Abstract
Fission yeast replication checkpoint kinases Rad3p and Cds1p are essential for maintaining cell viability after transient treatment with hydroxyurea (HU), an agent that blocks DNA replication. Although current studies have focused on the cyclin-dependent protein kinase Cdc2p that is regulated by these checkpoint kinases, other aspects of their functions at the onset of S phase arrest have not been fully understood. In this study, we use genome-wide DNA microarray analyses to show that HU-induced change of expression profiles in synchronized G2 cells occurs specifically at the onset of S phase arrest. Induction of many core environmental stress response genes and repression of ribosomal genes happen during S phase arrest. Significantly, peak expression level of the MluI-like cell cycle box (MCB)-cluster (G1) genes is maintained at the onset of S phase arrest in a Rad3p- and Cds1p-dependent manner. Expression level maintenance of the MCB-cluster is mediated through the accumulation of Rep2p, a putative transcriptional activator of the MBF complex. Conversely, the FKH-cluster (M) genes are repressed during the onset of S phase arrest in a Rad3p-dependent manner. Repression of the FKH-cluster genes is mediated through the decreased levels of one of the putative forkhead transcription factors, Sep1p, but not Fkh2p. Together, our results demonstrate that Rad3p and Cds1p modulate transcriptional response during the onset of S phase arrest.
INTRODUCTION
Cell cycle is a term describing a fundamental biological phenomenon whereby a cell duplicates its genome (DNA replication), segregates chromosomes (mitosis), and forms two identical daughter cells (cytokinesis). Orderly progression through cell cycle events is critical for the maintenance of genome integrity. Eukaryotic cells are equipped with a number of surveillance mechanisms known as checkpoints that monitor completion of various events to ensure a sequential progression through various stages of the cell cycle. Individual events within the cell cycle can be arrested at a checkpoint if an upstream event has not been completed or if damage to DNA is detected (Elledge, 1996; Carr, 1997; Matsuoka et al., 1998). A common downstream target for all checkpoints in the fission yeast Schizosaccharomyces pombe is cyclin-dependent protein kinase Cdc2p whose activity is inhibited by phosphorylation at the tyrosine-15 residue upon activation of checkpoints (Gould and Nurse, 1989; Rhind and Russell, 1998).
Checkpoints that monitor DNA damage and blocks to DNA replication seem to be conserved in most eukaryotes (Elledge, 1996; Carr, 1997; Matsuoka et al., 1998). In S. pombe, ATM-related kinase (Rad3p; Mec1p in Saccharomyces cerevisiae) is a component of a sensor complex including Rad1p, Rad9p, Rad17p, Rad26p, and Hus1p (Murray et al., 1991; Enoch et al., 1992; Rowley et al., 1992; al-Khodairy et al., 1994). When the complex detects DNA lesions, Rad3p activates CHEK1-like kinase (Chk1p) to arrest cells at the G2/M boundary (the G2/M checkpoint) (Walworth et al., 1993; Walworth and Bernards, 1996). Conversely, when the complex senses a stalled DNA replication fork, Rad3p activates CHEK2-like kinase (Cds1p; Rad53p in S. cerevisiae) to arrest cells at S phase (the intra-S phase checkpoint) (Murakami and Okayama, 1995; Lindsay et al., 1998). Both Cds1p and Chk1p phosphorylate/inhibit Cdc2p from causing cell cycle arrest at S phase and G2/M boundary, respectively (Boddy et al., 1998; Rhind and Russell, 1998). This study highlights the observation of a much higher level (∼25%) of “cut” phenotypes (assembly of septa in cells without completion of DNA synthesis and/or chromosome segregation) in the rad3− mutant than that (∼10%) seen in the cds1− upon hydroxyurea (HU) treatment (HU inhibits ribonucleotide reductase RNR causing depletion of intercellular dNTP and consequently inducing replication blocks). This indicates that Rad3p activates an additional pathway to prevent high frequency division-septum formation in a cds1+-independent manner.
Microarray analyses in fission yeast have highlighted several hundred genes that display cell cycle-stage–specific expression, many of which are involved in cell cycle regulation and/or cell cycle–specific events (Rustici et al., 2004; Oliva et al., 2005; Peng et al., 2005). Furthermore, several cell cycle–specific transcriptional activities that are responsible for the cell cycle-regulated expression patterns have also been identified, besides the previously characterized MBF activity (also known as DSC1, consisting of Cdc10p, Res1p, Res2p, and Rep2p) that regulates MluI-like cell cycle box (MCB) motif-containing genes (Lowndes et al., 1992; Caligiuri and Beach, 1993; Miyamoto et al., 1994; Sugiyama et al., 1994; Zhu et al., 1994; McInerny et al., 1995; Nakashima et al., 1995; Baum et al., 1997; White et al., 2001). Approximately 15 MCB motif-containing genes exhibited MBF-dependent expression and peaked at G1 phase, most of which are involved in DNA replication and are required for the G1/S transition (Rustici et al., 2004; Oliva et al., 2005; Peng et al., 2005). Many ACE2 motif-containing genes, encoding proteins involved in regulation of cytokinesis and cell separation, are regulated by a zinc-finger transcription factor Ace2p (Martin-Cuadrado et al., 2003), and they show a peak expression at a time interval that overlaps with expression of MCB genes owing to a short G1 phase in fission yeast. Expression of ace2+ is controlled by forkhead transcription factor Sep1p and Fkh2p (Zilahi et al., 2000; Buck et al., 2004) that at the same time regulates >50 other forkhead (FKH; also known as SFF) motif-containing genes, most of which encode proteins involved in mitosis, exit of mitosis, and cytokinesis.
In this study, we investigated the effect of HU-induced DNA replication block on genome-wide expression profiles. We show that dynamic expression profile changes of synchronized G2 cells in response to HU treatment can be clearly determined when expression levels of the synchronized cells supplemented with HU or without (control) were compared with a common reference. Expression patterns in synchronized cells treated with HU did not exhibit noticeable changes until encountering S phase arrest. Many core environmental stress response (CESR) genes (Chen et al., 2003) were found to change expression profiles at the onset of S phase arrest. Significantly, the peak expression levels of the MCB- and histone-cluster genes were maintained. The expression level maintenance correlated with the accumulation of Rep2p. Furthermore, overproduction of Rep2p alleviated rad3Δ or cds1Δ phenotypes. Conversely, the FKH- and ACE2-clusters seemed to be repressed during the S phase block. This repression depended on Rad3p function. Notably, in cds1Δ cells Chk1p could complement for repression of the FKH-clusters, but not for maintenance of peak expression levels of the MCB-clusters. Repression of the FKH-cluster was associated with the decreased Sep1 protein levels. Moreover, disruption of sep1Δ alleviated rad3Δ phenotype.
MATERIALS AND METHODS
Strains, Media, and Culture Manipulations
Strains used in this study are listed in Table 1. Construction of various strains using a polymerase chain reaction (PCR)-mediated gene disruption protocol (Bahler et al., 1998) is described in Supplemental Material.
Table 1.
List of S. pombe strains used in this study
| Strain | Relevant genotype | Comments |
|---|---|---|
| LJY1497 | leu1-32 h− | Laboratory stock |
| LJY188 | leu1-32 ura4-D18 h− | Laboratory stock |
| LJY1715 | rad3Δ::ura4+leu1-32 ura4-D18 h− | This study |
| LJY1687 | cds1Δ::ura4+leu1-32 ura4-D18 h− | Laboratory stock |
| LJY1669 | chk1Δ::ura4+leu1-32 ura4-D18 h− | Laboratory stock |
| LJY291 | cds1Δ:: ura4+chk1Δ::ura4+leu1-32 his3-D1 ura4-D18 h− | This study |
| LJY2254 | rep1Δ::ura4+ura4-D18 leu1-32 h− | This study |
| LJY2125 | rep2Δ::ura4+ura4-D18 leu1-32 h− | This study |
| LJY2255 | res1Δ::ura4+ura4-D18 leu1-32 h− | This study |
| LJY2135 | res2Δ::ura4+ura4-D18 leu1-32 h− | This study |
| LJY2123 | sep1Δ::ura4+leu1-32 ura4-D18 h− | Laboratory stock |
| LJY2264 | fkh2Δ::ura4+ura4-D18 leu1-32 h− | This study |
| LJY1603 | ace2Δ::ura4+ura4-D18 leu1-32 h− | This study |
| LJY275 | cdc10-V50 ura4-D18 h− | Laboratory stock |
| LJY172 | cdc25–22 ura4-D18 h+ | Laboratory stock |
| LJY2660 | cds1Δ::ura4+leu1-32 his3-D1 ura4-D18 ade6-M216 h+ | Laboratory stock |
| LJY2235 | fkh2+-3HA-6His::ura4+ura4-D18 leu1-32 h− | This study |
| LJY1842 | sep1+-3HA-6his::ura4+leu1-32 ura4-D18 h− | This study |
| LJY1859 | sep1+-3HA-6his::ura4+rad3Δ::LEU2 leu1-32 ura4-D18 h− | This study |
| LJY2650 | sep1+-3HA-6his::LEU2 cds1Δ::ura4+leu1-32 ura4-D18 h− | This study |
| LJY2646 | sep1+-3HA-6his::LEU2 chk1Δ::ura4+leu1-32 ura4-D18 h− | This study |
| LJY2337 | sep1+-3HA-6his::ura4+cdc25–22 leu1-32 ura4-D18 h− | This study |
| LJY2340 | rad3Δ::ura4+sep1Δ::LEU2 leu1-32 ura4-D18 h− | This study |
| LJY2655 | cds1Δ::ura4+sep1Δ::LEU2 leu1-32 ura4-D18 h− | This study |
| LJY2658 | chk1Δ::ura4+sep1Δ::LEU2 leu1-32 ura4-D18 h− | This study |
| LJY2031 | cdc10+-3HA-6His::ura4+ura4-D18 h− | This study |
| LJY2168 | res1+-3HA-6His::ura4+ura4-D18 leu1-32 h− | This study |
| LJY2112 | res2+-3HA-6His::ura4+ura4-D18 leu1-32 h− | This study |
| LJY1902 | rep2+-3HA-6His::ura4+ura4-D18 leu1-32 h− | This study |
| LJY2743 | rep2+-3HA-6His::ura4+rad3Δ::LEU2 ura4-D18 leu1-32 h− | This study |
| LJY2742 | rep2+-3HA-6his::ura4+cds1Δ::ura4+ura4 D-18 leu1-32 h− | This study |
| LJY2744 | rep2+-3HA-6His::ura4+chk1Δ::LEU2 ura4-D18 leu1-32 h− | This study |
| PN1942 | cig2Δ::ura4+ura4-D18 h− | P. Nurse |
Standard rich medium yeast extract plus supplements (YES) and minimal medium Edinburgh minimal medium in liquid or solid (supplemented with 1.5% Bacto agar) forms were used for growth (Moreno et al., 1991). In all assays, fresh log phase cells were used and supplemented with 8 mM HU when appropriate. Otherwise, the concentration of HU will be stated. Synchronization of cells was performed using either physical size-based separation by elutriation centrifugation as described previously (Peng et al., 2005) or cell cycle block–release through a cdc temperature-sensitive allele.
Plating assays was used for determination of HU hypersensitivities. In brief, overnight cultures at 30°C were inoculated into fresh YES medium and incubated for >1.5 generations. Log phase growth cells were collected, subjected to 10-fold serial dilutions, and spotted on YES plates containing 2.5–8 mM HU (Sigma-Aldrich, St. Louis, MO) or as otherwise indicated (no HU as control). For temporal treatment with HU, early log phase cultures were supplemented with 2.5–8 mM HU (no HU as control) and incubated for 3 h. Before dilution and spotting, cells were washed twice with YES without HU supplement. For viability tests, log-growth cultures were treated with HU at various dosages for 3 h. Subsequently, cells were washed twice and plated on YES media at varying dilutions. Colony-forming units were calculated based on at least three independent experiments.
Time course experiments were used for analysis of expression responses to HU treatment. Both synchronous and asynchronous cultures were applied. For analysis of asynchronous cultures, log phase growth cultures were treated with 8 mM HU and sampled as various time points for a period of 120 min. For analysis of synchronous cultures, elutriation centrifugation (Beckman Coulter, Fullerton, CA) was used to enrich small G2 cells. In brief, >5-l log phase growth cells were pumped at a speed of 100 ml/min into a chamber of the elutriator at a rotation rate of 3000–4000 rpm. Small-sized cells were collected at ∼8°C, and the cultures were adjusted to an OD of ∼0.15 before starting synchronous growth at 30°C with 8 mM HU supplement or without HU (as control).
Fluorescence Microscopic and Fluorescence-activated Cell Sorting (FACS) Analyses
Cells were fixed by adding one-tenth volume of concentrated formaldehyde (Sigma-Aldrich) for 10 min and washed with phosphate-buffered saline (PBS). DNA and cells walls were stained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) and Aniline blue (Sigma-Aldrich) for fluorescence microscopic analysis (Leica, Wetzlar, Germany), respectively.
To measure DNA content, cells were quickly spun out and resuspended in ice-cold 70% ethanol and digested with RNase A (F. Hoffman-La Roche, Basel, Switzerland) in 50 mM sodium citrate overnight. Then, they were briefly washed with PBS. DNA was stained with propidium iodine (Sigma-Aldrich). Fluorescence intensities of individual cells were measured by flow cytometry using a BD FACScan (BD Biosciences, Franklin Lakes, NJ).
Protein Analysis
Standard protein lysate preparation was used for Western analyses. Antibodies against HA (monoclonal; Santa Cruz Biotechnology, Santa Cruz, CA), Cdc2-Y15-P (monoclonal; Cell Signaling Technology, Beverly, MA), PSTAIR (polyclonal; Upstate, Charlottesville, VA), Cig2p (CIG 3A11/5; Abcam, Cambridge, United Kingdom), and β-tubulin (TAT1; from Dr. K. Gull, University of Oxford) were used according to manufacturer's recommendations. Anti-mouse or anti-rabbit IgG conjugated with horseradish peroxidase (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) were used as secondary antibodies. The chemiluminescence was developed and captured using the horseradish peroxidase system by enhanced chemiluminescence (GE Healthcare).
Extraction and Labeling of RNA and Hybridization of Microarrays
To extract total RNA, cells were quickly spun out and chilled in liquid nitrogen. Acid phenol was then added to the frozen cell pellet, which was incubated at 65°C for 15 min. The aqueous phase was recovered and extracted with phenol/chloroform (1:1). RNA was precipitated using isopropanol and quantified with a UV-spectrophotometer (Nano Drop, Wilmington, DE). Approximately 30 μg of total RNA was used to synthesize cDNA coupled with amino allyl-dUTP by reverse transcriptase (Superscript II; Invitrogen, Carlsbad, CA) according to manufacturer's instructions. cDNA was subsequently washed with Milli-Q water using Microcon YM-30 (Millipore, Billerica, MA). Approximately 1.5 μg of cDNA was coupled with Cy5- or Cy3 fluorescence dye for 1 h in dark and purified through a spin column (QIAGEN, Hilden, Germany) followed by washing in a Microcon YM-30. Cy5- and Cy3-labeled cDNA were pooled and hybridized to the spotted S. pombe oligonucleotide (oligo)-based DNA microarrays (Peng et al., 2005) by using MAUI hybridization system (BioMicro, Salt Lake City, UT) in Roche hybridization buffer (catalog no. 1063588; Roche Diagnostics) overnight. Hybridized slides were washed and scanned using a GenePix scanner (Molecular Devices, Sunnyvale, CA) at wavelength 635 and 532 μm with a resolution of 10 μm as described previously (Peng et al., 2005).
Acquisition and Normalization of Microarray Data
Intensities of individual microarray features based on TIFF-formatted images were measured by GenePix Pro4.0 software (Molecular Devices) and are reported in the GenePix Result files. To ensure well-measured features, the ratio (i.e., median of ratio) for individual features was only collected if its intensity in either channel was twofold or greater than that of the background. Measured ratios for every individual array were normalized using locally weighted linear regression and smooth scatter plot (LOWESS) (Cleveland, 1979; Yang et al., 2002) to remove intensity-dependent dye bias. An MA-plot was used to facilitate the detection of intensity-dependent patterns in log scale, where M = log2(Ratio) and A = ½log10(Cy3·Cy5). The LOWESS smoother, implemented in the statistical software package R (Ihaka and Gentleman, 1996), locally normalizes M in an A-dependent manner log2(Ratio′) = log2(Ratio) − L(A), where L(A) is the LOWESS fit to the MA-plot. The user defined parameters f (fraction of data used for smoothing at each point), n-steps (number of iterations used in LOWESS fit) are 0.5 and 3, respectively.
Analysis of Differential Gene Expressions
To analyze differential expressions in asynchronous log phase growth cultures, significant analysis of microarrays (SAM, version 1.21) (Tusher et al., 2001) was applied based on three independent repeats of expression profiles. To analyze differential expression in synchronous cultures, a meta-analysis after t test was used (see below).
Identification of Differentially Expressed Genes in HU-treated Synchronized Cultures.
Differentially expressed genes in synchronized wild-type cultures responding to HU treatment were identified using a pooled-variance t test followed by a nonparametric meta-analysis (Hedges and Olkin, 1985; Benjamini and Hochberg, 1995). In brief, we first estimated the variance of the individual gene expression error by pooling differences of the two wild-type strain repeats at all time points in both HU-treated and untreated expression profiles. The derived variance was then used to estimate the standardized change of expressions in response to HU at a given time point. The change should follow a central t distribution with (2n + 1) degrees of freedom, where n is the number of differences that were used to determine the variance. After having calculated the p values of expression change at each time point in response to HU, the overall p value of the change was estimated using the minimum of the p values obtained at all time points by applying the formula 1 − (1 − z)k, where z is the minimum p value, and k is the number of p values. See detailed description in Supplemental Material.
Identification of Kinase-dependent HU-responsive Genes.
The same approach was applied to identify kinase-dependent genes as for the identification of HU-responsive genes except that significant changes were based on the comparison between expression profiles of wild-type and a kinase mutant strain in response to HU.
Original data sets were submitted to the GEO database (accession no. GSE4284).
Statistical Methods
Standard binomial analysis was applied to examine nonrandom distribution in the enrichment test.
RESULTS
rad3Δ and cds1Δ Are Hypersensitive to Transient HU Exposure
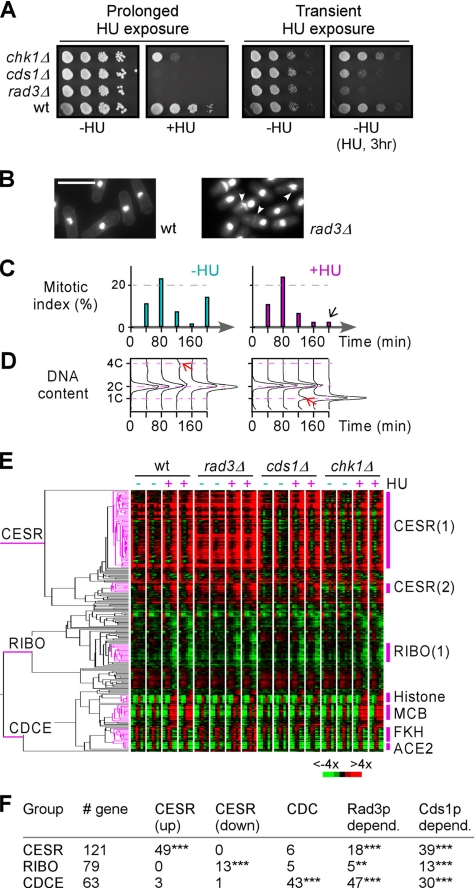
We have previously shown that eight individual protein kinase deletion mutants exhibited growth hypersensitivity on plates containing HU (Bimbo et al., 2005). Given that prolonged exposure to HU could induce DNA damage, the hypersensitivity upon HU exposure might not be a direct result of an impaired kinase function in restoring replication forks or replication checkpoint. To determine whether these kinases were involved in signaling DNA replication blocks, we investigated growth hypersensitivity to transient exposure to HU. Of the nine protein kinase mutants (in this study, we also included Rad3p, a protein kinase that contains a phosphatidylinositol 3- and 4-kinase catalytic domain), only rad3Δ and cds1Δ strains displayed hypersensitivity when subjected to HU treatment at an final concentration of 8 mM for 3 h (Figure 1A; data not shown). This hypersensitivity to transient HU treatment was seen at various test dosages of HU ranging from 2.5 to 8 mM tested (Supplemental Figure 1A). The remaining kinase mutant strains displayed a wild-type growth profile as exemplified using chk1Δ (Figure 1A). This indicates that only Rad3p and Cds1p out of a subset of nine HU-sensitive kinase mutants identified in the fission yeast genome seem to be the major protein kinases that are involved in signaling DNA-replication blocks.
Figure 1.
Phenotypic assessment and expression profiling of wild-type and kinase mutant cells upon HU treatment. (A) Plating assay for kinase mutant sensitivities to prolonged or transient exposure to HU. For prolonged HU treatment, ∼5 μl of 10-fold series diluted wild-type, rad3Δ, cds1Δ, and chk1Δ strains were spotted and grown on YES plates supplemented with 8 mM HU at 30°C. For transient HU treatment, cells were treated with 8 mM HU for 3 h and recovery on plate without supplement with HU. (B) Images of wild-type and rad3Δ cells after HU treatment for 3 h. Nuclei and division septa were stained using DAPI and Aniline blue, respectively. Arrowheads indicate cells with a cut phenotype. (C) Mitotic indexes of synchronized G2 cell cultures. Mitotic indexes or percentage of binucleate cells was determined in synchronized cultures at various time points as indicated after addition of HU (+HU) or without as control (−HU). (D) DNA content profiles in synchronized cultures. DNA contents in samples (C) were determined by FACS. Red arrows indicate a 4C-peak and a 1C-peak in cultures without and with HU supplement, respectively. (E) Hierarchical cluster analysis of expression profiles in various strains supplemented with or without HU. Expression profile clustering of the 263 differentially expressed genes from wild-type, rad3Δ, cds1Δ, and chk1Δ strains supplemented with or without HU. The dendrogram on the left depicts the structure of the clusters, and three groups—CESR, RIBO, and CDCE—are indicated. The phasogram on the right show the levels of gene expression at various time points as in C. Genes correspond to rows and time points form the columns. Color intensities reflect the magnitudes of induction (red) or repression (green) for each gene (see key at bottom right). Black indicates no change of expression levels; and gray refers to data not available. (F) Characterization of genes in CESR, RIBO, and CDCE groups. Numbers of genes belong to induced (up) CESR, repressed (down) CESR, and cell cycle–regulated (CDC), Rad3p-dependent, and Cds1p-dpenedent genes in three groups are indicated. Enrichment of particular gene types are indicated as *** and ** for p < 0.001 and p < 0.01, respectively.
Next, we investigated terminal phenotypes of rad3Δ and cds1Δ cells at various time points for a period of 3 h during the treatment with HU. In wild-type cells, activation of the DNA-replication checkpoint delayed the subsequent cell cycle progression. As a result, the number of mitotic cells was reduced to nearly zero in cultures after 3-h HU treatment (Supplemental Figure 1B). Defects in checkpoint kinases would lead to cells incapable of restoring replication forks. Although both rad3Δ and cds1Δ cells failed to recover after brief HU treatment, the number of cells exhibiting a cut phenotype was much more elevated in rad3Δ cells than in cds1Δ cells (Figure 1B and Supplemental Figure 1B). This result indicates that Rad3p plays an additional role in response to DNA replication blocks imposed by HU.
HU Treatment Induces S Phase-specific Expression Profile Changes
DNA microarray technology was used to analyze our mutants of interest by using oligo-based DNA microarrays (Peng et al., 2005). Given that S phase-arrested cells were accumulated during HU treatment, S phase-specific transcripts might be enriched as HU-induced genes in expression profiling by using log phase growth (asynchronized) cells. However, the enrichment of cell cycle stage–specific transcripts during HU treatment might not be a direct result of expression responses to HU. To eliminate the interference of cell cycle stage–specific transcripts, we investigated expression profiles using synchronized G2 cells that were generated through elutriation centrifugation. Synchronous cultures of wild-type, rad3Δ, cds1Δ, and chk1Δ strains were split into two aliquots: one aliquot served as a control, whereas HU was added to the other at the final concentration of 8 mM. Samples were taken at 40-min intervals for a period of 200 min for microarray analysis. At least two microarray hybridizations were performed for individual samples to ensure reproducibility. The mitotic indexes of synchronous cultures indicated that the first wave of mitosis peaked within 40 to 80 min, and a second wave occurred at ∼200 min after release of synchronous cultures (i.e., a trough of mitotic indexes at 160 min was obvious) (Figure 1C). Interestingly, HU treatment did not affect the first peak of mitosis. However, the cresting of mitotic indexes into the second peak at ∼200 min was delayed or abolished (Figure 1C) except in rad3Δ (data not shown). FACS analysis of DNA content detected a 4C-peak at 120 min, indicative of an S phase peak in synchronous cultures without HU (Figure 1D). Conversely, a 1C-peak first occurred at 120 min, coincident with the occurrence of a 4C-peak in HU-treated cultures, indicating the beginning of the S phase arrest. Thus, these time course experiments using synchronous cultures were able to cover a complete length of the cell cycle and detect the onset of an S phase arrest (i.e., at ∼120 min) after HU treatment, which coincided with the peak of S phase in cultures without HU.
To classify differentially expressed genes in response to HU treatment, we first used standard t test with pooled variances, and subsequently we applied nonparametric meta-analysis (Hedges and Olkin, 1985; Benjamini and Hochberg, 1995) on expression profiles of wild-type synchronized cultures supplemented with and without HU (see Materials and Methods). As a result, ∼260 genes were found to exhibit significant alteration of expression patterns (p < 0.05) in at least one time point out of five tested and to have dynamic expression level changes of twofold or greater in response to HU treatment (Supplemental Table 1). Expression responses were found to occur at or after S phase (i.e., 120 min) in all genes except for one that showed significant change at 40 min. This result indicates that DNA replication block imposed by HU treatment triggers expression response at the onset of S phase arrest.
Next, we investigated expression profiles of asynchronized cultures in responses to HU for comparison with those of synchronized cells. To this end, log phase growth cells supplemented with 8 mM HU were sampled at 0, 10, 30, 60, and 120 min. More than 90% cells exhibited 1C-DNA content at 2 h after HU treatment, indicating an early S phase arrest (Supplemental Figure 1C). Microarray hybridizations of three independently conducted time course experiments were carried out for reproducibility. SAM analysis (Tusher et al., 2001) identified ∼90 genes whose expression levels changed twofold or greater with a false discovery rate of <0.03%. Majority (66 genes) were found to overlap with the 263 HU response genes based on analyses of synchronized cultures. This result suggests that cell cycle stage–specific expression responses were more easily discernible in synchronized cells compared with asynchronized cells. The remaining 24 genes might represent the response of asynchronized cells, several of which encode proteins that are involved in oxidative stress (GO:0016491; p < 0.005).
Expression Profile Changes of the CESR Genes at the Onset of S Phase Arrest Is Partly Dependent on the Function of Rad3p and Cds1p
To investigate whether DNA replication checkpoint kinases Rad3p and Cds1p were involved in modulation of expression in response to HU treatment, we investigated alterations of expression profiles upon HU treatment in rad3Δ and cds1Δ strains (see Materials and Methods) with the chk1Δ strain was included as a negative control. Of ∼260 HU response genes, 41% (109 genes) were found to exhibit significant alterations (p < 0.05) of their expression response profiles in mutant strains, indicating that replication checkpoint kinases modulate expression response of those genes at the onset of S phase arrest. Among the 109 genes, 70 were rad3+ dependent, 81 were cds1+ dependent, and only four were chk1+ dependent, suggesting that Rad3p and Cds1p are the major kinases that mediate expression in responses to HU treatment. This result is consistent with the notion that Chk1p is not a major player in DNA replication checkpoint function.
Approximately 260 gene expression profiles of synchronized wild-type, rad3Δ, cds1Δ, and chk1Δ cells, supplemented with HU or without HU, were hierarchically clustered and revealed three groups: the CESR, ribosome biogenesis (RIBO), and cell division cycle-specific expression (CDCE) according to the expression patterns of the enriched genes (Figure 1, E and F). The CESR group enriched for induced CESR genes (Chen et al., 2003) (p < 0.001). Alternatively, the RIBO group enriched for repressed CESR genes or genes involved in ribosome biogenesis (p < 0.001). The CDCE group enriched cell cycle-regulated genes as identified within a list of ∼170 genes whose oscillated expression patterns were confirmed by three independent studies (Rustici et al., 2004; Oliva et al., 2005; Peng et al., 2005). All three groups were found to be enriched for rad3- or cds1-dependent HU response genes, indicating that replication checkpoint kinases Rad3p and Cds1p modulate expression of both stress responsive genes and cell cycle-regulated genes at the onset of S phase arrest (Figure 1F).
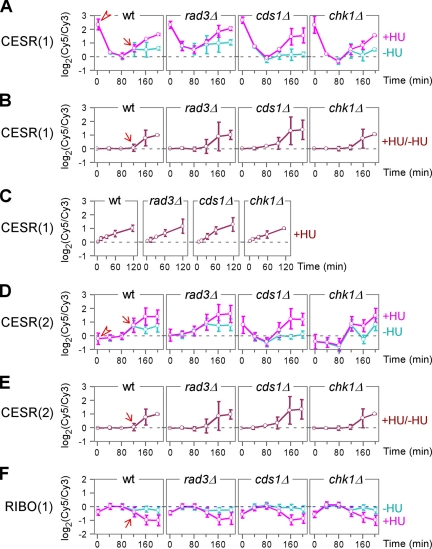
Seven highly coregulated gene clusters (>8 genes with minimal correlation of 85%) were apparent in the three groups (Figure 1E). Two clusters were found in the CESR group (Supplemental Figure 2). Although genes in both clusters were induced at the onset of S phase arrest, expression levels of the large cluster were highly elevated just before addition of HU (i.e., 0 min), suggesting that they are most likely induced by the synchronization procedure (Figure 2A, arrowhead). This effect quickly decayed in 40 min as the expression level dropped to reference levels (i.e., levels in asynchronized wild-type cells). However, expression levels of the small cluster were not affected by the synchronization (Figure 2D, arrowhead). This difference was not obvious from a comparison of expression levels of HU-treated cells to those of untreated control cells (Figure 2, B and E). Nevertheless, it could still identify S phase–specific expression profile changes (Figure 2, B and E, arrow). In contrast, it would not be possible to reveal the cell cycle stage–specific change in expression profiles by using asynchronized cells (Figure 2C). A coregulated cluster in the RIBO group was found with an apparent repression of the members at the onset of S phase (Figure 2F).
Figure 2.
Average expression profiles of highly coregulated clusters CESR(1), CESR(2), and RIBO(1). The x- and y-axis indicate expression ratios and time points after HU treatment, respectively. Arrowheads indicate expression levels in cells before HU addition. An arrow indicates the time of expression level changes. (A–C) Average expression profiles of the CESR(1)-cluster. Expression levels of the CESR(1)-cluster in synchronized cells supplemented with HU (+HU) and without (−HU) are separated compared with a common reference (A). Expression levels in HU-treated samples as in (A) are directly compared with those in the control (B). Expression levels of the CESR(1)-cluster in asynchronized cells supplemented with HU are compared with those without HU (C). (D and E) Average expression profiles of the CESR(2)-cluster. Comparisons of expression levels in D and E are the same as described in A and B, respectively. (F) Average expression profiles of the RIBO(1)-cluster. Comparisons of expression levels is the same as described in A.
Four more highly coregulated clusters were found in the CDCE group, namely, the MCB-, the histone-, the FKH-, and the ACE2-clusters, so named for their enriched promoter motifs. Regulation of the expression of genes belonging to these clusters, whose transcript levels fluctuate based on the cell cycle, are described below.
Peak Expression Levels of the MCB-Cluster Are Maintained at the Onset of Phase Arrest in a rad3+- or cds1+-dependent Manner
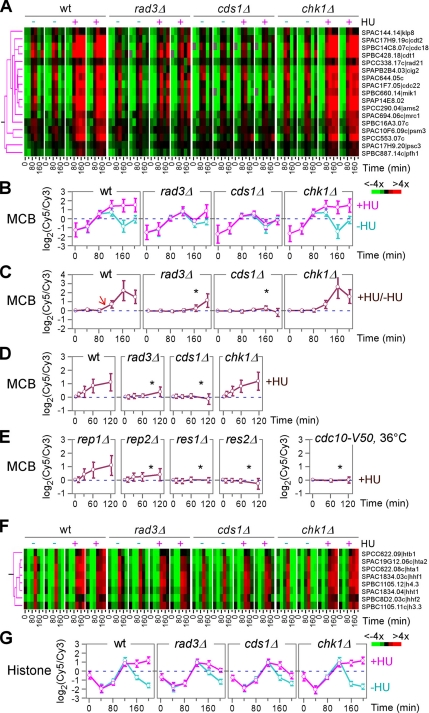
Expression profiles of the MCB-cluster peaked at G1 (between 80 and 120 min) and subsequently were induced again at 200 min in synchronized wild-type cells unsupplemented with HU, consistent with our previous study (Peng et al., 2005). Similar cell cycle-regulated expression patterns were seen in rad3Δ, cds1Δ, and chk1Δ strains without HU supplement, consistent with the notion that the checkpoint kinases are not involved in regulation of cell cycle–specific expressions. Significantly, the peak expression levels of the MCB-cluster were maintained in synchronized wild-type cultures when supplemented with HU (Figure 3, A and B). This expression level maintenance, however, was diminished in rad3Δ or cds1Δ strains, indicating a requirement for the replication checkpoint function. A wild-type expression pattern of the MCB-cluster was seen in chk1Δ strain, consistent with the notion that Chk1p is not involved in replication checkpoint signaling.
Figure 3.
The MCB- and histone-clusters. (A) Magnified view of the MCB-cluster as in Figure 2B. Individual genes in the cluster are shown on the right. (B–E) Average expression profiles of the MCB-cluster. The x- and y-axis indicate expression ratios and time points after HU treatment, respectively. An arrow indicates the time of expression level changes. Asterisks indicate the defect in expression induction. Expression levels of the MCB-cluster in synchronized cells supplemented with HU (+HU) and without (−HU) are separated compared with a common reference (B). Expression levels in HU treated samples as in B are directly compared with those in the control (C). Expression levels of the MCB-cluster in asynchronized cells supplemented with HU are compared with those without HU (D and E). (F) Magnified view of the histone-cluster as in Figure 2B. Individual genes in the cluster are shown on the right. (G) Average expression profiles of the histone-cluster. The x- and y-axis indicate expression ratios and time points after HU treatment, respectively. Expression levels of the histone-cluster in synchronized cells supplemented with HU (+HU) and without (−HU) are separated compared with a common reference.
Direct comparisons of expression levels in HU-treated synchronized cells to those in synchronized control (HU-untreated) cells have revealed S phase-specific expression profile changes (Figure 3C, arrow). However, it could not reveal the peak level maintenance of the MCB-cluster expression. In profiles of asynchronous cells, cell cycle stage–specific changes were unclear (Figure 3D). Despite this, alterations of wild-type expression profiles in rad3Δ and cds1 were obvious (Figure 3D, asterisk). This result indicates that asynchronized cells can be applied for studies of profile alteration in various mutant strains.
MCB-cluster genes are known to be regulated by the MBF complex composed of Cdc10p, Rep2p, Res1p, and Res2p. To investigate the function of individual components on transcriptional regulation of the MCB-cluster in response to HU, we performed expression profiling in cdc10-V50, rep2Δ, res1Δ, and res2Δ strains. The rep1Δ strain bearing a deletion allele of the meiotic-specific MBF component was included as a negative control. Upon HU treatment, expression profiles of res1Δ, res2Δ, rep2Δ, and cdc10− strains showed no induction of the MCB-cluster expression except for rep1Δ, indicating that all MBF components are essential for expression responses of the MCB-cluster (Figure 3E, asterisk). This result is consistent with other studies indicating that all MBF components are important for induction of MCB motif-containing genes (White et al., 2001).
The cluster of eight histone genes displayed an expression profile similar to that of the MCB-cluster except for an ∼20-min delay (Figure 3, F and G). The similarity in expression profiles of the two clusters (namely, MCB- and histone-clusters) at the onset of the S phase arrest suggests that MBF might also be involved in regulation of histone gene expression, as MCB-motifs are found in promoters of histone genes. However, the identity of transcriptional regulators that directly regulate histone genes remains unclear.
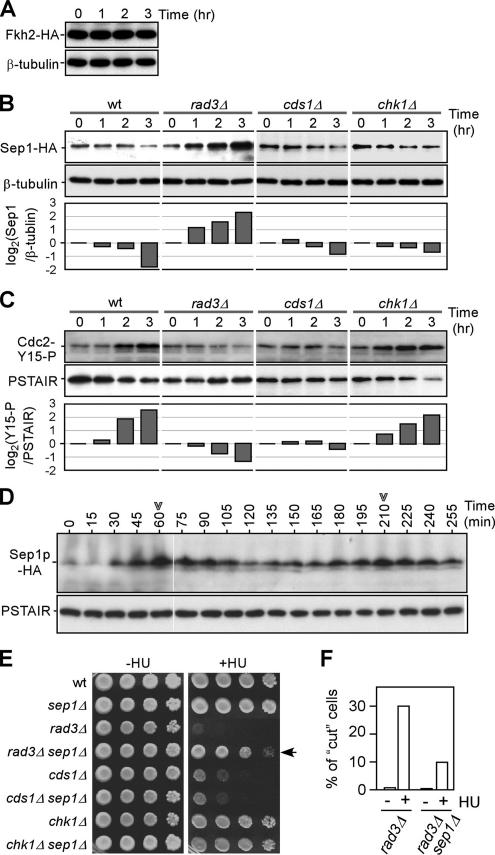
Maintenance of Peak Expression Levels for the MCB-Cluster Is Mediated through the Accumulation of Rep2p
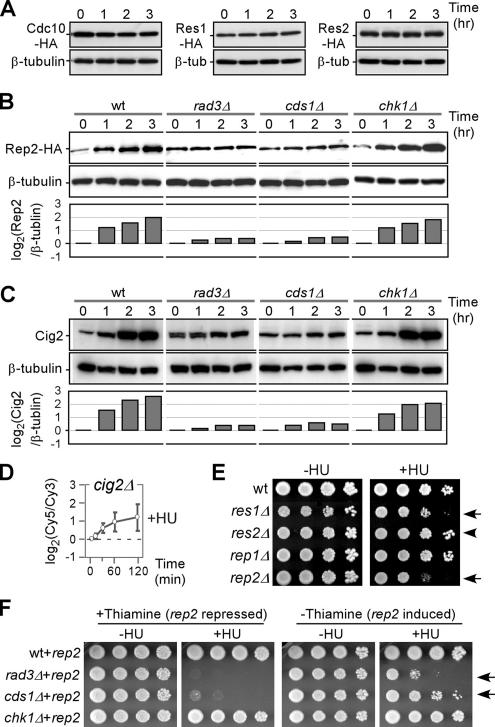
Strains bearing either one of cdc10−, rep2Δ, res1Δ, or res2Δ alleles could abolish the induction of the MCB-cluster in response to HU (Figure 3E, asterisk). We thus investigated protein levels of individual MBF components in cells when treated with HU. To this end, cells bearing a sole copy of 3HA-tagged Cdc10p, Rep2, Res1p, and Res2p were treated with HU and sampled at 1-h intervals for a period of 3 h. Western blot analysis indicated that levels of Cdc10p, Res1p, and Res2p proteins were unchanged after HU treatment (Figure 4A). Significantly, however, Rep2p level was increased approximately four-fold, suggesting that Rep2p was accumulated at the onset of S phase arrest (Figure 4B). This accumulation was absent in rad3Δ and cds1Δ strains, correlating with the observation that maintenance of peak expression level of the MCB-cluster requires the function of Rad3p and Cds1p.
Figure 4.
Regulation of protein levels of individual MBF factors Cdc10, Res1p, Res2p, and Rep2p upon HU treatment. (A–C) Cells treated with 8 mM HU were sampled at various time points as indicated for Western blot analysis. Strains bearing a sole copy of cdc10+-3HA, res1+-3HA, and res2+-3HA were used for analysis of protein levels after HU treatment in A, and strains bearing a sole copy of rep2+-3HA were used as indicated in B. The tubulin levels were used as loading control. Membranes in B were stripped and reblotted for determination of Cig2p levels. Individual bands were quantified using ImageJ software (National Institutes of Health, Bethesda, MD). Fold changes of Rep2p and Cig2p levels are plotted after 0-h normalization. (D) Average expression profiles of the MCB-cluster in cig2Δ cells. The x- and y-axis indicate expression ratios and time points after HU treatment, respectively. (E) Plating assays for HU susceptibility. Approximately 5 μl of 10-fold series diluted samples were inoculated onto plates supplemented with 5 mM HU and grown at 30°C for ∼2–3 d. Arrows and arrowhead indicate strains with and without noticeable sensitivities to HU, respectively. (F) Plating assays for sensitivity to HU. ∼5 μl of 10-fold series diluted samples were inoculated onto plates containing 2.5 mM HU and supplemented with thiamine (repressed for rep2+) or without thiamine (induced for rep2+). Arrows indicate that phenotypes of rad3Δ and cds1Δ strains are alleviated when rep2+ is over produced.
The cyclin Cig2p is thought to be involved in regulating MBF activities (Ayte et al., 2001) and its level increases upon HU treatment (Zarzov et al., 2002). We thus investigated Cig2p levels in wild-type, rad3Δ, cds1Δ, and chk1Δ strains upon HU treatment. Notably, Cig2 level increase was Rad3p or Cds1p dependent, almost identical to the pattern of Rep2p (Figure 4C). We next examined gene expression profile patterns of cig2Δ cells in response to HU treatment. Surprisingly, disruption of cig2 did not weaken the expression response of the MCB-cluster (Figure 4D). This result indicates that accumulation of Cig2p is not essential for the maintenance of peak level expression of the MCB-cluster genes.
We next assessed cells bearing one of the res1Δ, res2Δ, and rep2Δ alleles for susceptibility to HU treatment. Series diluted cells were inoculated on plates supplemented with 5 mM HU and cells bearing the rep2Δ allele grew the slowest (Figure 4E, arrow), consistent with its role in HU response. It is also worth noting that res1Δ cells displayed noticeable sensitivity to HU (Figure 4E, arrow), suggesting that maintenance of peak level expression of the MCB-cluster is part of the replication checkpoint function. Consistent with this, cells bearing deletion allele of res2, a negative regulator of the MCB-cluster, exhibited no sensitivity to HU (Figure 4E, arrowhead).
We judged that if Rep2p accumulation was part of the replication checkpoint function in affecting resistance to HU treatment, overproduction of Rep2p might alleviate the checkpoint defect in rad3Δ or cds1Δ strains. Thus, we constructed a plasmid containing the rep2+ gene under the control of the nmt1 promoter and transformed this recombinant DNA into wild type, rad3Δ, cds1Δ, and chk1Δ strains. The rep2+-bearing plasmid could alleviate rad3Δ or cds1Δ phenotype only when induced (growth in media lacking thiamine) (Figure 4F, arrow), indicating that overexpression of Rep2p can partially suppress the HU sensitivity of checkpoint mutants, which suggests that HU sensitivity of checkpoint mutants is due, in part, to reduced Rep2p levels.
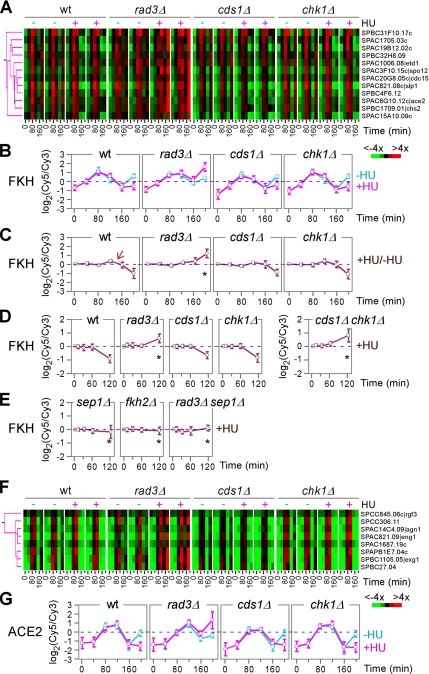
Repression of the FKH-Cluster during the Onset of S Phase Arrest Depends on the Function of Rad3p
FKH motif-containing genes are known to exhibit cell cycle–specific expression with a peak level at M phase (Rustici et al., 2004; Oliva et al., 2005; Peng et al., 2005). Many of the FKH motif-containing genes encode proteins involved in mitosis, mitotic exit, and cytokinesis. Consistent with this, expression profiles of the FKH-cluster without HU treatment showed a peak at ∼80 min with a subsequent peak around ∼200 min, coincident with the peaks of mitosis (Figure 5, A and B). HU treatment did not affect the first expression peak of the FKH-cluster in wild-type and various kinase mutant strains. In contrast, the second peak was delayed or abolished in wild-type and chk1Δ strains, suggesting a robust replication checkpoint response during the onset of S phase arrest. Significantly, this repression of the FKH-cluster was abolished only in the rad3Δ strain, thus accounting for its high level of cut phenotype, which is not seen in the cds1Δ strain, owing to the redundant function of Chk1p (see below).
Figure 5.
The FKH- and ACE2-clusters. (A) Magnified view of the FKH-cluster as in Figure 2B. Individual genes in the cluster are shown on the right. (B–E) Average expression profiles of the FKH-cluster. The x- and y-axis indicate expression ratios and time points after HU treatment, respectively. An arrow indicates the time of expression level changes. Asterisks indicate the defect in expression induction. Expression levels of the FKH-cluster in synchronized cells supplemented with HU (+HU) and without (−HU) are separated compared with a common reference (B). Expression levels in HU-treated samples as in B are directly compared with those in the control (C). Expression levels of the FKH-cluster in asynchronized cells supplemented with HU are compared with those without HU (D and E). (F) Magnified view of the ACE2-cluster as in Figure 2B. Individual genes in the cluster are shown on the right. (G) Average expression profiles of the ACE2-cluster. The x- and y-axis indicate expression ratios and time points after HU treatment, respectively. Expression levels of the histone-cluster in synchronized cells supplemented with HU (+HU) and without (−HU) are separated compared with a common reference.
The rad3+-dependent repression of the KFH-cluster was also apparent in expression profiles in which expression levels of synchronous cells supplemented with HU were directly compared with those of untreated samples (Figure 5C) or simply in profiles of asynchronous cultures upon HU treatment (Figure 5D, asterisk). It has been reported that Chk1p can complement for the defect in the cds1Δ strain during S phase (Brondello et al., 1999). To investigate the possibility that Chk1p could counteract the defect in repression of the FKH-cluster in cds1Δ strain, we constructed a strain bearing both cds1Δ and chk1Δ alleles for expression profiling. Indeed, expression profiles of the FKH-cluster failed to be repressed in the cds1Δ chk1Δ strain, and they were almost identical to those seen in rad3Δ, indicating that Chk1p complements for a lack of Cds1p function related to repression of the FKH-cluster in a cds1Δ strain (Figure 5D, asterisk).
FKH motif-containing genes are thought to be regulated by the forkhead transcription factors Sep1p and Fkh2p (Buck et al., 2004). To investigate the transcriptional regulation of the FKH-cluster by Sep1p and/or Fkh2p, we performed chromatin immunoprecipitation (ChIP) assays for promoter binding activities. Promoter sequences of the FKH-cluster genes were significantly enriched in immunoprecipitated protein–DNA complexes but not in whole cell extracts, supporting the notion that Sep1p and Fkh2p regulate FKH motif-containing genes (Supplemental Figure 3).
It has been shown that disruption of sep1 abolishes the cell cycle–oscillated expression of the FKH-cluster genes (Rustici et al., 2004). To investigate the effect of Sep1p and Fkh2p on transcriptional regulation of the FKH-cluster in response to HU, we performed expression profiling in sep1Δ and fkh2Δ strains. Deletion of either sep1+ or fkh2+ was sufficient to abolish the expression response pattern of the FKH-cluster (Figure 5E, asterisk), suggesting a collaborative function between Sep1p and Fkh2p in regulation of the FKH-cluster genes. In addition, the elevated level of the FKH-cluster expression at 120 min in rad3Δ strain was alleviated when sep1+ was disrupted (Figure 5E, asterisk).
Expression profiles of the ACE2-cluster were reminiscent to those of the FKH-cluster except for an ∼20-min delay (Figure 5, F and G). Given that ace2+ is one of the FKH-cluster genes, profile changes of the ACE2-cluster may reflect those of the FKH-cluster. Consistent with this, the repression of the ACE2-cluster was diminished in sep1Δ or fkh2Δ strains. Alternatively, repression of the FKH-cluster was not much affected in the ace2Δ strain (data not shown).
Repression of the FKH-Cluster Is Mediated through the Decreased Sep1 Protein Levels
Next, we investigated the changes of Sep1p and Fkh2p levels upon HU treatment. To this end, strains bearing sep1+-3HA and fkh2+-3HA were treated with HU and levels of HA-tagged proteins were determined using Western blot analysis. Fkh2p levels seemed to be unchanged upon HU treatment (Figure 6A). In contrast, Sep1p levels were dramatically decreased by approximately fourfold (Figure 6B). Significantly, this decrease was diminished in the rad3Δ strain, but not in cds1Δ, correlating well with the decrease of the expression level of the FKH-cluster. Under the same conditions, Cdc2p was found to be inactivated in wild-type and chk1Δ strains but not in rad3Δ and cds1Δ strains (Figure 6C), consistent with the notion that both Rad3p and Cds1p are the major players of the DNA replication checkpoint. Furthermore, we showed that protein levels of Sep1p oscillated along with the cell cycle with a peak at M phase (Figure 6D), correlating with the peak level expression of the FKH-cluster genes.
Figure 6.
Regulation of protein levels of forkhead transcription factors Sep1p and Fkh2p upon HU treatment. (A–C) Cells treated with 8 mM HU were sampled at various time points as indicated for Western blot analysis. Strains bearing a sole copy of fhk2+-3HA and sep1+-3HA were used for analysis of Fkh2p and Sep1p levels after HU treatment in A and B, respectively. Tubulin levels were used as loading control. Membranes in B were stripped and reblotted for determination of Cdc2-Y15-P levels. Levels of PSATIR were used as loading control. Individual bands are quantified using ImageJ software. Fold-changes of Sep1p and Cdc2-Y15-P levels are plotted after 0-h normalization. (D) Levels of Sep1p are oscillated along cell cycle. Cells bearing a sole copy of sep1+-3HA in the cdc25-22 background were synchronized by temperature shift. G2-arrested cells were released to permissive temperature and sampled at various time points for Western blot analysis of Sep1p levels. Arrowheads indicate the peak levels of Sep1p coincident with peak of mitotic indexes. (E) Plating assays for sensitivity to HU. Approximately 5 μl of 10-fold series diluted samples were inoculated onto plates containing 2.5 mM HU. An arrow indicates that phenotype of rad3Δ strain is alleviated when sep1Δ is in the background. (F) Frequencies of HU-induced cut phenotype. The cut phenotype was examined after rad3Δ and rad3Δ sep1Δ cells were challenged with 8 mM HU for 3 h. Numbers of cells with cut phenotype are countered from >500 cells.
To test whether deprivation of Sep1p is part of the Rad3p functions in cells upon HU treatment, we thus investigated the viability of HU-treated rad3Δ strain in the sep1Δ background. To this end, we performed growth assays on kinase deletion strains with a sep1Δ background on plates supplemented with HU. Significantly, disruption of sep1 could rescue the growth defect of the rad3Δ strain but not cds1Δ (Figure 6E). This result suggests that down-regulation of Sep1p at the onset of S phase arrest is affected by Rad3p. Consistent with this, the elevated frequency of the cut phenotype was also found to be reduced in the rad3Δ sep1Δ strain (Figure 6F).
DISCUSSION
Out of nine kinase deletion strains that are sensitive to prolonged exposure to HU, we have shown that only rad3Δ and cds1Δ strains display hypersensitivity to transient exposure. This result indicates that Rad3p and Cds1p are the major players in mediating DNA replication checkpoint signaling.
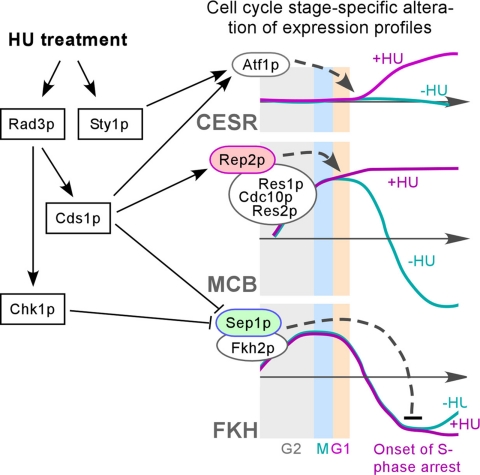
In this study, we have shown that cell cycle stage–specific changes of expression profile can be clearly revealed upon examination of expression levels of synchronized cells supplemented with HU or without (control) compared with a common reference. On HU treatment, expression profile changes occur only at the onset of S phase arrest, suggesting a cell cycle stage–specific expression response. Approximately 50 CESR genes are induced at the onset of S phase arrest imposed by HU treatment (Figure 2, A–E). Expression of these CESR genes is largely not cell cycle specific. This result indicates that inhibition of RNR by HU triggers stress responses at the onset of S phase arrest. Rad3p and Cds1p are partly involved in expression response of these CESR genes (Figure 1F). It has been proposed that Rad3p also modulates expression responses of CESR genes, mediating through Aft1p (human ATF2 homologue), in response to ionizing radiation in S. pombe (Watson et al., 2004). It is therefore likely that Atf1p is activated by both the Rad3p–Cds1p pathways and the Sty1p-mediated stress response pathway at the onset of S phase arrest (Figure 7). Similarly, S. cerevisiae Atf1p homolog Atf1p and Atf2p, have been shown to be involved in response to HU (Dubacq et al., 2006).
Figure 7.
Schematic model for HU-induced expression responses. Kinases Rad3p, Cds1p, Chk1p, and Sty1p are shown in rectangles and transcription factors are in ovals. Kinases directly or indirectly regulate transcription factors. Arrows and blunted lines indicate positive and negative regulations, respectively. Expression profiles of the CESR-, MCB-, and FKH-clusters in presence (+HU) and absence (−HU) of HU are shown.
Besides CESR genes, many non-CESR genes such as cell cycle–regulated MCB-, histone-, FKH-, and ACE2-cluster genes displayed significant expression profile alteration during the onset of S phase arrest imposed by HU treatment. The Rad3p–Cds1p pathway apparently also plays a major role in expression response of these cell cycle-regulated genes (Figure 1F).
Many MCB-cluster genes such as cdc22, cdc18, cdt1, cig2, and mik1 are essential for DNA replication. The MCB-cluster expression with a peak level at G1 phase is known to be controlled by the MBF complex that consists of Cdc10p, Rep2p, Res1p, and Res2p. At the onset of S phase arrest, we show that the peak expression level of the MCB-cluster is maintained in a rad3+- or cds1+-dependent manner (Figure 3, A and B). The MBF factors regulate the MCB-cluster in concert and disruption of any one of the MBF components is sufficient to abolish expression responses of the MCB-cluster. The B-type cyclin Cig2p has been proposed to be involved in a positive feedback regulation of the MBF factors (Ayte et al., 2001) and to be stabilized upon HU treatment (Zarzov et al., 2002). However, disruption of cig2 is not sufficient to diminish expression response patterns of the MCB-cluster (Figure 4D). Given that Cdc13p is known to substitute Cig2p function in cig2Δ cells (Zarzov et al., 2002), we thus could not exclude the involvement of cyclin function in regulation of MCB-cluster genes at the onset of S phase arrest. In this study, we show that Rep2p, like Cig2p, is also accumulated/stabilized upon HU treatment but not other MBF components (Figure 4B). However, unlike cig2, disruption of rep2 is sufficient to abolish the expression response of the MCB-cluster (Figure 3E), suggesting that peak level maintenance of the MCB-cluster expression is mediated through Rep2p (Figure 7). In agreement with this, cells bearing a rep2Δ allele exhibit apparent hypersensitivity to HU (Figure 4D).
It is intriguing to realize that peak expression levels of the MCB-cluster are maintained at the onset of the S phase arrest. This is probably the mechanism whereby the Rad3–Cds1p pathway maintains high competence potential for resumption of DNA replication in S phase–arrested cells. Two genes cdc18+ and cdt1+ of the MCB-cluster encode proteins involved in initiation of DNA replication (Kelly et al., 1993; Hofmann and Beach, 1994). High-level expression of these genes could lead to rereplication at G2 phase (Yanow et al., 2001). To prevent this from happening, we observed that the level of Cig2p (likely Cdc13p when in absence of Cig2p), a negative regulator of the Cdc18p, remains high at S phase block. It is not clear now how other MCB-cluster genes could be involved in restoring stalled replication forks. However, it is interesting to note that rep2+ overproduction could alleviate rad3Δ or cds1Δ phenotypes.
The observation of accumulation of Rep2p but not other MBF components upon HU-treatment indicates that Rep2p plays a key role in expression regulation of the MCB-cluster genes at the onset of S phase arrest. Indeed, Rep2 protein levels seem to oscillate during cell cycle with a peak at G1 phase and Rep2 protein is less stable than other MBF components (Chu and Liu, unpublished data). However, there is no evidence to suggest that phosphorylation of Rep2p enhances its stability at the onset of S phase arrest in a Rad3p–Cds1p-dependent manner. Clearly, further investigations are required to uncover additional components that link the Rad3p–Cds1p pathway and stabilization of Rep2p.
The histone-cluster contains eight histone genes, of nine in the genome. Expression profiles of the histone genes are almost identical to those of the MCB-cluster except for a ∼20-min delay (Figure 3, F and G). This profile similarity suggests that components of the MBF complex and/or products of the MCB-cluster are involved in transcriptional regulation of the histones at the onset of S phase. However, factors that directly regulate histone transcription remain unidentified.
Many M phase genes are known to encode proteins involved in mitosis (e.g., etd1+ and spo12+), mitotic exit (e.g., slp1+), and cytokinesis (e.g., cdc15+). Most of these M phase genes possess FKH motifs and thus form the FKH-cluster that is proposed to be regulated by the forkhead transcription factors Sep1p and Fkh2p (Buck et al., 2004). Apparently, down-regulation of these M phase FKH-cluster genes would presumably prevent untimely mitosis and cytokinesis. Indeed, the FKH-cluster genes are repressed during the onset of S phase arrest (Figure 5, A and B). Strikingly, this repression depends largely on Rad3p but not Cds1p, correlating with the frequency of cut phenotypes in rad3Δ and cds1Δ strains. This is owing to the redundant function of Chk1p in cds1Δ cells (Brondello et al., 1999). However, it worth noting that Chk1p does not complement the function for maintenance of peak level expression of the MCB-cluster in the cds1Δ strain. Similarly, Chk1p does not complement for the hypersensitivity of the cds1Δ strain to transient exposure to HU.
We show that forkhead transcription factors Sep1p and Fhk2p bind to the promoters of the FKH-cluster genes in ChIP assays (Supplemental Figure 3). Disruption of either sep1+ or fkh2+ is sufficient to abolish the expression response of the FKH-cluster upon HU treatment (Figure 5E), indicating two factors must act in concert. Significantly, only Sep1p is deprivated during S phase block but not Fkh2p (Figure 6, A and B), indicating that the Rad3p-dependent repression of the FKH-cluster is mediated via deprivation of Sep1p. Consistent with this, levels of Sep1p oscillate along the cell cycle with a peak at M phase coincident with a peak of Cdc2p activity (Figure 6D).
Analysis of genetic interaction indicates that disruption of sep1+ can alleviate rad3Δ phenotype, consistent with that Sep1p mediates the Rad3p-dependent expression response of the FKH-cluster upon HU treatment. Interestingly, Chk1p can complement for repression of the FKH-cluster in cds1Δ strain (Figure 5D) but not for maintenance of peak level expression of the MCB-cluster (Figure 3, A–D). Thus, we propose that during S phase arrest, both Rad3p–Cds1p and Rad3p–Chk1p pathways play a role in deprivation of Sep1p resulting in down-regulation of the FKH-cluster (Figure 7). Regulation of expression response of the ACE2-cluster at S phase arrest may simply be a secondary effect owing to ace2+ transcription being under the regulation of the Sep1p and Fkh2p.
Proteins involved in DNA replication checkpoint signaling are evolutionary conserved from mammalian systems to S. cerevisiae and S. pombe and play a major role in restoring stalled replication forks (Matsuoka et al., 1998). Upon S phase block, replication checkpoint Mec1p–Rad53p pathway in S. cerevisiae inactivates Cdc28p by tyrosine 19 phosphorylation (equivalent of tyrosine 15 in Cdc2p) (Krishnan et al., 2004), just as in S. pombe (Figure 6C). It has been shown that Mec1 and RAD53 also play a role in preventing precocious chromosome segregation. In this study, we show that Rad3p and Cds1p or Chk1p are involved in down-regulation of the FKH-cluster mediated through deprivation of Sep1p (Figure 7). Evidently, further investigations are required to establish the link between the Rad3p–Cds1p pathway and transcriptional activities and/or to uncover components that link the accumulation/deprivation of transcription factors and thus modulate gene expressions at the onset of S phase arrest.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. T. Liu for encouragement and support throughout the course of this study. We also thank L. Miller (Genome Institute of Singapore) for manufacturing S. pombe DNA microarrays, K. Gull for the TAT1 antibodies, and P. Nurse (The Rockefeller University) for the cig2Δ strain. We appreciate M. Balasubramanian, P. Robson, T. Lufkin, V. M. D'souza, and members of J. Liu's laboratory for discussion and critical reading of the manuscript. We acknowledge the comments by the anonymous reviewers that significantly improved the manuscript. This work was supported by the Agency for Science, Technology and Research (A-STAR), Singapore.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-10-0928) on March 1, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- al-Khodairy F., Fotou E., Sheldrick K. S., Griffiths D. J., Lehmann A. R., Carr A. M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayte J., Schweitzer C., Zarzov P., Nurse P., DeCaprio J. A. Feedback regulation of the MBF transcription factor by cyclin Cig2. Nat. Cell Biol. 2001;3:1043–1050. doi: 10.1038/ncb1201-1043. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Baum B., Wuarin J., Nurse P. Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J. 1997;16:4676–4688. doi: 10.1093/emboj/16.15.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Bimbo A., et al. Systematic deletion analysis of fission yeast protein kinases. Eukaryot. Cell. 2005;4:799–813. doi: 10.1128/EC.4.4.799-813.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Furnari B., Mondesert O., Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- Brondello J. M., Boddy M. N., Furnari B., Russell P. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol. Cell Biol. 1999;19:4262–4269. doi: 10.1128/mcb.19.6.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck V., Ng S. S., Ruiz-Garcia A. B., Papadopoulou K., Bhatti S., Samuel J. M., Anderson M., Millar J. B., McInerny C. J. Fkh2p and Sep1p regulate mitotic gene transcription in fission yeast. J. Cell Sci. 2004;117:5623–5632. doi: 10.1242/jcs.01473. [DOI] [PubMed] [Google Scholar]
- Caligiuri M., Beach D. Sct1 functions in partnership with Cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell. 1993;72:607–619. doi: 10.1016/0092-8674(93)90079-6. [DOI] [PubMed] [Google Scholar]
- Carr A. M. Control of cell cycle arrest by the Mec1sc/Rad3sp DNA structure checkpoint pathway. Curr. Opin. Genet. Dev. 1997;7:93–98. doi: 10.1016/s0959-437x(97)80115-3. [DOI] [PubMed] [Google Scholar]
- Chen D., Toone W. M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bahler J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland W. S. Robust locally weighted regression and smoothing scatterplots. Biol. Psychiatry. 1979;44:775–777. [Google Scholar]
- Dubacq C., Chevalier A., Courbeyrette R., Petat C., Gidrol X., Mann C. Role of the iron mobilization and oxidative stress regulons in the genomic response of yeast to hydroxyurea. Mol. Genet. Genomics. 2006;275:114–124. doi: 10.1007/s00438-005-0077-5. [DOI] [PubMed] [Google Scholar]
- Elledge S. J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Enoch T., Carr A. M., Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Gould K. L., Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- Hedges L. V., Olkin I. Statistical methods for meta-analysis: Academic Press. 1985 [Google Scholar]
- Hofmann J. F., Beach D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J. 1994;13:425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R., Gentleman R. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- Kelly T. J., Martin G. S., Forsburg S. L., Stephen R. J., Russo A., Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Krishnan V., Nirantar S., Crasta K., Cheng A. Y., Surana U. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell. 2004;16:687–700. doi: 10.1016/j.molcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lindsay H. D., Griffiths D. J., Edwards R. J., Christensen P. U., Murray J. M., Osman F., Walworth N., Carr A. M. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes N. F., McInerny C. J., Johnson A. L., Fantes P. A., Johnston L. H. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+ Nature. 1992;355:449–453. doi: 10.1038/355449a0. [DOI] [PubMed] [Google Scholar]
- Martin-Cuadrado A. B., Duenas E., Sipiczki M., Vazquez de Aldana C. R., del Rey F. The endo-beta-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J. Cell Sci. 2003;116:1689–1698. doi: 10.1242/jcs.00377. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Huang M., Elledge S. J. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- McInerny C. J., Kersey P. J., Creanor J., Fantes P. A. Positive and negative roles for cdc10 in cell cycle gene expression. Nucleic Acids Res. 1995;23:4761–4768. doi: 10.1093/nar/23.23.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto M., Tanaka K., Okayama H. res2+, a new member of the cdc10+/SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J. 1994;13:1873–1880. doi: 10.1002/j.1460-2075.1994.tb06456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murakami H., Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- Murray J. M., Carr A. M., Lehmann A. R., Watts F. Z. Cloning and characterisation of the rad9 DNA repair gene from Schizosaccharomyces pombe. Nucleic Acids Res. 1991;19:3525–3531. doi: 10.1093/nar/19.13.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N., Tanaka K., Sturm S., Okayama H. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J. 1995;14:4794–4802. doi: 10.1002/j.1460-2075.1995.tb00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva A., Rosebrock A., Ferrezuelo F., Pyne S., Chen H., Skiena S., Futcher B., Leatherwood J. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3:e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., et al. Identification of cell cycle-regulated genes in fission yeast. Mol. Biol. Cell. 2005;16:1026–1042. doi: 10.1091/mbc.E04-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N., Russell P. Tyrosine phosphorylation of cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol. Cell Biol. 1998;18:3782–3787. doi: 10.1128/mcb.18.7.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley R., Subramani S., Young P. G. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustici G., Mata J., Kivinen K., Lio P., Penkett C. J., Burns G., Hayles J., Brazma A., Nurse P., Bahler J. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- Sugiyama A., Tanaka K., Okazaki K., Nojima H., Okayama H. A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J. 1994;13:1881–1887. doi: 10.1002/j.1460-2075.1994.tb06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth N. C., Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- Walworth N., Davey S., Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- Watson A., Mata J., Bahler J., Carr A., Humphrey T. Global gene expression responses of fission yeast to ionizing radiation. Mol. Biol. Cell. 2004;15:851–860. doi: 10.1091/mbc.E03-08-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S., Khaliq F., Sotiriou S., McInerny C. J. The role of DSC1 components cdc10+, rep1+ and rep2+ in MCB gene transcription at the mitotic G1-S boundary in fission yeast. Curr. Genet. 2001;40:251–259. doi: 10.1007/s002940100248. [DOI] [PubMed] [Google Scholar]
- Yang Y. H., Dudoit S., Luu P., Lin D. M., Peng V., Ngai J., Speed T. P. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanow S. K., Lygerou Z., Nurse P. Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 2001;20:4648–4656. doi: 10.1093/emboj/20.17.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzov P., Decottignies A., Baldacci G., Nurse P. G(1)/S CDK is inhibited to restrain mitotic onset when DNA replication is blocked in fission yeast. EMBO J. 2002;21:3370–3376. doi: 10.1093/emboj/cdf346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Takeda T., Nasmyth K., Jones N. pct1+, which encodes a new DNA-binding partner of p85cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev. 1994;8:885–898. doi: 10.1101/gad.8.8.885. [DOI] [PubMed] [Google Scholar]
- Zilahi E., Salimova E., Simanis V., Sipiczki M. The S. pombe sep1 gene encodes a nuclear protein that is required for periodic expression of the cdc15 gene. FEBS Lett. 2000;481:105–108. doi: 10.1016/s0014-5793(00)01990-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.