Abstract
In Drosophila, the as yet uncloned heterochromatic locus flamenco (flam) controls mobilization of the endogenous retrovirus gypsy through the repeat-associated small interfering (rasi) RNA silencing pathway. Restrictive alleles (flamR) downregulate accumulation of gypsy transcripts in the somatic follicular epithelium of the ovary. In contrast, permissive alleles (flamP) are unable to repress gypsy. DIP1, the closest transcription unit to a flam-insertional mutation, was considered as a good candidate to be a gypsy regulator, since it encodes a dsRNA-binding protein. To further characterize the locus we analyzed P-induced flam mutants and generated new mutations by transposon mobilization. We show that flam is required somatically for morphogenesis of the follicular epithelium, the tissue where gypsy is repressed. This developmental activity is necessary to control gypsy and another retroelement, ZAM. We also show that flam is not DIP1, as none of the new permissive mutants affect the DIP1 coding sequence. In addition, two deletions removing DIP1 coding sequences do not affect any of the flamenco functions. Our results suggest that flamenco extends proximally to DIP1, spanning >130 kb of transposon-rich heterochromatin. We propose a model explaining the multiple functions of this large heterochromatic locus.
TRANSPOSABLE elements constitute an important part of eukaryotic genomes. Among them retrotransposons, which replicate by transcription of an RNA intermediate, subsequent reverse transcription, and insertion of new copies elsewhere in the genome, make up at least 45% of the human genome (Lander et al. 2001). Mobilization of retrotransposons is tightly controlled so as not to be overly deleterious. Drosophila offers an unique system to study interactions between these parasite-like elements and their host given the wealth of genetic tools available in this organism. In Drosophila, retroelements are silent most of the time. However, several strains are known in which massive mobilization of one or several elements occurs or has occurred (Mével-Ninio et al. 1989; Pasyukova et al. 1997; Desset et al. 2003).
For the past decade our laboratory has been engaged in studying the control of the mobilization of gypsy, an endogenous retrovirus of Drosophila melanogaster (Bucheton 1995; Prud'homme et al. 1995). Endogenous retroviruses can be considered as a particular class of long terminal repeat (LTR)-containing elements. They are inserted in the genome of their host and therefore transmitted through the germline. Some of them, including gypsy, can produce infectious particles and consequently infect horizontally new individuals (Kim et al. 1994a; Song et al. 1994). The genetic organization and replication cycle of gypsy is similar to that of vertebrate retroviruses (Marlor et al. 1986; Avedisov and Ilyin 1994; Pelisson et al. 1994).
D. melanogaster strains generally contain very few copies of active gypsy located in euchromatin. Genetic analysis of gypsy proviral amplification has shown that it is controlled by an as-yet-uncloned locus, flamenco (flam), located in the centromeric heterochromatin of the X chromosome (Prud'homme et al. 1995; Robert et al. 2001). Two classes of alleles, restrictive and permissive, are present in natural populations in about the same proportions (Pelisson et al. 1997). Restrictive alleles (flamR) are dominant and repress mobilization of functional gypsy proviruses. Therefore, strains containing flamR alleles are stable with regard to gypsy copy number. Conversely, permissive alleles (flamP) allow a high level of gypsy replicative transposition in homozygous strains (Prud'homme et al. 1995). However, most of the permissive stocks are stable because they do not contain functional gypsy proviruses (Kim et al. 1994b; Pelisson et al. 1997). In homozygous permissive females, the transcripts of active gypsy proviruses and virus-like particles accumulate in the somatic follicular epithelium (FE) surrounding germinal cells of the ovary (Pelisson et al. 1994; Lecher et al. 1997). It has been shown that restrictive flamR alleles repress gypsy transcript accumulation through a novel repeat-associated small interfering (rasi) RNA pathway, which is piwi-dependent (Sarot et al. 2004; Pelisson et al. 2007), similar to the recently described pathway, which controls repetitive sequences in Drosophila (Saito et al. 2006; Vagin et al. 2006).
When this study began, one unique flam mutant [flampy+(P)] was known. It was generated by insertion of the P{lyB} transposon into the 20A region of the Drosophila genome (Robert et al. 2001). Genetic and molecular studies of flam have been hindered because this insertion is located in an area of heterochromatin close to a sequencing gap of unknown size. The genomic DNA extending proximally to the P{lyB} insert (toward the centromere) consists mostly of defective repeated elements, especially truncated retroviral sequences. As such, no transcriptional unit corresponding to a single-copy gene was found. Distal to the P{lyB} insert, three different transcription units have been characterized. DIP1, the closest to the P{lyB} insertion site, encodes a nuclear protein containing two double-stranded RNA-binding domains (DeSousa et al. 2003) and therefore may be considered as a good candidate to function as a gypsy regulator.
In this study, we attempted to further characterize the flam locus by analyzing new insertional mutations and creating new genome rearrangements by transposon mobilization. This study reveals that, besides its function in gypsy repression in follicle cells, flam also controls FE morphogenesis. Moreover, the induction of flam mutants by insertions or deletions not only results in both female sterility and gypsy derepression but also relieves the repression of the ZAM retroelement. Our data also indicate that flam is not DIP1 but a distinct multifunctional heterochromatic locus that extends proximally to DIP1 over >130 kb.
MATERIALS AND METHODS
Drosophila strains:
Flies were grown at 25° on standard Drosophila medium. The strain wOR(P) carrying a natural permissive flam allele has been previously described (Sarot et al. 2004). The BG, KG, and EY collections from which the flamBG(P), flamKG(P), and flamEY(R) stocks were obtained are described in Bellen et al. (2004). PEY(R) is the y w parental strain of flamEY(R). GE89(R) is a deletion derivative of the insertional mutant G1261 sold by the Genexel company. G1261 contains the EP transposon (Rorth 1996) inserted at exactly the same position as EY in the flamEY(R) mutant. The strains of the laboratory collection were previously described, including the sample of deletions Df(1)lx (Prud'homme et al. 1995; Robert et al. 2001). The ZAM-lacZ tester strain (Desset et al. 1999) was kindly provided by C. Vaury.
Northern blots:
Ovarian gypsy rasiRNAs were detected using a low-molecular-weight Northern blotting method and a small sense RNA probe that were described previously (Pelisson et al. 2007). The amount and integrity of the RNA were checked both by staining the gels with 2 mg/ml ethidium bromide and by reprobing the unstripped membranes with an end-labeled oligonucleotide complementary to the abundant mir-13b microRNA (5′-ACTCGTCAAAATGGCTGTGATA-3′).
Rescue experiments:
Cosmid 7a is issued from the NotBamNot-CoSper genomic library, kindly provided by J. Tamkun. It consists of 35 kb of genomic sequence including the whole DIP1 sequence and 29 kb extending upstream of the DIP1 transcription start site (see Robert et al. 2001). An AvrII–BciVI 8-kb fragment of cosmid 7a, potentially able to produce all DIP1 transcripts, was subcloned in the HpaI and SpeI sites of the pCaSpeR4 vector to generate transgenic lines.
DIP1-b and DIP1-c cDNAs were issued from an ovarian cDNA library (Stroumbakis et al. 1994) and from the Berkeley Drosophila Genome Project (BDGP) (EST LD14381), respectively. The NotI–XbaI digestion fragment of DIP1-b and the NotI–SmaI digestion fragment of DIP1-c, both cloned into the pUAST vector, were subcloned into the NotI–XbaI and NotI–HpaI sites of the pCaSpeR-HS vector, respectively. The sterility of flampy+(P) mutant females is thermosensitive, partial rescue being observed when L3 and pupal stages are raised at 18° (15 adult progeny per female at 18° for 9-day egg laying). For this reason, our attempt to rescue the sterility was performed by heat-shocking transgenic females at these developmental stages for 1 hr/day at 37°.
Ovary DAPI staining:
Ovaries were stained with the nuclear dye DAPI for the examination of ovary morphology. They were fixed in 4% formaldehyde, 1× PBS, 1% Triton (PBT) for 10 min and rinsed twice in PBT. Then, they were placed for 5 min in PBT with 1 μg/ml of DAPI (Sigma, St. Louis). After three rinses they were mounted in PBS:glycerol (1:1). Samples were examined by confocal microscopy using a Leica SP2 UV microscope.
Permissivity and instability assays:
The reporter constructs for gypsy (p#12) or ZAM (pZAM-lacZ) expression in ovaries were described in Sarot et al. (2004) and Desset et al. (2003), respectively. They consist of the gypsy or ZAM promoters fused to the Escherichia coli lacZ gene. Permissive w flamOR(P)/w flamOR(P) females homozygous for the #12 transgene were crossed to males under test. The flam restrictive character is semidominant and daughters do not express high levels of β-galactosidase in their FE cells if their father carries a flam restrictive allele. In the same way, XU/XU; ZAM-LacZ/ZAM-LacZ females were crossed to males of lines containing various flam alleles. Coloration develops in all egg chambers only if the father carries a U allele. Histochemical staining of β-galactosidase was performed as described in Sarot et al. (2004).
Germline clonal analysis:
To obtain mosaic females with germline cells homozygous for flamBG(P), we induced mitotic crossing over in w ovo+ flamBG(P)/ovoD1 v flam+ flies according to Perrimon and Gans (1983). Early larvae were irradiated with 1000 rad of X radiation from an X-ray source. Adult females were mated to w flamBG(P)/Y males and tested for fertility. Confirmation that the female germline clones were flamBG(P) was obtained by examining the male progeny for the white marker. Ovaries were fixed, stained for DAPI, and examined.
P-element excision analysis:
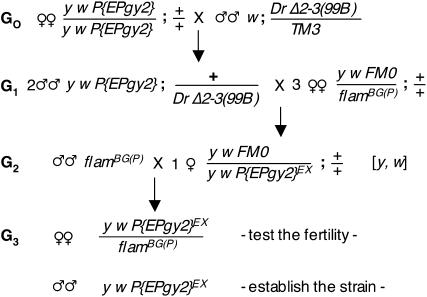
As shown in Figure 3, mobilization of the P{EPgy2} transposon was induced in flamEY(R) G1 males that had received the Δ2-3(99B) source of transposase. These males were crossed to FM0/flamBG(P) females and independent excision events were isolated in G2 as FM0/flamEY females having lost the w and/or y markers. Viability of the excised alleles as well as their ability to complement flamBG(P) sterility was tested in G3. All 130 tested alleles were viable.
Figure 3.—
Mating scheme used to select for excisions of the P{EPgy2} transposon from the flamEY(R) strain. FM0 and TM3 are balancer chromosomes that are transmitted through meiosis with little or no recombination with the homologous chromosome. They carry the B and Ser markers, respectively, which, like Dr, are semidominant and can therefore be used to recognize and select for the appropriate chromosome combination in heterozygous flies. Mobilization of the P{EPgy2} transposon was induced in flamEY(R) G1 males that had received the Δ2-3(99B) source of transposase. These males were then crossed to FM0/flamBG(P) females and independent excision events were isolated in G2 as FM0/flamEY females not expressing the w and/or y markers that indicate the presence of the P{EPgy2} insert. Viability of the excised alleles as well as their ability to complement flamBG(P) sterility was tested in G3. All 130 tested alleles were viable.
Molecular analysis of the excisants:
Deletions of the P{EPgy2} transposon were analyzed by amplification assays of genomic DNA using the oligonucleotide primers oligo a (DIP1 sense orientation), GAGGAAACGCAAAACTCTTGTGAA, and oligo b (DIP1 antisense orientation), TGAACACTTTCACTTTGTGCGCTGCAA (see Figure 1). Deletions of sequences flanking the P{EPgy2} transposon were tested by amplification of flanking DNA fragments. We used the following pairs of primers: oligo a and oligo c, TAAAAAACTTAAGTAGGGATGTCAAAAT; and oligo b and oligo d, TGCAGTACGATTCT TTCTGTGACCTG (see Figure 1). Controls were performed using DNA from the strain PEY(R), which is the flamEY(R) parent. Deletions present in flamEY4(R) and flamGE89(R) were analyzed by sequencing PCR products. Analysis of 6 kb of DNA sequence proximal and distal to the P{EPgy2} insertion site was performed by amplification of overlapping DNA sequences using appropriate pairs of oligonucleotide primers (oligo sequences are available upon request). Although the DNA sequence proximal to the P{EPgy2} insertion site consists of repeated sequences, the arrangement of the repeated DNA is unique in the genome and a unique amplification product was always obtained. The genomic sequence flanking proximally the deletion in flamEY18(P) was determined by sequencing of the inverse PCR product. A detailed protocol is available on the BDGP webpage at http://www.fruitfly.org/about/methods/inverse.pcr.html. Genomic DNA was digested with HpaII and the flanking sequence was amplified with the PlacI CACCCAAGGCTCTGCTCCCACATT and PwhtI GTAACGCTAATCACTCCGAACAGGTCACA primers. Database searches were carried out by BLAST search using the last release database at the National Center for Biotechnology Information (NCBI) and at FlyBase.
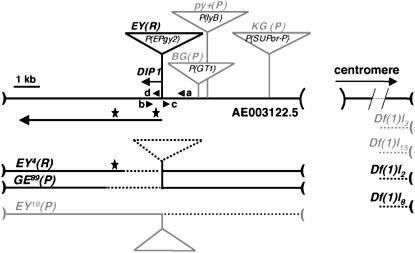
Figure 1.—
Structure of the DIP1-flamenco region showing the positions of P-element-induced mutations and X-ray-induced deletions. The horizontal thick line represents sequenced DNA from scaffold AE003122.5, which extends between the P{lyB} insertion and a sequencing gap of unknown length (thin dotted lines). Positions of the P-element insertions within the region are indicated by a stalk and a triangle (not to scale). Insertions of P{lyB}, P{GT1}, and P{SUPor-P} generated, respectively, the flampy+(P), flamBG(P), and flamKG(P) female permissive and sterile alleles (shown shaded). Arrowheads a, b, c, and d indicate the positions and orientations of the primers used to analyze the excision events. The bent arrow indicates the DIP1 transcription initiation site as deduced from the longest 5′ cDNA sequence (GenBank accession no. BI242976). Position and size of the DIP1 transcription unit are indicated by a thick arrow. The two stars above the arrow indicate the position of the two DIP1 translation start sites. The P{EPgy2} insertion, 14 bp downstream of the DIP1 transcription start site, generated the flamEY(R) fertile allele. Below, the maps of deletions EY4(R), GE89(R), and EY18(P) are shown. Thick dotted lines indicate deletions. Distal deletions EY4(R) and GE89(R) are still fertile, while EY18(P) is female sterile (shown shaded). Also shown at the right are the maps of X-ray-induced deletions (Robert et al. 2001). The extremities of these deletions are not determined but their nearest breakpoint is located in the sequencing gap, >130 kb away from the P[lyB] insertion point.
RESULTS
P-element-induced flam mutants are both female sterile and permissive for gypsy expression:
Permissive and restrictive flam alleles coexist in natural populations (Pelisson et al. 1997). Derepression of gypsy, in permissive females, occurs in the somatic FE (Pelisson et al. 1994). Thus, we wondered whether flam also plays a role in FE formation. We first analyzed females from the wOR(P) strain that carry a natural permissive allele. These females are normally fertile and examination of their ovaries, after DAPI staining, did not reveal any defects (data not shown). Accordingly, either flam has no function in the FE or the gypsy control function can be inactivated independently from a putative ovarian function.
Next, we examined the only available flam mutant, flampy+(P), resulting from insertion of the P{lyB} transposon into region 20A1-3 of the restrictive FM7c balancer chromosome (Robert et al. 2001). We eliminated the sn female-sterile mutation present on FM7c by recombination with the FM7a balancer chromosome and found that homozygous flampy+(P) females did not lay any eggs (Table 1). Subsequently, we checked that the sterility phenotype of flampy+(P) females was actually caused by the P{lyB} insertion. Thirty excision events of the P{lyB} transposon were studied by Southern blot analysis as described in Robert et al. (2001). In 20 of them, either precise or imprecise excision was associated with simultaneous reversion toward the restrictive character and restoration of fertility, indicating that the P{lyB} insertion was responsible for both mutant phenotypes and had taken place in a noncoding region (data not shown). The 10 excisants that remained permissive were still female sterile. This further strengthened the link between the two flam mutant phenotypes: sterility and loss of gypsy control.
TABLE 1.
Complementation tests of flam mutants
| Genotype | Average no. of eggs laid/female/day | % sterilitya | % fertilityb |
|---|---|---|---|
| flampy+(P)/flampy+(P) | 0 | 100 | 0 |
| flamBG(P)/flamBG(P) | 0 | 100 | 0 |
| flamKG(P)/flamKG(P) | 0.41 | 98.5 | 0.23 |
| flampy+(P)/flamBG(P) | 0.09 | 99.7 | 0.3 |
| flampy+(P)/flamKG(P) | 12 | 43 | 17.3 |
| flamBG(P)/flamKG(P) | 0.85 | 97.2 | 0 |
| flamBG(P)/Def(1)l3 | 1.4 | 95 | 0.08 |
| flamBG(P)/Def(1)l15 | 0.37 | 98 | 0.13 |
| flampy+(P)/flamEY7(P) | 15 | 40.3 | 14 |
| flamBG(P)/flamEY7(P) | 0 | 100 | 0 |
| flamBG(P)/flamEY2(P) | 0 | 100 | 0 |
| flamBG(P)/flamEY18(P) | 0 | 100 | 0 |
[1 − (no. of eggs laid/mutant female/no. of eggs laid/wild-type sibling)] × 100. At least 100 3-day-old females were tested over a period of 9 days, at 22°.
[no. of adult progeny/mutant female (9-day egg laying)/no. of adult progeny/wild-type sibling] × 100.
We then tested two additional mutant strains for the permissivity and female sterility characteristics: BG02658 contains the P{GT1} element (Lukacsovich et al. 2001) inserted 338 bp distally to the P{lyB} insertion site and KG0476 carries the P{SUPor-P} element (Roseman et al. 1995), 2.4 kb proximal to P{lyB} (Figure 1). The permissive/restrictive character was tested by monitoring the expression of a gypsy-lacZ transcriptional fusion in the FE (Pelisson et al. 1994). We found that both mutant lines are permissive and therefore we renamed them flamBG(P) and flamKG(P). Most interestingly, both homozygous flamBG(P) and flamKG(P) females are sterile. This sterility is not fully complemented by flampy+(P) (Table 1). flampy+(P)/flamBG(P) females produce very few eggs. There is partial complementation between flampy+(P) and flamKG(P) alleles. The number of eggs laid by flampy+(P)/flamKG(P) females is about half the number laid by wild-type sibling sisters. However, a high proportion of these eggs do not develop into adulthood. We could not test whether the permissivity of flamBG(P) and flamKG(P) females is caused by the insertions, as the flamBG(P) parental strain (Iso2A; Iso3A; Bellen et al. 2004) already carries a natural permissive allele and the flamKG(P) parental strain is not available. From these experiments we conclude that the flam locus is required for female fertility. The three insertions present in flampy+(P), flamBG(P), and flamKG(P) disrupt this oogenic function and, at least in the case of flampy+(P), also generate permissivity.
P-element-induced flam mutations reduce the accumulation of gypsy rasiRNAs in the ovary:
We previously reported that antisense RNAs, 25–30 nucleotides in length, complementary to gypsy, accumulate in ovaries of restrictive females, while the amount of such RNAs is dramatically reduced in females carrying a natural permissive allele (Sarot et al. 2004; Pelisson et al. 2007). These results strongly suggested that gypsy regulation is mediated by a rasiRNA pathway. We thus wondered whether in P-induced flam mutants the amount of gypsy rasiRNAs is affected. Comparison of the levels of gypsy rasiRNAs in flampy+(P)/flampy+(P) mutant and flamFM7c(R)/flampy+(P) wild-type ovaries revealed that the flam mutation event resulted in a dramatic reduction of the amount of gypsy rasiRNAs (Figure 2). This effect does not result from the lack of development of flampy+(P) ovaries. Indeed, as we show below, homozygous flampy+(P) females, although they do not lay any egg, develop almost normal ovaries. The same significant reduction of gypsy rasiRNAs was also observed in a much less sterile genetic combination, flampy+(P)/flamKG(P) (Figure 2 and Table 2). We conclude that flam is likely required for the accumulation of most, if not all, gypsy rasiRNAs in the ovary.
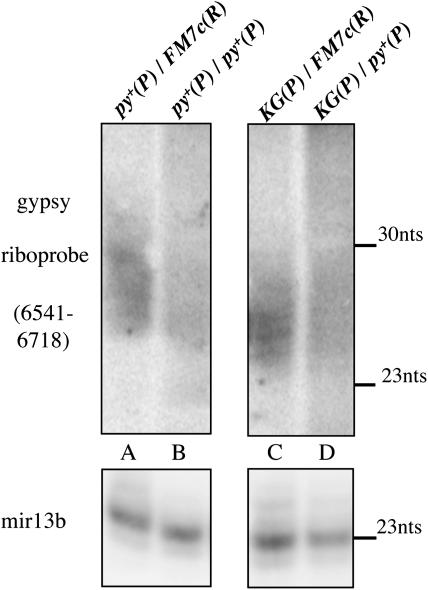
Figure 2.—
P-element-induced flam mutations reduce the accumulation of gypsy rasiRNAs in the ovary. Low molecular weight-enriched RNA extracted from ovaries was hybridized (top) with a sense gypsy RNA probe (coordinates 6541–6718 in the gypsy sequence, accession no. M12027) and reprobed (bottom) with the end-labeled oligonucleotide complementary to the mir-13b miRNA. The position of the mir-13b 23-nt major band is indicated, as well as that of the 30-nt band of the Decade size marker (Ambion, Austin, TX). Antisense rasiRNAs, complementary to the gypsy probe were mostly detected in both genotypes containing the parental FM7c(R) restrictive chromosome, as compared with those containing its py+(P) mutant derivative.
TABLE 2.
Mutations or reversions at the flam locus induce the simultaneous loss or gain of female fertility and ability to control gypsy and ZAM
| flam allele | Complementation of BG(P) sterility | gypsy control | ZAM control |
|---|---|---|---|
| KG(P) | − | P | U |
| Iso2A; Iso3Aa | + | P | S |
| BG(P) | − | P | U |
| FM7c(R)b | + | R | S |
| py+(P) | − | P | U |
| FM7c,679-5b(R)b | + | R | S |
| 413(NP)c | + | R | S |
| l2, l8 | + | R | S |
| l3, l15 | − | P | U |
| PEY(R)d | + | R | S |
| EY(R) | + | R | S |
| EY4, EYne | + | R | S |
| EY2, EY7, EY13, EY18 | − | P | U |
| GE89(R) | + | R | S |
P, permissive; R, restrictive; U, unstable; S, stable.
An isogenized stock from which BG(P) originates (Bellen et al. 2004). It carries a spontaneous permissive allele.
The FM7c(R) and FM7c,679-5b(R) X chromosomes, respectively, contain the parent and an excision revertant of the py+(P) mutant (Robert et al. 2001).
The 413(NP) X chromosome, restrictive for gypsy expression, has been used to generate the set of lx deficiencies (Prud'homme et al. 1995; Robert et al. 2001).
The X parental chromosome of EY(R).
EYn stands for any of the 25 excision derivatives that remained fertile.
The flam locus spans a large region of heterochromatin:
To characterize the flam locus, we analyzed a series of deletions previously generated from a restrictive chromosome (Prud'homme et al. 1995). The distal breakpoints of all these deletions are located >130 kb proximal to the P{lyB} insertion site (Robert et al. 2001; Figure 1). Two of them, Df(1)l3 and Df(1)l15, have been shown previously to delete the ability of flam to repress gypsy expression and mobilization (Robert et al. 2001). Here we show that these deficiencies also delete the flam ovarian function since neither of them complements flamBG(P) sterility (Table 1). In contrast, two other deficiencies, Df(1)l2 and Df(1)l8, which remained restrictive, do complement flamBG(P) sterility (Table 2). These results indicate that both functions of flam are disrupted by deficiencies located >130 kb proximal to the P{lyB} insert.
DIP1 transgenes do not rescue flam functions:
The three inserts present in flampy+(P), flamBG(P), and flamKG(P) are located very close to the DIP1 transcription start site (1.7 kb in flampy+(P), 1.360 kb in flamBG(P), and 4.1 kb in flamKG(P); see Figure 1), suggesting that the flam mutant phenotypes displayed by these strains might result from DIP1 inactivation. However, previous studies have shown that the DIP1 transcription patterns of females carrying naturally permissive vs. restrictive flam alleles were not obviously different (Robert et al. 2001). This pattern is not affected in homozygous flampy+(P) mutant females either (data not shown).
We have previously shown that an 8-kb long genomic DNA transgene, potentially able to produce all DIP1 transcripts, cannot rescue the gypsy regulatory function (Robert et al. 2001). The same was true for cosmid 7a, a larger genomic construct, extending 29 kb upstream of the DIP1 transcription start site (Robert et al. 2001). Here, we performed appropriate crosses to yield homozygous flamBG(P) or flampy+(P) females, carrying one or two copies of either transgene. Four independent transgenic lines carrying the 8-kb transgene and one line carrying cosmid 7a were tested. In none of these cases was the flam sterility phenotype rescued. We obtained the same negative results by heat-shocking at the L3-pupal stages two heat-shock constructs able to express one of the two longest DIP1 isoforms, DIP1-b and DIP1-c (see materials and methods).
DIP1 deletions do not affect flam functions:
We analyzed a new strain, EY02625 (renamed flamEY(R)), containing the P{EPgy2} transposon inserted 14 bp inside the 5′ end of the longest DIP1 cDNA (Bellen et al. 2004; Figure 1). We found that flamEY(R) is not a flam mutant. It remained female fertile and restrictive like its parent PEY (Table 2). This result suggests that the region into which P{EPgy2} is inserted does not belong to the flam locus.
The possibility still exists, however, that the P{EPgy2} transposon does not inactivate DIP1. With the aim of inactivating this gene, we induced P-element imprecise excisions (see Figure 3). We obtained 130 independent excision derivatives of flamEY(R) and analyzed them for their ability to complement the sterility of flamBG(P). For 30 of these derivatives we also amplified by PCR the genomic DNA flanking the P{EPgy2} transposon to molecularly characterize the excisions.
All derivatives were viable. For most of the 30 lines analyzed by PCR, a partial deletion inside the transposon was found, while flanking sequences were unaffected. Four excisants, flamEY2(P), flamEY7(P), flamEY13(P), and flamEY18(P), were homozygous female-sterile mutants that did not complement flamBG(P) sterility (Table 1). Interestingly, these female-sterile excisants also simultaneously acquired the flam permissive character (Table 2). We analyzed by PCR 6 kb of sequence on each side of the initial insert. On the DIP1 side, none of the four sterile and permissive excisants exhibited any difference from the wild-type sequence. On the proximal side, except for flamEY18(P), we did not detect any alteration either. We assume that in these strains the mutational event that led to the flam mutant phenotype has taken place more proximally. The derivative flamEY18(P) is different. In this case, the P{EPgy2} insert is still present, but proximally a large deletion, 133.9 kb long, has occurred (Figure 1). Indeed, the sequence of a 720-bp long genomic DNA fragment, flanking proximally the deletion, hits perfectly to Drosophila genomic sequences located 133.9 kb proximal to the EY insert (see supplemental data at http://www.genetics.org/supplemental/).
One derivative, flamEY4(R) is particularly interesting. Proximally, the sequence is not affected. However, distally, 1.28 kb of the DIP1 transcribed sequence has been deleted, removing the first putative translation start site (Figure 1). Nevertheless, flamEY4(R) is fertile and restrictive like its parent. We also obtained a larger deletion, flamGE89(R), generated from the mutant G1261 available from the Genexel company (see materials and methods). As in flamEY4(R), the sequence is not affected proximally, but distally 1.9 kb of the DIP1 transcribed sequence is deleted, removing the two putative DIP1 translation start sites (Figure 1). Assuming that the following in-frame ATG could be used to initiate translation, flamGE89(R) would produce truncated isoforms lacking the nuclear localization signal and a complete NH2 terminal domain. The DIP1-a, -b, and -c isoforms would be truncated by 69, 156, and 169 amino acids, respectively. Nevertheless, flamGE89(R) is fertile and weakly restrictive like its parent (Table 2).
flam is necessary for ovarian germline stem cell differentiation and division:
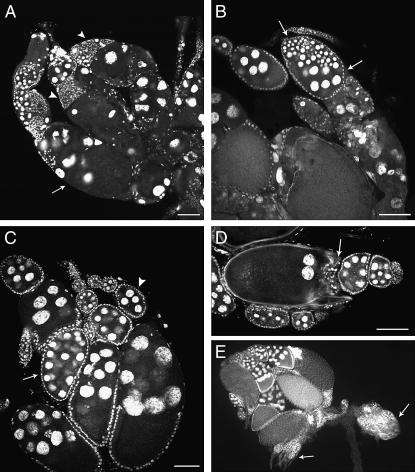
Females of flam mutants exhibit atrophic ovaries, flamBG(P) females showing the most severe phenotype. All ovaries of 24-hr-old flamBG(P) females contain rudimentary ovarioles within which differentiation and division of germline cells is affected (Figure 4B). When differentiation takes place, ovaries become increasingly disorganized. Ovarioles contain less than three egg chambers and most egg chambers are characterized by a reduced number of degenerating nurse cells (Figure 4D). Cysts containing as few as one single nurse cell are frequently observed, suggesting that the ability of the differentiated cystoblasts to divide has been affected. The phenotypes presented by flampy+(P) (Figure 5C) and flamKG(P) (Figure 4C) mutant females are not as severe. Nevertheless, flamKG(P), like flamBG(P) females, develop ovarioles that may be devoid of differentiating germline cells (Figure 4, B and C).
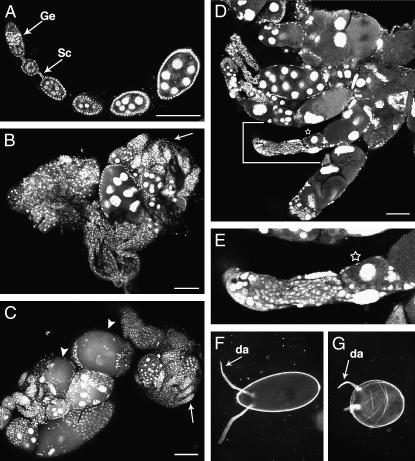
Figure 4.—
flam mutations affect germline cell development. Ovaries were stained with DAPI. (A) A 24-hr-old wild-type ovariole contains a long string of developing egg chambers produced continuously by the germarium (Ge) and connected to each other by stalk cells (Sc). (B) flamBG(P) and (C) flamKG(P) are typical 24-hr-old ovaries. They are very small and contain a number of rudimentary ovarioles within which no differentiation of germ cells has taken place (arrows). In C two egg chambers exhibit gaps in the FE (arrowheads). (D) flamBG(P) 48-hr-old ovarioles are severely dystrophic. Ovarioles contain two to three completely disorganized egg chambers. These chambers contain a reduced number of abnormally shaped nuclei. (E) Enlargement of the portion of ovariole shown in D, delimited by brackets. The star indicates an egg chamber containing a single nurse cell-like nucleus. (F) A wild-type mature egg. (G) A mutant flamBG(P) mature egg lacks polarity and has abnormal dorsal appendages (da). Bars, 80 μm.
Figure 5.—
Soma-dependent FE defects of flam mutants. (A) In 6-day-old flamBG(P) ovaries, follicle cells form aggregates (arrowheads) while large gaps are seen elsewhere in the FE (arrow). (B) Forty-eight-hour-old flamKG(P) ovarioles. Follicle cells fail to invaginate in the germarium, resulting in a compound egg chamber with nurse cells that present various degrees of polyploidization (arrows). (C) A 3-day-old flampy+(P) ovary. Abnormalities are frequently observed during ovarian development. Here a compound egg chamber is present (arrow). Also present is an egg chamber with a deficit in nurse cell number (arrowhead). (D) Stage 11 egg chamber from a 3-day-old flampy+(P) female in which follicle cells have not migrated properly, resulting in nurse cells being pinched off into the ooplasm. Follicle cells accumulate at the anterior of the egg (arrow). (E) A pair of ovaries containing ovarioles with homozygous flamBG(P) germline clones and ovarioles from ovoD1 flam+ germline cells. The ovoD1 flam+ germline cells are arrested at the beginning of oogenesis because of the ovoD1 mutation (arrows). By contrast, in ovarioles in the left ovary, containing flamBG(P) germline clones, germline stem cells divided normally. Bars, 80 μm.
The very rare mature eggs that are formed in flamBG(P) females present obvious defects in shape and dorsal appendage formation (Figure 4G). They are smaller than wild-type eggs and appear to lack dorsal–ventral and anterior–posterior polarities. Their dorsal appendages are shortened and abnormally shaped. Such defects could result from abnormal signaling from the FE.
flam is somatically required for the formation of the follicular epithelium:
All three flampy+(P), flamBG(P), and flamKG(P) alleles cause FE aberrations. In 6-day-old flamBG(P) females (Figure 5A), the FE is completely disorganized. Large aggregates of cells can be observed between egg chambers while large gaps are formed above dying nurse cells. In some flamKG(P) and flampy+(P) mutant germaria, follicle cells probably fail to invaginate between the developing cysts. This results in composite egg chambers in which several germ cell syncytia are surrounded by a single follicle cell layer (Figure 5, B and C). Similar defects have been described for ovaries lacking the fs(1)Yb, N, Dl, da, or brn functions (Ruohola et al. 1991; Goode et al. 1992; Cummings and Cronmiller 1994; Johnson et al. 1995). Defects in the FE of flampy+(P) females are not as severe as those presented by flamBG(P) or flamKG(P) females. Sometimes the backward migration of the columnar follicle cells is defective, with the result that a couple of nurse cells may be pinched off into the ooplasm (Figure 5D). Although a few mature egg chambers of normal appearance are found in flampy+(P) ovaries, they must be defective since no eggs are laid.
To determine if flam is needed in the germline or the soma, we produced mosaic females with germline cells homozygous for flamBG(P) (the most severe flam mutation), while the somatic tissue carried one wild-type allele (Perrimon and Gans 1983). We obtained fertile adult females that developed several completely normal ovarioles (Figure 5E). This result demonstrates that oogenesis occurs normally in flamBG(P) germline clones. The effect of flam mutations on germ cell differentiation described above must therefore reflect interactions between the germline and the defective ovarian somatic cells. Eggs produced from homozygous flamBG(P) germ cell clones gave rise to normal adult flies. This result demonstrates that expression of flam in the germline, if any, does not provide an essential maternal contribution for embryogenesis.
flam also controls the expression of the ZAM retroelement:
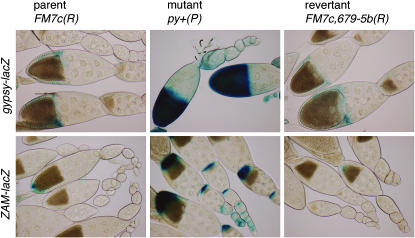
The control of ZAM, another retroelement from D. melanogaster, appears to display strikingly similar features to that of gypsy (Desset et al. 1999, 2003). ZAM expression is tightly repressed in most Drosophila strains (referred to as stable or S lines). However, in the Rev line, referred to as an unstable or U line, this control is disrupted, resulting in a high mobilization rate. The genetic determinant controlling ZAM mobilization is localized to the 20A region, the same region where flam maps (Desset et al. 2003). Thus, we asked whether the flam mutants described above also disrupt ZAM control. We monitored ZAM expression with a ZAM-LacZ reporter containing the full-length LTR of ZAM. In the U/U background, ZAM-lacZ is expressed in the posterior follicle cells of all egg chambers, starting at stage 1, while in the U/S background, it is expressed only in stage 10 egg chambers (Desset et al. 2003). As shown in Table 2 and Figure 6, loss or restoration of the female fertility function was always correlated with the corresponding loss or restoration not only of gypsy repression, but also of ZAM repression.
Figure 6.—
flam controls the expression of both gypsy and ZAM retroelements. The parental FM7c(R) allele is restrictive, stable for ZAM expression (S), and female fertile (see Table 2). It represses the β-galactosidase expression driven by gypsy as well as ZAM LTRs. The flam female-sterile mutant allele, py+(P), caused by insertion of the P{lyB} element into the flam locus of the FM7c chromosome, no longer represses the gypsy and ZAM reporters. In contrast, repression and fertility are restored in females carrying the revertant allele FM7c,679-5b(R), generated by excision of the P{lyB} element.
We also tested deficiencies Df(1)l3 and Df(1)l15 (Figure 1), which are permissive and sterile. We observed that ZAM control was lost in both deletions. By contrast, deficiencies Df(1)l2 and Df(1)l8, which are restrictive and fertile, retained the ability to repress ZAM (Table 2). These results strongly suggest that Flam is necessary for three functions: ovary development, gypsy control, and ZAM control.
DISCUSSION
A strategy used by retroelements to gain access to the germinal cells is to be expressed specifically in the somatic tissue adjacent to the germline (Lock et al. 1988; Godwin et al. 1995). In D. melanogaster, gypsy clearly relies on its somatic expression in the FE to increase its copy number inside the germline (Chalvet et al. 1999). Indeed, not only are gypsy transcripts observed in FE cells, but also virus-like particles (Lecher et al. 1997). These are subsequently transferred to the oocyte before new copies of gypsy are inserted into the germline. This invasion is controlled by the flam locus. Here we show that flam, which represses the expression of gypsy in the FE, is required for morphogenesis of the FE and that it also controls the expression, in this tissue, of at least one other retroelement, ZAM.
flam fertility function is correlated with the ability to control gypsy and ZAM:
Both permissive and restrictive flam alleles are present in natural populations (Pelisson et al. 1997), suggesting that they represent two functional allelic forms. We show in this article that permissive phenotypes can also be induced by mutations at the flam locus. Rearrangements at this locus, over a range of at least 130 kb, cause female sterility and simultaneously transform “restrictive-stable” alleles into “permissive-unstable” ones (i.e., unable to repress both gypsy and ZAM). Although flam mutations cause defects in both the somatic and the germline components of the ovary, we demonstrate by germline clonal analysis that somatic expression of flam is sufficient to support oogenesis. This result indicates that the effect of flam mutations on female germ cell differentiation and division is most likely the consequence of aberrant somatic ovarian development. Thus, flam appears to be a large locus, at least 130 kb long, whose integrity is required for normal FE development and for control of the gypsy and ZAM retroelements.
DIP1 does not encode the Flamenco function:
The two following observations led to the hypothesis that the flam function might be performed by DIP1. First, P-element insertions inactivating the flam restrictive function are all located upstream and close to the transcription initiation site of DIP1. Second, flam-dependent RNA silencing of gypsy (Sarot et al. 2004) might well be operated by the DIP1 protein that encodes two double-stranded RNA-binding domains (dsRBD) and binds dsRNA, a typical trigger for RNA silencing (Desousa et al. 2003).
However, our present results do not support this hypothesis. On the contrary, they suggest that flam is a large gene that extends proximal to DIP1. Indeed, the large P{EPgy2} transposon, present at the DIP1 transcription start site, does not affect flam functions. The same is true for the deletions present in flamEY4(R) and flamGE89(R) that remove 1.28 and 1.9 kb, respectively, of DIP1 transcribed sequences. Yet, in flamGE89(R) the deletion removes the two translation start sites, presumably preventing translation of any of the known DIP1 isoforms. Conversely, all rearrangements occurring upstream of the DIP1 transcription start site, located from a few kilobases up to >130 kb away, exhibit the flam mutant phenotypes. Identifying a transcription unit in this region has been, up to now, hindered by its repetitive nature and the presence of a sequencing gap of unknown size.
A model for the control of retroelements by flamenco:
How could the flam heterochromatic gene perform such different functions such as the repression of retroelements and the control of FE development? Although both functions were always simultaneously affected by the rearrangements described in this article, they may be uncoupled from each other in natural allelic variants. Moreover, gypsy and ZAM retroelements can also be controlled independently from each other. Indeed, all the known natural flam alleles that are permissive for gypsy expression are stable, i.e., able to repress ZAM. Symmetrically, the only spontaneous allele, known to derepress ZAM, restricts gypsy expression (Desset et al. 2003). Furthermore, examples have been described of a permissive-stable strain becoming spontaneously restrictive stable (Prud'homme et al. 1995) and of a restrictive-stable strain becoming restrictive unstable (Desset et al. 2003). A model for flamenco should therefore explain both types of mutational events: on the one hand, the simultaneous loss of all functions, including fertility, resulting from genomic rearrangements at the flam locus, and, on the other hand, gain or loss of the ability to control either of the retroelements without loss of female fertility. The model should also take into account the fact that the flam locus of restrictive strains contains the genetic determinant(s) for the production of rasiRNAs that target any fragment of the gypsy genomic RNA (Sarot et al. 2004; Pelisson et al. 2007). At the molecular level, the recent D. melanogaster genome assembly (BDGP release 5) extends now 180 kb proximally to the P{lyB} insert. Most of this sequence consists of nested transposon remnants. Among them, it was of special interest to find two blocks of gypsy sequences, 2.4 and 3.2 kb long, that altogether might accommodate most, if not all, of the gypsy rasiRNAs. In addition, this 180-kb fragment also contains a 5-kb block homologous to ZAM.
flam might be an RNA-silencing gene. Indeed, we know that a number of genes required for RNA interference (RNAi) are also involved in oocyte development (Chekulaeva and Ephrussi 2004). However, this hypothesis does not explain how three functions—FE development, regulation of the expression of gypsy, and ZAM—can be controlled by the same RNA silencing pathway independent of each other.
Alternatively, flam might correspond to heterochromatic proviral sequences, producing autonomously the RNA trigger(s) required for silencing of the homologous retrotransposon(s). This process could be similar to that reported for Mu killer, a naturally occurring transposon derivative in maize responsible for the silencing of the MuDr transposon family (Slotkin et al. 2005). Unlike Mu killer, the two putative gypsy triggers, found in the 180-kb sequence of release 5, do not have hairpin structures. The hypothesis of autonomously regulated RNA silencing triggers does not explain why the mutational insertions would abolish simultaneously the expression of all triggers. It does not explain either the sterility phenotype or why these sequences have to be localized at the flam locus rather than somewhere else in the genome.
Analyzing the genomic DNA sequence of the flam locus reveals striking features of this 180-kb heterochromatic chromosome region. Nearly all the retroelements present in the locus are oriented toward the telomere. We, thus, suggest a third hypothesis, assuming that the whole flam region is transcribed toward the centromere under the control of a single promoter. This promoter would cotranscribe a fertility gene and proviral sequences, among which are gypsy and ZAM, in an antisense orientation. This organization would be similar to that of the Parp gene, which spans >150 kb of centromeric heterochromatin and contains large introns accumulating various transposable elements, nearly all oriented opposite to the gene (Tulin et al. 2002). This hypothesis explains why P-induced female-sterile flam mutations also result in the failure of retroelement control: reducing the transcription level of this fertility gene would at the same time reduce the amount of RNA silencing trigger(s). This hypothesis also explains why such a variety of fertile natural flam alleles exist that are able to control or not able to control the expression of one or the other retrovirus, gypsy and ZAM. Such control would depend on the polymorphic presence, in putative flam introns, of gypsy and/or ZAM retroviral sequences. According to this hypothesis, the flam locus should be able to control the expression of other transposable elements.
We cannot exclude the possibility that this putative transcription unit does not contain any coding exon and that the sterility observed, in flam-mutant females, is the indirect consequence of high levels of transposition, resulting from the lack of rasiRNAs. This would be reminiscent of the indirect effect of mutants of the rasiRNA pathway on the specification of embryonic axis in Drosophila (Klattenhoff et al. (2007). Whatever the case, the localization of a transcribed locus in the pericentromeric heterochromatin could represent one of the ways used by organisms to defend themselves against retroviral invasion: the expression of such a locus in the very tissue used by retroviruses to gain access to the germline (here the FE) would be essential for triggering the repression of retroviral replication.
Acknowledgments
We thank Nicole Prud'homme who initiated the study of the sterility phenotype of flamenco. We are grateful for Valérie Robert for obtaining the hsp-DIP1-b and hsp-DIP1-c transgenes. We are thankful to Geneviève Payen for performing the inverse PCR experiment. We thank Nicole Lautredou for excellent assistance in confocal microscopy. This work was supported by grants from the Association pour la Recherche sur le Cancer, from the European Union (Silencing in Different Organisms network), and from the Natural Science and Engineering Research council of Canada.
References
- Avedisov, S. N., and Y. V. Ilyin, 1994. Identification of spliced RNA species of Drosophila melanogaster gypsy retrotransposon. New evidence for retroviral nature of the gypsy element. FEBS Lett. 350: 147–150. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton, A., 1995. The relationship between the flamenco gene and gypsy in Drosophila: how to tame a retrovirus. Trends Genet. 11: 349–353. [DOI] [PubMed] [Google Scholar]
- Chalvet, F., L. Teysset, C. Terzian, N. Prud'homme, P. Santamaria et al., 1999. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 18: 2659–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva, M., and A. Ephrussi, 2004. Drosophila development: RNA interference ab ovo. Curr. Biol. 14: R428–R430. [DOI] [PubMed] [Google Scholar]
- Cummings, C. A., and C. Cronmiller, 1994. The daughterless gene functions together with Notch and Delta in the control of ovarian follicle development in Drosophila. Development 120: 381–394. [DOI] [PubMed] [Google Scholar]
- DeSousa, D., M. Mukhopadhyay, P. Pelka, X. Zhao, B. K. Dey et al., 2003. A novel double-stranded RNA-binding protein, disco interacting protein 1 (DIP1), contributes to cell fate decisions during Drosophila development. J. Biol. Chem. 278: 38040–38050. [DOI] [PubMed] [Google Scholar]
- Desset, S., C. Conte, P. Dimitri, V. Calco, B. Dastugue et al., 1999. Mobilization of two retroelements, ZAM and Idefix, in a novel unstable line of Drosophila melanogaster. Mol. Biol. Evol. 16: 54–66. [DOI] [PubMed] [Google Scholar]
- Desset, S., C. Meignin, B. Dastugue and C. Vaury, 2003. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 164: 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin, A. K., P. D. Miller, L. A. Getts, K. Jackson, G. Sonoda et al., 1995. Retroviral-like sequences specifically expressed in the rat ovary detect genetic differences between normal and transformed rat ovarian surface epithelial cells. Endocrinology 136: 4640–4649. [DOI] [PubMed] [Google Scholar]
- Goode, S., D. Wright and A. P. Mahowald, 1992. The neurogenic locus brainiac cooperates with the Drosophila EGF receptor to establish the ovarian follicle and to determine its dorsal-ventral polarity. Development 116: 177–192. [DOI] [PubMed] [Google Scholar]
- Johnson, E., S. Wayne and R. Nagoshi, 1995. fs (1) Yb is required for ovary follicle cell differentiation in Drosophila melanogaster and has genetic interactions with the Notch group of neurogenic genes. Genetics 140: 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, A., C. Terzian, P. Santamaria, A. Pelisson, N. Purd'homme et al., 1994. a Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 91: 1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, A. I., N. V. Lyubomirskaya, E. S. Belyaeva, N. G. Shostack and Y. V. Ilyin, 1994. b The introduction of a transpositionally active copy of retrotransposon GYPSY into the stable strain of Drosophila melanogaster causes genetic instability. Mol. Gen. Genet. 242: 472–477. [DOI] [PubMed] [Google Scholar]
- Klattenhoff, C., D. P. Bratu, N. McGinnis-Schultz, B. S. Koppetsch, H. A. Cook et al., 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis dpecification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12: 45–55. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody et al., 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lecher, P., A. Bucheton and A. Pélisson, 1997. Expression of the Drosophila retrovirus gypsy as ultrastructurally detectable particles in the ovaries of flies carrying a permissive flamenco allele. J. Gen. Virol. 78: 2379–2388. [DOI] [PubMed] [Google Scholar]
- Lock, L. F., E. Keshet, D. J. Gilbert, N. A. Jenkins and N. G. Copeland, 1988. Studies of the mechanism of spontaneous germline ecotropic provirus acquisition in mice. EMBO J. 7: 4169–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacsovich, T., Z. Asztalos, W. Awano, K. Baba, S. Kondo et al., 2001. Dual-tagging gene trap of novel genes in Drosophila melanogaster. Genetics 157: 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlor, R. L., S. M. Parkhurst and V. G. Corces, 1986. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol. Cell. Biol. 6: 1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mével-Ninio, M., M. C. Mariol and M. Gans, 1989. Mobilization of the gypsy and copia retrotransposons in Drosophila melanogaster induces reversion of the ovoD dominant female-sterile mutations: molecular analysis of revertant alleles. EMBO J. 8: 1549–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova, E., S. Nuzhdin, W. Li and A. J. Flavell, 1997. Germ line transposition of the copia retrotransposon in Drosophila melanogaster is restricted to males by tissue-specific control of copia RNA levels. Mol. Gen. Genet. 255: 115–124. [DOI] [PubMed] [Google Scholar]
- Pelisson, A., S. U. Song, N. Prud'homme, P. A. Smith, A. Bucheton et al., 1994. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 13: 4401–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson, A., L. Teysset, F. Chalvet, A. Kim, N. Prud'homme et al., 1997. About the origin of retroviruses and the co-evolution of the gypsy retrovirus with the Drosophila flamenco host gene. Genetica 100: 29–37. [PubMed] [Google Scholar]
- Pelisson, A., E. Sarot, G. Payen-Groschene and A. Bucheton, 2007. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary-sense gypsy transcripts in somatic cells of the Drosophila ovary. J. Virol. 81: 1951–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon, N., and M. Gans, 1983. Clonal analysis of the tissue specificity of recessive female-sterile mutations of Drosophila melanogaster using a dominant female-sterile mutation Fs(1)K1237. Dev. Biol. 100: 365–373. [DOI] [PubMed] [Google Scholar]
- Prud'homme, N., M. Gans, M. Masson, C. Terzian and A. Bucheton, 1995. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139: 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, V., N. Prud'homme, A. Kim, A. Bucheton and A. Pelisson, 2001. Characterization of the flamenco region of the Drosophila melanogaster genome. Genetics 158: 701–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth, P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman, R. R., E. A. Johnson, C. K. Rodesch, M. Bjerke, R. N. Nagoshi et al., 1995. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruohola, H., K. A. Bremer, D. Baker, J. R. Swedlow, L. Y. Jan et al., 1991. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell 66: 433–449. [DOI] [PubMed] [Google Scholar]
- Saito, K., K. M. Nishida, T. Mori, Y. Kawamura, K. Miyoshi et al., 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20: 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot, E., G. Payen-Groschene, A. Bucheton and A. Pelisson, 2004. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166: 1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin, R. K., M. Freeling and D. Lisch, 2005. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat. Genet. 37: 641–644. [DOI] [PubMed] [Google Scholar]
- Song, S. U., T. Gerasimova, M. Kurkulos, J. D. Boeke and V. G. Corces, 1994. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 8: 2046–2057. [DOI] [PubMed] [Google Scholar]
- Stroumbakis, N. D., Z. Li and P. P. Tolias, 1994. RNA- and single-stranded DNA-binding (SSB) proteins expressed during Drosophila melanogaster oogenesis: a homolog of bacterial and eukaryotic mitochondrial SSBs. Gene 143: 171–177. [DOI] [PubMed] [Google Scholar]
- Tulin, A., D. Stewart and A. C. Spradling, 2002. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16: 2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, V. V., A. Sigova, C. Li, H. Seitz, V. Gvozdev et al., 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. [DOI] [PubMed] [Google Scholar]