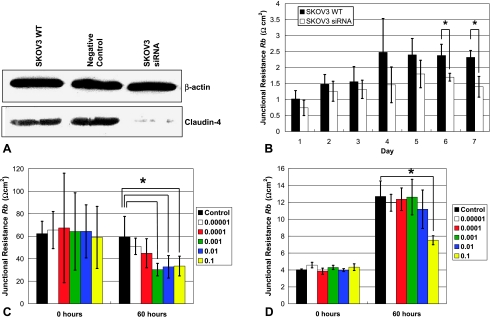
Claudin-4 silencing was performed by introducing a homologous double-stranded RNA into the ovarian cancer cell line, SKOV-3. SKOV-3 cells (2 x 105) were cultured in T25 flasks, after which they were transfected with claudin-4 siRNA duplex (Silencer Predesigned siRNA) or control RNA duplex (Silencer Negative Control) using the siRNA transfection kit (Silencer siRNA Transfection II) according to the manufacturer's instructions (all products from Ambion, Austin, TX). Cells were treated for 48 hours in the transfection reagent to allow for maximum claudin-4 silencing, after which they were placed in cell culture medium and used for ECIS experiments within 24 hours. A Western blot was performed to confirm claudin-4 silencing.
For ECIS measurements, 9 x 104 cells were plated into each well (1 cm in height, 0.9 cm2 in area) of an electrode array. Each electrode array consisted of eight wells; 0.4 ml of medium was used per well. Cell density was controlled at 1 x 105 cells/cm2. After 48 hours in culture, the confluence and viability of the cell monolayer were confirmed by light microscopy and electrically. The experimental conditions were then imposed and transepithelial impedance data were obtained from cell-covered electrode arrays. The electrode arrays, lock-in amplifier, and software for the ECIS were obtained from Applied Biophysics (Troy, NY). Attached cells on the electrode will act as insulating particles; accordingly, current will flow through cell-substrate spaces beneath the cell that can eventually lead to the paracellular space. Changes in cell morphology manifest as changes in impedance as the paracellular and/or cell-substrate spaces changes, and the contribution of each of these is accounted for separately in the ECIS system model. We applied a 1-V AC signal at 4000 Hz to the samples through a 1-µω resistor and monitored transepithelial impedance changes. The current of 1 µA across the cell layer had no detectable deleterious effect on the cells. The impedance of each well was measured at 2-minute intervals; impedance for both a cell-free and a cell-covered electrode was measured at frequencies from 25 to 60 kHz. Comparing the experimental data from cell-covered electrodes with the calculated values obtained from the model and the impedance data from the cell-free electrode, we determined the parameter Rb as described previously [34,35].
Statistical Analysis
Survival data were collected by chart review and archived under protocols approved by the institutional review board. qRT-PCR data were tested for normality by using the Kolmogorov-Smirnov method and failed (P < .001); therefore, nonparametric methods (Mann-Whitney test, Kruskal-Wallis, Spearman's rank correlation) were used for statistical analysis. Using a subset of 56 grade 3, advancedstage (stages 3 or 4) samples, claudin-4 expression was correlated with survival, chemosensitivity, optimal debulking status, primary versus recurrent tumors, and short-term versus long-term survival. ECIS data were tested for normality vy using the Kolmogorov-Smirnov method. Oneway analysis of variance and the Bonferroni t test were used to compare the effect of various concentrations of C-CPE on the parameter Rb. All statistical testing was performed by using software provided by Statview (SAS, Cary, NC), Minitab (State College, PA), or SigmaStat (Point Richmond, CA), and statistical significance was defined as α < .05.
Results
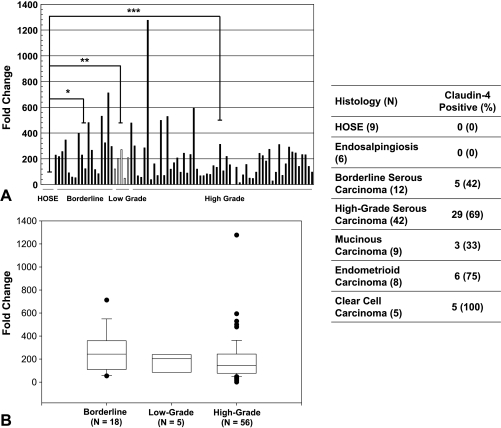
As shown in Figure 1A, significantly higher claudin-4 expression was found in borderline (n = 18, P = .003), lowgrade (n = 5, P = .02), and high-grade cancers (n = 56, P < .001) when compared with HOSE (n = 4). However, there was no difference in claudin-4 expression between borderline, low-grade, and high-grade serous carcinomas (P = .14, Figure 1B). IHC data are summarized in Figure 1 and representative slides are shown in Figure 2. Claudin-4 staining intensity was generally strong in endometrioid carcinomas, strong to intermediate in high-grade serous, borderline serous, and mucinous carcinomas, and weak in clear cell carcinomas. Claudin-4 expression by qRT-PCR and IHC were compared in a subset of 42 grade 3, advanced-stage (stages 3 or 4) serous cancers and found to have a statistically significant relationship (P = .04).
Figure 1.
(A) qRT-PCR was used to compare claudin-4 expression in 79 serous ovarian cancer tissue samples of various grades to HOSE tissue. Claudin-4 expression was significantly higher in borderline, low-grade, and high-grade cancers when compared to HOSE. *P = .003, **P = .02, ***P < .001; (B) expression was not significantly different between high-grade and borderline or low-grade cancers (P = .14). Table: Using IHC, 76 ovarian cancer tissue samples of various histologic types were stained for claudin-4 and compared to nine HOSE and six endosalpingiosis samples. Slides were scored as follows: 0, absent/trace staining; 1, minimal staining; 2, moderate staining; 3, strong staining. Samples were categorized into “claudin-4-negative” (absent/trace staining, score 0) and “claudin-4-positive” (any degree of staining, scores 1–3) groups for comparison and statistical analysis.
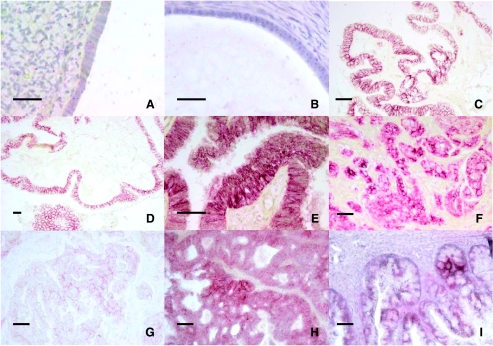
Figure 2.
Representative claudin-4-immunostained IHC slides from various histologic types of ovarian tissue. (A) Normal ovarian surface epithelium, (B) endosalpingiosis, (C and D) borderline serous carcinoma, (E and F) high-grade serous carcinoma, (G) clear cell carcinoma, (H) endometrioid carcinoma, and (I) mucinous carcinoma. Scale bar = 20 µm.
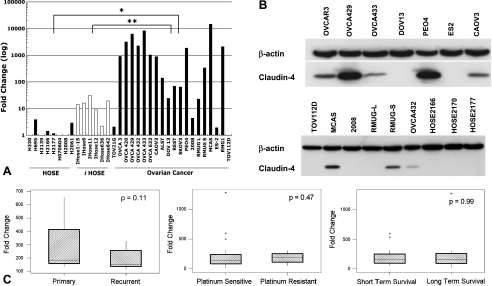
Claudin-4 expression was also found to be significantly higher in ovarian cancer cell lines when compared to HOSE (P < .001) and immortalized HOSE (P = .04) (Figure 3A). Nine of 14 serous (TOV21G to 2008), two of three mucinous (RMUGL to MCAS), and one of two clear cell (ES2, RMG1) cancer cell lines exhibited greater than 100-fold increased expression compared to HOSE. Similarly, Western blot (Figure 3B) showed claudin-4 expression in 8 of the 13 ovarian cancer cell lines, whereas expression was absent in all three HOSE cell lines. In these ovarian cancer cell lines, Western blot claudin-4 expression positively correlated with qRT-PCR claudin-4 expression (R = 0.70, P = .01).
Figure 3.
(A) qRT-PCR was used to compare claudin-4 expression in 20 ovarian cancer cell lines to eight HOSE and six immortalized HOSE cell lines. *P < .001, **P = .04. (B) Claudin-4 expression, as determined by Western blot, in ovarian cancer and HOSE cell lines. β-Actin was used as a loading control. (C) Claudin-4 expression, as determined by qRT-PCR, was compared in a subset of high-grade advanced-stage tissue samples. There was no statistically significant difference between paired primary and recurrent tumors, platinum-sensitive and platinum-resistant tumors, or short-term (less than 24 months) and long-term (60 months or more) survivors.
Claudin-4 expression was correlated to a number of clinically important outcomes in a subset of high-grade, advanced-stage serous ovarian cancer samples. Cox proportional hazards testing did not reveal any significant association between claudin-4 expression and survival (P = .21). Claudin-4-positive and claudin-4-negative IHC groups were compared by using Kaplan-Meier methods (curves not shown) and no difference in survival was found (log rank test, P = .42). As depicted in Figure 3C, claudin-4 expression was not different between paired primary and recurrent tumors (n = 7, P = .11), platinum-sensitive and platinum-resistant tumors (n = 36, P = .47), or short-term survivors (less than 24 months) and long-term survivors (60 months or more) (n = 35, P = .99). No statistically significant relationship was found between claudin-4 expression (as assessed by IHC) and optimal debulking (n = 42, Fisher exact test, P = .71) or platinum chemosensitivity (n = 30, Fisher exact test, P = .67). Twelve of the 18 borderline carcinomas were clinically stage I, whereas almost all of the high-grade carcinomas were stage III or IV. There was no difference in claudin-4 expression based on stage in borderline carcinomas (analysis of variance, P = .99) or high-grade carcinomas (Fisher exact test, P = .38).
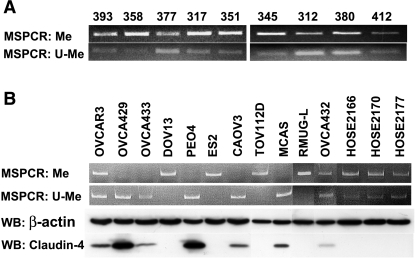
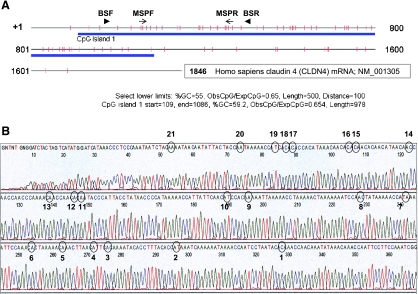
The methylation status of CLDN4 was determined in nine ovarian cancer tissue samples (Figure 4A), all of which exhibited both methylated and unmethylated PCR products. All nine of these samples overexpressed claudin-4 compared to HOSE (qRT-PCR fold change ∼80–200, per Figure 1A). Seven of the 11 ovarian cancer cell lines exhibited an unmethylated CLDN4 allele, and each of these expressed claudin-4. In contrast, in all four cancer cell lines with a methylated allele only, claudin-4 expression was absent. The three HOSE cell lines exhibited both methylated and unmethylated PCR products—none of them expressed claudin-4. DOV13 and TOV112D, neither of which expressed claudin-4 (per Figure 3B) and both of which had a methylated CLDN4 allele only, were treated with 5-AZA, an inhibitor of DNA methylation. 5-AZA treatment increased claudin-4 expression, as demonstrated by qRT-PCR, and was accompanied by the appearance of a new unmethylated band on MSPCR in both cell lines (Figure 5). To confirm the completeness of the bisulfite modification and the accuracy of MSPCR, bisulfite sequencing was performed on OVCA429, a cancer cell line with an unmethylated CLDN4 allele only (per Figure 4B). Figure 6A shows the location of the CpG island and MSPCR and bisulfite-sequencing primers. The bisulfite-sequenced fragment contained 21 CpG dinucleo tides, all of which were unmethylated (Figure 6B).
Figure 4.
MSPCR was performed and average of three to four times on genomic DNA obtained from (A) nine claudin-4-expressing ovarian cancer tissue samples (393, 358, 377, 317, 351, 345, 312, 380, 412), (B) 11 ovarian cancer cell lines (OVCAR3, OVCA429, OVCA433, DOV13, PEO4, ES2, CAOV3, TOV112D, MCAS, RMUG-L, OVCA432) and three HOSE cell lines (HOSE2166, HOSE2170, HOSE2177). Also shown in (B), the methylation status of CLDN4 was compared to its protein expression in the cell lines. β-Actin was used as a loading control. Me, methylated primer; U-Me, unmethylated primer; WB, Western blot.
Figure 5.
Ovarian cancer cell lines DOV13 and TOV112D, neither of which expressed claudin-4, were treated with 5-AZA (1 µmol/l), an inhibitor of DNA methylation. Treatment with 5-AZA resulted in the appearance of new unmethylated MSPCR products (inset) and corresponded with increased expression of claudin-4, as assessed by qRT-PCR (bar graph). The experiments were performed in duplicate. Also shown, MSPCR resulted in a methylated product for the methylated control DNA; PCR products were absent for unmodified DNA (which had not undergone bisulfite treatment). Me, methylated primer; U-Me, unmethylated primer.
Figure 6.
(A) The schematic diagram represents claudin-4 mRNA: +1, transcription initiation site; | (vertical marks), CpG dinucleotide; blue band, CpG island identified using CpG Island Searcher software (http://ccnt.hsc.usc.edu/cpgislands/) and the displayed criteria; → (arrowhead), forward (BSF, bp 156–180) and reverse (BSR, bp 468–491) primer locations for the bisulfite sequencing; ▸ (arrows), forward (MSPF, bp 233–256) and reverse (MSPR, bp 432–452) primer locations for MSPCR. (B) The ovarian cancer cell line OVCA429, which exhibited a hypomethylated CLDN4 allele only (per Figure 4B), underwent bisulfite sequencing. Complete hypomethylation was demonstrated at all 21 CpG dinucleotides (circled and numbered) in the sequenced fragment.
Using ECIS, transepithelial impedance was measured and junctional (paracellular) resistance Rb was calculated in nine ovarian cancer cell lines with varying levels of claudin-4 overexpression (qRT-PCR fold change range, 2–2120). There was a positive correlation between Rb and claudin-4 (R = 0.85, P < .001). Using siRNA, claudin-4 expression was silenced in the cancer cell line SKOV3 (Figure 7A) and Rb was determined during 7 days in culture. As shown in Figure 7B, Rb was lower in the claudin-4 silenced SKOV3 compared with the wild-type SKOV3, and this was found to be statistically significant on days 6 and 7. Exposure of OVCA429 (highly claudin-4 overexpressing, per Figure 3A) to increasing concentrations of C-CPE did not result in any changes in cell morphology, nor did it result in cell cytotoxicity (data not shown). However, in OVCA429, C-CPE treatment resulted in a dose-dependent and statistically significant (P < .05) ∼50% decrease in Rb (compared to a control concentration of 0 µg/ml) after 60 hours at concentrations of 1 x 10-3, 1 x 10-2, and 1 x 10-1 µg/µl (Figure 7C). In contrast, when SKOV3 (lower claudin-4 overexpression) was treated with C-CPE, it remained relatively resistant to changes in Rb, except at the highest C-CPE concentration (1 x 10-1 µg/µl), when a significant ∼40% decrease in Rb was observed compared to control (Figure 7D).
Figure 7.
(A) Claudin-4 Western blot for SKOV3 WT, a negative control, and SKOV3 siRNA (claudin-4 silenced). β-Actin was used as a loading control. (B) Junctional (paracellular) resistance (Rb) was compared between SKOV3 WT (wild type) and SKOV3 siRNA (siRNA claudin-4 silenced) cell lines over 7 days in cell culture medium, *P < .05. Cell lines (C) OVCA429 and (D) SKOV3 were treated with increasing amounts of C-CPE (µg/µl), resulting in a dose-dependent decrease in Rb compared to control (cell culture medium only, C-CPE concentration of 0 µg/µl). Bars represent means ± SD. Measurements were repeated four to six times, *P < .05.
Discussion
This study sought to elucidate the patterns, mechanism, and significance of claudin-4 overexpression in EOC. Our results show that claudin-4 is overexpressed in almost all borderline and invasive serous ovarian carcinomas. Nonetheless, claudin-4 expression is not different between grades and stages and was absent in all the HOSE and endosalpingiosis samples. As some borderline carcinomas are thought to progress in a stepwise fashion toward their malignant counterparts [36], this raises the possibility that claudin-4 overexpression may be an early event in EOC tumorigenesis. Other investigators have come to similar conclusions, noting aberrant claudin expression in preinvasive as well as invasive carcinomas of the pancreas, breast, and cervix [13,16,37]. Our current findings expand on those of Rangel et al. [6] and Zhu et al. [38], who reported on claudin-4 expression in various histologic subtypes of EOC, albeit in small numbers. Interestingly, the highest percentage of staining occurred in endometrioid and clear cell carcinomas, histologic subtypes thought to have a different pathogenesis than the more common serous carcinomas, again suggesting that claudin-4 overexpression may be present at the earliest stages of tumorigenesis.
Overexpression of claudin-4 may be mediated through multiple mechanisms, and gene amplification may be one of these. However, comparative genomic hybridization analysis of our oligonucleotide array did not show any changes in CLDN4 DNA copy number (data not shown). Methylation of 5′-cytosines in CpG islands is one of the most important epigenetic modulators of gene expression [39], and alterations in the methylation status of CLDN4 and the CLDN7 promoter have been associated with aberrant gene expression in pancreatic and breast carcinomas, respectively [13,21]. Using MSPCR, we found that all the ovarian cancer cell lines with a hypomethylated CLDN4 allele overexpressed claudin-4 and that treatment with a demethylating agent induced claudin-4 expression in cell lines not expressing claudin-4. Bilsufite sequencing was also performed, and confirmed the completeness of the bisulfite modification and the accuracy of the MSPCR results. Similar findings were reported in an elegant and recently published paper; namely, that promoter methylation and histone deacetylation contribute to claudin-4 silencing in ovarian cancer cells [40]. As cultured cells may have an altered methylation pattern compared to their tissue counterparts [13], we also examined ovarian cancer tissue samples and found both methylated and hypomethylated CLDN4 alleles in all the samples (all of which overexpressed claudin-4). Although the methylated PCR products in these tissue samples may be secondary to stromal cell contaminants harvested at the time of genomic DNA extraction, an alternative suggestion is that CLDN4 was only partially hypomethylated. This view is supported by Honda et al. [40], who showed that the inhibition of claudin-4 expression is related to the density of methylated CpG dinucleotides. Moreover, we observed both methylated and unmethylated CLDN4 alleles in the cancer cell lines OVCAR3 and OVCA432 and in the three HOSE cell lines. In contrast to OVCAR3 and OVCA432, the three HOSE cell lines did not express claudin-4. This observation may again be related to the relative density of DNA methylation, as noted above. Alternatively, the above-mentioned authors also found that although claudin-4 expression increased after treatment with inhibitors of DNA methylation and histone deacetylase, the mRNA and protein products did not correlate well. This suggests that other regulatory mechanisms may be involved in the expression of claudin-4 [40]. Lastly, it is interesting to note that whereas Honda et al. [40] characterized the methylation status of the CLDN4 promoter as well as some downstream elements, our MSPCR primers did not include the CLDN4 promoter region (Figure 6A); still, the methylation status of CLDN4 correlated with claudin-4 expression.
Although other investigators have shown that claudin-4 is overexpressed in ovarian cancer [3–7], the clinical and biological significance of this finding is yet to be fully elucidated. Claudin-4 overexpression has been associated with platinum resistance in ovarian cancer based on proteomic analysis [41], and moderate to strong claudin-4 staining was associated with decreased survival in gastric carcinomas [42]. Accordingly, we sought to compare claudin-4 expression to several clinically important outcomes. We found that claudin-4 expression did not correlate with survival, nor was expression different between primary and recurrent tumors. Consistent with this finding, claudin-4 expression was not different between chemoresistant and chemosensitive tumors, and expression did not correlate with the ability to optimally debulk tumor burden at the time of surgery. These findings suggest that although claudin-4 overexpression may have an important role in ovarian cancer, its overexpression is not a marker of increased tumor aggressiveness per se.
Claudin proteins form the backbone of TJ strands [9], and their existence is necessary and sufficient for TJ formation [8]. Yet, although claudin-4 predominantly localizes to the cell membrane in ovarian cancer, as shown in this study and by others [3,4,7], it is not clear whether this corresponds to the formation of functional TJs with the ability to regulate paracellular flow (barrier function). Although Zhu et al. [43] suggested that functional TJs exist in normal ovarian surface epithelium, using the transepithelial electric resistance (TER) method, Rangel et al. [6] suggested that claudin-4 overexpression in ovarian cancer does not correlate with the presence of functional TJs. TER measurements have been directly related to TJ barrier function [9]; however, TER data can be inconsistent, do not always correlate with TJ morphology, and do not separately account for the resistance due to cell-substrate contacts [9,44]. With this in mind, we used ECIS to measure TJ barrier function. ECIS accounts for the transepithelial current flowing through the paracellular and cell-substrate spaces separately and, as a result, individually determines the contribution of each of these currents to total transepithelial resistance [44]. Using ECIS, we found that as claudin-4 expression increased, so did the junctional (paracellular) resistance. Junctional resistance was also found to be significantly lower in claudin-4 silenced SKOV3 cells compared to the wild type. Lastly, treatment of claudin-4-expressing ovarian cancer cell lines with a C-terminal fragment of C. perfringens enterotoxin (C-CPE), which binds claudin-4, resulted in decreased junctional resistance in a dose- and claudin-4-dependent fashion, and this was not due to cell cytotoxicity. These findings suggest that claudin-4 contributes to the formation of functional TJs and is consistent with the observation that epithelial ovarian carcinomas, as opposed to most other cancers, acquire more stable and more differentiated epithelial characteristics and morphology as tumorigenesis proceeds [45].
Jang et al. [46] observed that pretreatment with paclitaxel decreased cell density and improved subsequent drug penetration and accumulation in the tumor core. Accordingly, the ability of cytotoxic drugs to diffuse paracellularly through the interstitium deep into the tumor core may be important, especially for IP drug delivery, when drugs are delivered without primary access to the vascular compartment and thus largely depend on surface diffusion [25]. C-CPE has been used in a noncytotoxic manner to target claudin-4, disrupt TJ barrier function, and hence improve drug delivery in the small bowel [23]. Our demonstration that C-CPE decreased TJ barrier function (junctional resistance) in ovarian cancer cells in a dose- and claudin-4-dependent fashion raises the possibility that C-CPE may play a future role in drug delivery strategies. C-CPE may potentially be used as an absorption or diffusion-enhancing agent, or, alternatively, by acting as a claudin-4-targeting fusion molecule [27]. Lastly, although claudins are known to localize exclusively to the TJ [9], using ECIS, we also found that C-CPE affected the cell-substrate separation (data not shown). Whether this observation is due to mechanical alterations in the cell cytoskeleton or changes in cell signaling secondary to TJ disruption is a current area of investigation by our group.
In summary, our results confirm that claudin-4 is overexpressed in a variety of EOCs and that this is related to CLDN4 hypomethylation. Furthermore, claudin-4 overexpression in EOC may be an early event and is not a marker of increased tumor aggressiveness or poor prognosis. Lastly, it appears that claudin-4 overexpression corresponds with the existence of functional TJs, and that C-CPE, which targets claudin-4, may be used to diminish TJ barrier function. Future studies examining the biologic response of ovarian cancer to C-CPE, alone and in combination with cytotoxic drugs, and studies using animal models are currently under way to further evaluate and expand on these findings.
Abbreviations
- CPE
Clostridium perfringens enterotoxin
- C-CPE
C-terminal fragment of Clostridium perfringens enterotoxin
- ECIS
electric cell-substrate impedance sensing
- EOC
epithelial ovarian cancer
- HOSE
human ovarian surface epithelium
- IP
intraperitoneal
- Rb
junctional (paracellular) resistance
Footnotes
This study was supported by The Harold Amos Faculty Development Program of the Robert Wood Johnson Foundation (E.G.), R33CA103595 and the Ovarian Cancer SPORE P50CA105009 (S.M.) from The National Institutes of Health, Department of Health and Human Services, The Gillette Center For Women's Cancers, Adler Foundation, Inc., Edgar Astrove Fund, The Ovarian Cancer Research Fund, Inc., The Morse Family Fund, The Natalie Pihl Fund (B.L., J.K, M.A., J.H., R.B., S.M., E.G.), The Ruth N. White Research Fellowship (B.L.), R37AI19844 from The National Institute of Allergy and Infectious Diseases, National Institutes of Health (J.S., B.M.), and the Robert and Deborah First Fund (S.M.).
References
- 1.Vignati S, Albertini V, Rinaldi A, Kwee I, Riva C, Oldrini R, Capella C, Bertoni F, Carbone GM, Catapano CV. Cellular and molecular consequences of peroxisome proliferator-activated receptorgamma activation in ovarian cancer cells. Neoplasia. 2006;8:851–861. doi: 10.1593/neo.06433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonome T, Lee JY, Park DC, Radonovich M, Pise-Masison C, Brady J, Gardner GJ, Hao K, Wong WH, Barrett JC, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–10612. doi: 10.1158/0008-5472.CAN-05-2240. [DOI] [PubMed] [Google Scholar]
- 3.Hibbs K, Skubitz KM, Pambuccian SE, Casey RC, Burleson KM, Oegema TR, Jr, Thiele JJ, Grindle SM, Bliss RL, Skubitz AP. Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol. 2004;165:397–414. doi: 10.1016/S0002-9440(10)63306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281–6287. [PubMed] [Google Scholar]
- 5.Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Au JL, Rosen DG, Liu J, Hellstrom I, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- 6.Rangel LB, Agarwal R, D'Souza T, Pizer ES, Alo PL, Lancaster WD, Gregoire L, Schwartz DR, Cho KR, Morin PJ. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 2003;9:2567–2575. [PubMed] [Google Scholar]
- 7.Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, et al. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer. 2004;112:14–25. doi: 10.1002/ijc.20408. [DOI] [PubMed] [Google Scholar]
- 8.Sawada N, Murata M, Kikuchi K, Osanai M, Tobioka H, Kojima T, Chiba H. Tight junctions and human diseases. Med Electron Microsc. 2003;36:147–156. doi: 10.1007/s00795-003-0219-y. [DOI] [PubMed] [Google Scholar]
- 9.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 10.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 11.Brehm R, Ruttinger C, Fischer P, Gashaw I, Winterhager E, Kliesch S, Bohle RM, Steger K, Bergmann M. Transition from preinvasive carcinoma in situ to seminoma is accompanied by a reduction of connexin 43 expression in Sertoli cells and germ cells. Neoplasia. 2006;8:499–509. doi: 10.1593/neo.05847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swisshelm K, Macek R, Kubbies M. Role of claudins in tumorigenesis. Adv Drug Deliv Rev. 2005;57:919–928. doi: 10.1016/j.addr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Kominsky SL, Argani P, Korz D, Evron E, Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP, Sukumar S. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–2033. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 14.Kominsky SL, Vali M, Korz D, Gabig TG, Weitzman SA, Argani P, Sukumar S. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol. 2004;164:1627–1633. doi: 10.1016/S0002-9440(10)63721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long H, Crean CD, Lee WH, Cummings OW, Gabig TG. Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res. 2001;61:7878–7881. [PubMed] [Google Scholar]
- 16.Nichols LS, Ashfaq R, Iacobuzio-Donahue CA. Claudin 4 protein expression in primary and metastatic pancreatic cancer: support for use as a therapeutic target. Am J Clin Pathol. 2004;121:226–230. doi: 10.1309/K144-PHVD-DUPD-D401. [DOI] [PubMed] [Google Scholar]
- 17.Michl P, Barth C, Buchholz M, Lerch MM, Rolke M, Holzmann KH, Menke A, Fensterer H, Giehl K, Lohr M, et al. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 2003;63:6265–6271. [PubMed] [Google Scholar]
- 18.Soini Y, Tommola S, Helin H, Martikainen P. Claudins 1, 3, 4 and 5 in gastric carcinoma, loss of claudin expression associates with the diffuse subtype. Virchows Arch. 2006;448:52–58. doi: 10.1007/s00428-005-0011-6. [DOI] [PubMed] [Google Scholar]
- 19.Agarwal R, D'Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005;65:7378–7385. doi: 10.1158/0008-5472.CAN-05-1036. [DOI] [PubMed] [Google Scholar]
- 20.Aravindakshan J, Chen X, Sairam MR. Differential expression of claudin family proteins in mouse ovarian serous papillary epithelial adenoma in aging FSH receptor-deficient mutants. Neoplasia. 2006;8:984–994. doi: 10.1593/neo.06529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C, Goggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–4166. [PubMed] [Google Scholar]
- 22.Wallace FM, Mach AS, Keller AM, Lindsay JA. Evidence for Clostridium perfringens enterotoxin (CPE) inducing a mitogenic and cytokine response in vitro and a cytokine response in vivo. Curr Microbiol. 1999;38:96–100. doi: 10.1007/s002849900410. [DOI] [PubMed] [Google Scholar]
- 23.Kondoh M, Masuyama A, Takahashi A, Asano N, Mizuguchi H, Koizumi N, Fujii M, Hayakawa T, Horiguchi Y, Watanbe Y. A novel strategy for the enhancement of drug absorption using a claudin modulator. Mol Pharmacol. 2005;67:749–756. doi: 10.1124/mol.104.008375. [DOI] [PubMed] [Google Scholar]
- 24.Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier. J Cell Biol. 1999;147:195–204. doi: 10.1083/jcb.147.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markman M. Intraperitoneal antineoplastic drug delivery: rationale and results. Lancet Oncol. 2003;4:277–283. doi: 10.1016/s1470-2045(03)01074-x. [DOI] [PubMed] [Google Scholar]
- 26.Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, Copeland LJ, Walker JL, Burger RA. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 27.Ebihara C, Kondoh M, Hasuike N, Harada M, Mizuguchi H, Horiguchi Y, Fujii M, Watanabe Y. Preparation of a claudin-targeting molecule using a C-terminal fragment of Clostridium perfringens enterotoxin. J Pharmacol Exp Ther. 2006;316:255–260. doi: 10.1124/jpet.105.093351. [DOI] [PubMed] [Google Scholar]
- 28.Tsao SW, Mok SC, Fey EG, Fletcher JA, Wan TS, Chew EC, Muto MG, Knapp RC, Berkowitz RS. Characterization of human ovarian surface epithelial cells immortalized by human papilloma viral oncogenes (HPV-E6E7 ORFs) Exp Cell Res. 1995;218:499–507. doi: 10.1006/excr.1995.1184. [DOI] [PubMed] [Google Scholar]
- 29.Kim JH, Skates SJ, Uede T, Wong KK, Schorge JO, Feltmate CM, Berkowitz RS, Cramer DW, Mok SC. Osteopontin as a potential diagnostic biomarker for ovarian cancer. JAMA. 2002;287:1671–1679. doi: 10.1001/jama.287.13.1671. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuda H, Ito YM, Ohashi Y, Wong KK, Hashiguchi Y, Welch WR, Berkowitz RS, Birrer MJ, Mok SC. Identification of overexpression and amplification of ABCF2 in clear cell ovarian adenocarcinomas by cDNA microarray analyses. Clin Cancer Res. 2005;11:2880–2888. doi: 10.1158/1078-0432.CCR-05-0751. [DOI] [PubMed] [Google Scholar]
- 33.Kokai-Kun JF, McClane BA. Deletion analysis of the Clostridium perfringens enterotoxin. Infect Immun. 1997;65:1014–1022. doi: 10.1128/iai.65.3.1014-1022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giaever I, Keese CR. Micromotion of mammalian cells measured electrically. Proc Natl Acad Sci USA. 1991;88:7896–7900. doi: 10.1073/pnas.88.17.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo CM, Keese CR, Giaever I. Impedance analysis of MDCK cells measured by electric cell-substrate impedance sensing. Biophys J. 1995;69:2800–2807. doi: 10.1016/S0006-3495(95)80153-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih Ie M, Kurman RJ. Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am J Pathol. 2004;164:1511–1518. doi: 10.1016/s0002-9440(10)63708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobel G, Paska C, Szabo I, Kiss A, Kadar A, Schaff Z. Increased expression of claudins in cervical squamous intraepithelial neoplasia and invasive carcinoma. Hum Pathol. 2005;36:162–169. doi: 10.1016/j.humpath.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Brannstrom M, Janson PO, Sundfeldt K. Differences in expression patterns of the tight junction proteins, claudin 1, 3, 4 and 5, in human ovarian surface epithelium as compared to epithelia in inclusion cysts and epithelial ovarian tumours. Int J Cancer. 2006;118:1884–1891. doi: 10.1002/ijc.21506. [DOI] [PubMed] [Google Scholar]
- 39.Esteller M. Relevance of DNA methylation in the management of cancer. Lancet Oncol. 2003;4:351–358. doi: 10.1016/s1470-2045(03)01115-x. [DOI] [PubMed] [Google Scholar]
- 40.Honda H, Pazin MJ, Ji H, Wernyj RP, Morin PJ. Crucial roles of Sp1 and epigenetic modifications in the regulation of the CLDN4 promoter in ovarian cancer cells. J Biol Chem. 2006;281:21433–21444. doi: 10.1074/jbc.M603767200. [DOI] [PubMed] [Google Scholar]
- 41.Stewart JJ, White JT, Yan X, Collins S, Drescher CW, Urban ND, Hood L, Lin B. Proteins associated with cisplatin resistance in ovarian cancer cells identified by quantitative proteomic technology and integrated with mRNA expression levels. Mol Cell Proteomics. 2006;5:433–443. doi: 10.1074/mcp.M500140-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Resnick MB, Gavilanez M, Newton E, Konkin T, Bhattacharya B, Britt DE, Sabo E, Moss SF. Claudin expression in gastric adenocarcinomas: a tissue microarray study with prognostic correlation. Hum Pathol. 2005;36:886–892. doi: 10.1016/j.humpath.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Y, Maric J, Nilsson M, Brannstrom M, Janson PO, Sundfeldt K. Formation and barrier function of tight junctions in human ovarian surface epithelium. Biol Reprod. 2004;71:53–59. doi: 10.1095/biolreprod.103.022913. [DOI] [PubMed] [Google Scholar]
- 44.Lo CM, Keese CR, Giaever I. Cell-substrate contact: another factor may influence transepithelial electrical resistance of cell layers cultured on permeable filters. Exp Cell Res. 1999;250:576–580. doi: 10.1006/excr.1999.4538. [DOI] [PubMed] [Google Scholar]
- 45.Sundfeldt K. Cell-cell adhesion in the normal ovary and ovarian tumors of epithelial origin; an exception to the rule. Mol Cell Endocrinol. 2003;202:89–96. doi: 10.1016/s0303-7207(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 46.Jang SH, Wientjes MG, Au JL. Enhancement of paclitaxel delivery to solid tumors by apoptosis-inducing pretreatment: effect of treatment schedule. J Pharmacol Exp Ther. 2001;296:1035–1042. [PubMed] [Google Scholar]