Abstract
Like other cell populations, undifferentiated human embryonic stem cells (hESCs) express a characteristic set of proteins and mRNA that is unique to the cells regardless of culture conditions, number of passages and methods of propagation. We have sought to identify a small set of markers that would serve as a reliable indicator of the balance of undifferentiated and differentiated cells in hESC populations. Markers of undifferentiated cells should be rapidly down-regulated as the cells differentiate to form embryoid bodies (EBs), while markers that are absent or low during the undifferentiated state but are induced as hESCs differentiate could be used to assess the presence of differentiated cells in the cultures. In this manuscript we describe a list of markers that reliably distinguish undifferentiated and differentiated cells. An initial list of approximately 150 genes was generated by scanning published MPSS, EST scan and microarray datasets. From this list, a subset of 109 genes was selected that included 55 candidate markers of undifferentiated cells, 46 markers of hESC derivatives, 4 germ cell markers and 4 trophoblast markers. Expression of these candidate marker genes was analyzed in undifferentiated hESCs and differentiating EB populations in four different lines by immunocytochemistry, RT-PCR, microarray analysis and quantitative RT-PCR (qPCR). We show that qPCR with as few as 12 selected genes can reliably distinguish differentiated cells from undifferentiated hESC populations.
Keywords: Human embryonic stem cell, Embryoid body, Differentiation
Introduction
Currently, more than 100 distinct human embryonic stem cell (hESC) lines have been derived and efforts at new derivations are ongoing. Approximately 20 lines from the 78 derivations undertaken before August 9, 2001 are available in sufficient quantities for general research use (NIH stem cell registry, http://stemcells.nih.gov/research/registry). Of these, only a small subset of lines is available for detailed characterization [1-8]. As expected, various hESC lines have a number of similarities. For example, undifferentiated hESCs are similar in expressing surface antigens and markers characteristic of the undifferentiated ESC state including Oct4 (POU5F1), Nanog, UTF1, DPPA5, TERT, gap junction proteins, SSEA and TRA antigens [1-8]. hESCs are also similar in their ability to proliferate and differentiate into cell types of the three germ layers in vitro and in vivo [9-16]. Properties of hESCs have also been compared using microarray, EST scan, SAGE and MPSS [4; 17-27]. These studies suggest that it is likely that markers shared by hESC lines but absent in other cell populations exist.
Although these studies have highlighted similarities among hESC lines and markers that distinguish them from mouse ESCs, it is likely that differences also exist. These include potential differences in methylation patterns [28; 29], likely HLA differences [25], allelic differences, variability of X-inactivation and adaptation of cells to different culture conditions [17, 2; 30; 31]. Indeed, important differences between hESC lines in growth rates, methods of propagation and karyotype have been reported using a variety of different techniques, suggesting that while shared markers may exist, care will be needed to identify them. Identifying such shared markers, however, remains an easier task than the technically challenging experiments of direct comparing hESCs under identical culture conditions-experiments that are being undertaken at the Stem Cell Center at the NIH (Dr. McKay) and at the International Stem Cell Initiative (Dr. Andrews) to identify fundamental differences among cell lines. Such experiments are beyond the scope of our laboratories. The available data, however, indicate that identifying a common pattern of gene expression that is conserved independent of culture conditions and reflects the fundamental properties of hESC is possible.
Several other experiments suggest that a unique molecular signature can be defined to distinguish undifferentiated hESCs from their differentiated progeny, and that this signature can be used to define the states of hESCs [17-19]. These experiments used MPSS, EST scan, and microarray data to suggest that a large pool of potential markers that could distinguish embryoid bodies (EBs) and other differentiated cells from hESCs exist. We have reasoned, therefore, that at the current level of resolution of techniques, it is possible to identify a core set of genes that are conserved and/or required to maintain hESC identity. These genes should be expressed irrespective of the conditions of culture, numbers of passages and methods of propagation as long as undifferentiated hESCs are present. Moreover, these core markers should be present regardless of the methods of hESC derivation and ethnic phenotypes of the blastocysts used. If present at lower levels, they should be detectable by RT-PCR as well as by SAGE/MPSS, and if robust by SAGE and microarray. Furthermore, if the expression of such genes is examined in EBs, then a subset of markers that are rapidly downregulated or rapidly induced as cells differentiate can be identified [19]. A combination of such markers can serve to reliably assess the states of ESCs and EBs.
To test this hypothesis we have performed a meta-analysis of published reports examining hESCs and EBs, and identified a list of potential markers. We tested a substantial number of these markers by quantitative RT-PCR (qPCR) and immunocytochemistry and identified a combination of markers to distinguish hESCs from EBs.
Materials and Methods
hESC culture and in vitro differentiation via EB
hESC lines BG01, BG02, BG03 and I6 were either maintained on inactivated mouse embryonic fibroblast (MEF) feeder cells in medium comprised of Dulbecco’s Modified Eagle’s Medium/Ham’s F12 supplemented with 20% knockout serum replacement, 2 mM non-essential amino acids, 2 mM L-glutamine, 50 μg/ml Penn-Strep (all from Invitrogen; Carlsbad, CA; http://www.invitrogen.com), 0.1 mM β-mercaptoethanol (Specialty Media; Phillipsburg, NJ; http://www.specialtymedia.com), and 4 ng/ml of basic fibroblast growth factor (bFGF, Sigma; St. Louis, MO; http://www.sigmaaldrich.com), or on fibronectin (BD Biosciences, Bedford, MA;http://www.bdbiosciences.com) coated dishes in medium conditioned with MEF for 24 hours as previously described [4; 25].
Differentiation via EB formation was performed as preciously described [4]. Briefly, hESCs were dissociated into small clumps by collagenase IV (Sigma) and grown as floating spheres in hESC medium without bFGF for up to 2 weeks, with a medium change every second day.
RT-PCR and qPCR analysis
Total RNA was extracted from undifferentiated hESCs or EBs (7-day and 14-day) using RNA STAT-60 (Tel-Test Inc., Friendswood, TX). cDNA was synthesized by using a reverse transcription kit (RETROscript, Ambion, Austin, TX) according to the manufacturer’s recommendations. The PCR primers are listed in Supplementary Table 1.
Real-time qPCR was used to quantify the levels of mRNA expression of 12 selected genes (Oct4, Nanog, Sox2, UTF1, DPPA5, Lin41, Sox1, DCN, H19, IGF2, GATA4 and Hand1) in hESCs or EBs at different times of differentiation. PCR reactions were carried out by DNA Engine Opticon Fluorescence Detection System (MJ Research, Waltham, MA) using a DyNAmo SYBR Green qPCR kit according to the manufacturer’s instructions. The content of selected genes was normalized to the content of 18S-rRNA and standard curves were generated using 10 to 1000 ng cDNA per 20 μl reaction volume. All PCR products were checked by melting curve analysis to exclude the possibility of multiple products or incorrect product size. PCR analyses were conducted in triplicate for each sample.
Immunocytochemistry
Immunocytochemistry and staining procedures were as described previously [32]. Briefly, hESCs grown either on MEF feeder cells or under feeder-free conditions, 7-day and 14-day EBs either attached or floating were fixed with 2% paraformaldehyde for half an hour. Parts of EBs were embedded in O.C.T. Blocks were cut on a cryostat to obtain 8 μm sections. Fixed cells and sections were blocked in blocking buffer (5% goat serum, 1% BSA, 0.1% Triton X-100) for 1 hour followed by incubation with the primary antibody at 4°C overnight. Appropriately coupled secondary antibodies (Molecular Probes) were used for single and double labeling. All secondary antibodies were tested for cross reactivity and non-specific immunoreactivity.
The following antibodies were used: Nanog (1:1000, R&D Systems AF1997), ITGB1 (CD29/Integrin β1, 1:1000, Chemicon MAB1951), CDH1 (E-Cadherin, 1:2500, R&D Systems MAB1838), PODXL (Podocalyxin, 1:500, R&D Systems MAB1658), Sox2 (1:1000, R&D Systems MAB2018), Oct4 (1:1000, R&D Systems AF1759), Brachyury (1:1000, R&D Systems AF2085), ACTC (Cardiac Actin, 1:50, Research Diagnostics, Inc., PRO61075), GATA4 (1:100, Santa Cruz sc-25310), GATA6 (1:100, Santa Cruz sc-9055), AFP (alpha-fetoprotein, 1:500, Sigma A8452) and TUBB3 (βIII tubulin 1:2000, Sigma T8660). Bis-benzamide (Dapi, 1:1000, Sigma) was used to identify the nuclei. Images were captured on an Olympus fluorescence microscope.
Microarray analysis using BeadArray platform
RNAs from undifferentiated BG01, BG02 and BG03 cells and the matched 14-day EBs were hybridized to prototype Illumina RefSeq-8 BeadChips, which contained more than 24,000 genes [33]. A detailed description of the BeadChip system has been provided elsewhere [33]. Samples were coded and run in duplicates and the results were analyzed using standard bioinformatics tools and the Bead Studio, a tool kit developed by Illumina. A detailed analysis of these and other samples will be reported elsewhere [Jeanne Loring, Burnham Institute, personal communication].
Results
Meta-analysis procedure
A large dataset on gene expression of undifferentiated hESCs and EBs that differentiated from them using large scale array experiments including MPSS, EST enumeration and microarrays has been generated and described by a variety of investigators [17-24]. To generate a list of genes characteristic of undifferentiated hESCs and differentiated EBs, we examined three published reports on gene expression in hESCs and EBs. These were: 1) a list of ninety-two genes identified as “stemness” genes which are expressed at high levels in six hESC lines as assessed by a 16,695 seventy base pair oligonucleotide microarray [18], 2) a comprehensive list of genes common to undifferentiated hESCs obtained by MPSS analysis using pooled hESCs samples [17], and 3) a large list of genes that are highly expressed in differentiated EBs detected by both MPSS and EST scan [19] (A complete list of EST scan data are obtained from http://www.ncbi.nlm.nih.gov/UniGene/library.cgi?all=yes&ORG=Hs&LID=14183 http://www.ncbi.nlm.nih.gov/UniGene/library.cgi?all=yes&ORG=Hs&LID=14184).
The criteria for selection were that the levels of expression should differ between hESCs and EBs by at least three fold by array data or by five fold for EST scanning and MPSS, the levels of expression should be high (>10 tpm for MPSS, >5 by EST scan) and that the genes must be detected by at least one independent method and in more than one hESC line. One exception to these selection criteria is that we included some genes that were not detected by MPSS or EST scanning but are used as ESC marker by convention, and a few genes that were highly expressed in both hESCs and EBs or expressed at higher level in EBs but were reported as ESC specific genes by microarray analysis when compared to human universal RNA [18] (Table 1), or because of being a potential cell surface marker. A list of several hundred genes generated after this selection was pruned to a list of about one hundred by examining published literature with the goal of including known genes with approximately half of them being candidate genes highly expressed in undifferentiated hESCs (but not in EBs or down-regulated in EBs), and the other half being those that are expressed in EBs (but lower or absent in hESCs). We did not include any unknown genes even though many of them were significantly differentially expressed in hESCs and EBs [19].
Table 1.
hESC specific markers
| Gene symbol | Gene ID (Locus ID) | UniGene | Description | Tpm (hESC) | Tpm (hEB) | Micro-array (hESC) | Micro-array (hEB) | EST scan (hESC) | EST scan (hEB) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Stem cell marker | ||||||||||
| LIN28 | 79727 | Hs.86154 | lin-28 homolog (C. elegans) | 1962 | 178 | 3944.1 | 1421.6 | 25 | 13 | 24 |
| DNMT3B | 1789 | Hs.251673 | DNA (cytosine-5-)-methyltransferase 3 beta | 1274 | 56 | 3391 | 274.5 | 58 | 3 | 24 |
| CLDN6 | 9074 | Hs. 533779 | claudin 6 | 1063 | 151 | 2247.2 | 581.1 | 14 | 0 | 24 |
| IFITM1 | 8519 | Hs.458414 | interferon induced transmembrane protein 1 (9-27) | 920 | 221 | 8844 | 2188.5 | 4 | 1 | 4 |
| POU5F1 | 5460 | Hs.249184 | POU domain, class 5, transcription factor 1 | 658 | 20 | 694.4 | 28.9 | 23 | 1 | 41 |
| ITGB1 | 3688 | Hs.429052 | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | 545 | 2246 | 3726.7 | 8211.2 | 16 | 70 | |
| SFRP2 | 6423 | Hs.481022 | secreted frizzled-related protein 2 | 505 | 31 | 1353.9 | 93.1 | 7 | 1 | 4 |
| GJA1 | 2697 | Hs.74471 | gap junction protein, alpha 1, 43kDa (connexin 43) | 452 | 406 | 2404.4 | 2926.1 | 27 | 24 | 8 |
| SOX2 | 6657 | Hs.518438 | SRY (sex determining region Y)-box 2 | 344 | 11 | N.D. | N.D. | 11 | 1 | 42 |
| CD9 | 928 | Hs.114286 | CD9 antigen (p24) | 221 | 13 | 2930.8 | 1267.2 | 1 | 0 | 43 |
| GAL | 51083 | Hs.278959 | galanin | 221 | 28 | 1043.3 | 97.5 | N.D. | N.D. | 44 |
| LIN41 | Non | Hs.517884 | LOC131405: similar to abnormal cell LINeage LIN-41, heterochronic gene; Drosophila dappled/ vertebrate TRipartite Motif protein related; B-box zinc finger, Filamin and NHL repeat containing protein (123.8 kD) (lin-41) | 165 | 41 | N.D. | N.D. | 3 | 2 | 45 |
| IMP-2 | 10644 | Hs.35354 | IGF-II mRNA-binding protein 2 | 140 | 133 | 1206 | 1163.2 | 5 | 8 | 4 |
| LECT1 | 11061 | Hs.421391 | leukocyte cell derived chemotaxin 1 | 116 | 3 | N.D. | N.D. | 4 | 0 | 4 |
| ZNF206 | 84891 | Hs.334515 | Zinc finger protein 206 | 109 | 0 | 1504.5 | 71.3 | 26 | 0 | |
| GABRB3 | 2562 | Hs.302352 | gamma-aminobutyric acid (GABA) A receptor, beta 3 | 96 | 7 | 482.7 | 28 | 5 | 1 | |
| GYLTL1B | 120071 | Hs.86543 | glycosyltransferase-like 1B | 92 | 0 | 1066.6 | 69.3 | 4 | 1 | |
| UTF1 | 8433 | Hs.458406 | undifferentiated embryonic cell transcription factor 1 | 90 | 0 | 1444.1 | 81.3 | N.D. | N.D. | 46 |
| NR6A1 | 2649 | Hs.20131 | nuclear receptor subfamily 6, group A, member 1 | 79 | 17 | N.D. | N.D. | N.D. | N.D. | 47 |
| LEFTY1 | 10637 | Hs.278239 | left-right determination, factor 1 | 72 | 0 | 6579.6 | 73.1 | 4 | 0 | |
| SCGB3A2 | 117156 | Hs.483765 | Secretoglobin, family 3A, member 2 | 65 | 0 | 890.5 | 12.2 | 1 | 0 | |
| KIT | 3815 | Hs.479754 | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene | 46 | 5 | 188.9 | 42.2 | 1 | 0 | |
| CKMT1 | 1159 | Hs.425633 | creatine kinase, mitochondrial 1 (ubiquitous) | 42 | 0 | 742.4 | 46.2 | 1 | 0 | |
| TDGF1 | 6997 | Hs.385870 | teratocarcinoma-derived growth factor 1 | 38 | 0 | 3169 | 200.2 | 16 | 1 | 18 |
| FOXD3 | 27022 | Hs.546573 | forkhead box D3 | 37 | 0 | N.D. | N.D. | N.D. | N.D. | 48 |
| DIAPH2 | 1730 | Hs.226483 | diaphanous homolog 2 (Drosophila) | 27 | 4 | 1971.1 | 261.4 | N.D. | N.D. | |
| NUMB | 8650 | Hs.509909 | numb homolog (Drosophila) | 24 | 1 | 618.3 | 778.9 | 0 | 4 | |
| CER1 | 9350 | Hs.248204 | cerberus 1 homolog, cysteine knot superfamily (Xenopus laevis) | 23 | 0 | 151.5 | 23.4 | N.D. | N.D. | |
| PMAIP1 | 5366 | Hs.96 | Phorbol-12-myristate-13-acetate-induced protein 1 | 23 | 1 | 1601.5 | 336.6 | 4 | 0 | |
| CDH1 | 999 | Hs.461086 | cadherin 1, type 1, E-cadherin (epithelial) | 20 | 0 | 1972.9 | 793.1 | 11 | 0 | |
| LEFTY2 | 7044 | Hs.520187 | Left-right determination factor 2 (LEFTY2) | 16 | 4 | 944.1 | 100.1 | 1 | 0 | |
| NANOG | 79923 | Hs.329296 | Nanog homeobox | 16 | 0 | 56.9 | 10.7 | 1 | 0 | 36, 49 |
| SMAD2 | 4087 | Hs.12253, Hs.465061 | SMAD, mothers against DPP homolog 2 (Drosophila) | 13 | 0 | 332.8 | 397.4 | 1 | 1 | |
| BRIX | 55299 | Hs.38114 | BRIX | 9 | 0 | 1833.6 | 1261.5 | 3 | 3 | 4 |
| REST | 5978 | Hs.401145 | RE1-silencing transcription factor | 6 | 0 | 18.5 | 15.4 | 1 | 1 | |
| ZFP42 (Rex1) | 132625 | Hs.335787 | zinc finger protein 42 | 5 | 0 | 159.3 | 34.5 | 2 | 0 | 50 |
| EDNRB | 1910 | Hs.82002 | endothelin receptor type B | 0 | 0 | 346.8 | 119.8 | 0 | 1 | |
| PTEN | 5728 | Hs.500466 | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 0 | 0 | 138.4 | 125.6 | 1 | 0 | |
| NOG | 9241 | Hs.248201 | noggin | 0 | 0 | N.D. | N.D. | N.D. | N.D. | |
| GDF3 | 9573 | Hs.86232 | growth differentiation factor 3 | N.D. | N.D. | 236.5 | 27.2 | N.D. | N.D. | 24 |
| GBX2 | 2637 | Hs.184945 | gastrulation brain homeo box 2 | N.D. | N.D. | 44.5 | 14.7 | N.D. | N.D. | |
| TFCP2L1 | 29842 | Hs.156471 | transcription factor CP2-like 1 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | |
| COMMD3 | 23412 | Hs.534398 | COMM domain containing 3 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | |
| TERT | 7015 | Hs.492203 | telomerase reverse transcriptase | N.D. | N.D. | N.D. | N.D. | 1 | 0 | |
| PODXL | 5420 | Hs.16426 | podocalyxin-like | N.D. | N.D. | 498.2 | 394.2 | 60 | 23 | 4 |
| FGF4 | 2249 | Hs.1755 | fibroblast growth factor 4 (heparin secretory transforming protein 1, Kaposi sarcoma oncogene) | N.D. | N.D. | N.D. | N.D. | 1 | 0 | 51 |
| NR5A2 | 2494 | Hs.33446 | nuclear receptor subfamily 5, group A, member 2 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | |
| IFITM2 | 10581 | Hs.174195 | interferon induced transmembrane protein 2 (1-8D) | N.D. | N.D. | 129.2 | 204 | 2 | 0 | |
| GRB7 | 2886 | Hs.86859 | growth factor receptor-bound protein 7 | N.D. | N.D. | 105.4 | 11.6 | 2 | 0 | |
| DPPA5 | 359942 | Hs.125331 | developmental pluripotency associated 5 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 52 |
| NODAL | 4838 | Hs.370414 | nodal homolog (mouse) | N.D. | N.D. | N.D. | N.D. | 1 | 0 | |
| LCK | 3932 | Hs.470627 | Lymphocyte-specific protein tyrosine kinase | N.D. | N.D. | 500 | 10.7 | N.D. | N.D. | |
| NTS | 4922 | Hs.80962 | Neurotensin | N.D. | N.D. | 1313 | 137.7 | N.D. | N.D. | |
| ITGB1BP3 | 9270 | Hs.135458 | Integrin beta 1 binding protein 3 | N.D. | N.D. | N.D. | N.D. | 2 | 0 | |
| CHST4 | 10164 | Hs.251383 | Carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 | N.D. | N.D. | N.D. | N.D. | 2 | 0 |
The final list of 109 potential markers included 55 stem cell markers in which most of them were highly expressed in undifferentiated hESCs. For potential markers of the EB state, 46 markers were selected including representative genes from each germ layer with 12 ectoderm, 15 mesoderm and 19 endoderm markers that are likely over-expressed in EBs. In addition, 4 trophoblast markers and 4 germ cell markers were included (Tables 1 and 2, Supplementary Table 1).
Table 2.
EB specific markers
| Gene symbol | Gene ID (Locus ID) | UniGene | Description | Tpm (hESCs) | Tpm (hEBs) | Micro-array (hEB) | Micro-array (hEB) | EST scan (hESC) | EST scan (hEB) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Ectoderm marker | ||||||||||
| VIM | 7431 | Hs.533317 | vimentin | 626 | 9881 | 6262.9 | 16146.1 | 38 | 114 | 53 |
| CRABP2 | 1382 | Hs.405662 | cellular retinoic acid binding protein 2 | 26 | 396 | 1549.9 | 3187.6 | 9 | 5 | 54 |
| SEMA3A | 10371 | Hs.252451 | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | 1 | 16 | 214.1 | 180.2 | 1 | 1 | 55 |
| MSI1 | 4440 | Hs.158311 | musashi homolog 1 (Drosophila) | 0 | 11 | 1968 | 787.6 | 5 | 1 | 56 |
| MAP2 | 4133 | Hs.368281 | microtubule-associated protein 2 | 0 | 10 | N.D. | N.D. | 1 | 1 | 57 |
| GFAP | 2670 | Hs.514227 | glial fibrillary acidic protein | 0 | 0 | 34.7 | 15 | N.D. | N.D. | 58 |
| OLIG2 | 10215 | Hs.176977 | oligodendrocyte lineage transcription factor 2 | 0 | 0 | 13.4 | 1.7 | N.D. | N.D. | 59 |
| SOX1 | 6656 | Hs.202526 | SRY (sex determining region Y)-box 1 | N.D. | N.D. | 21.2 | 10.1 | N.D. | N.D. | 60 |
| NES | 10763 | Hs.527971 | nestin | 386. | 50. | 2935.8 | 2471.3 | 4 | 6 | 61 |
| NEUROD1 | 4760 | Hs.72981 | neurogenic differentiation 1 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 62 |
| TH | 7054 | Hs.435609 | tyrosine hydroxylase | N.D. | N.D. | 158.4 | 107.8 | N.D. | N.D. | 63 |
| TUBB3 | 10381 | Hs.511743 | tubulin, beta 3 | N.D. | N.D. | N.D. | N.D. | 2 | 8 | 64 |
| Endoderm marker | ||||||||||
| FN1 | 2335 | Hs.203717 | fibronectin 1 | 3661 | 21250 | 3243.5 | 12913.2 | 34 | 351 | 65 |
| DCN | 1634 | Hs.156316 | decorin | 2 | 2008 | 95.6 | 1550.2 | 0 | 18 | 66 |
| H19 | 283120 | Hs.551588 | H19, imprinted maternally expressed untranslated mRNA | 64 | 2176 | N.D. | N.D. | 2 | 136 | |
| AFP | 174 | Hs.518808 | alpha-fetoprotein | 30 | 1829 | 1089.1 | 9765 | 0 | 5 | 67 |
| LAMB1 | 3912 | Hs.489646 | laminin, beta 1 | 186 | 614 | 126.7 | 504.2 | 15 | 19 | 68 |
| LAMC1 | 3915 | Hs.497039 | laminin, gamma 1 (formerly LAMB2) | 203 | 352 | 2098.4 | 4340.9 | 14 | 13 | 69 |
| BMP2 | 650 | Hs.73853 | bone morphogenetic protein 2 | 35 | 69 | 63.1 | 68.4 | 1 | 1 | 70 |
| SERPINA1 | 5265 | Hs.525557 | serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | 0 | 61 | 136.3 | 1045.2 | 1 | 1 | 71 |
| FLT1 | 2321 | Hs.507621 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 38 | 46 | N.D. | N.D. | 2 | 0 | 72 |
| ACVR1B | 91 | Hs.438918 | activin A receptor, type IB | 22 | 42 | 237.8 | 187.2 | 4 | 3 | 73 |
| GATA4 | 2626 | Hs.243987 | GATA binding protein 4 | 2 | 13 | 83.2 | 145.2 | 3 | 4 | 74 |
| GCG | 2641 | Hs.516494 | glucagon | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 75 |
| INS | 3630 | Hs.89832 | insulin | N.D. | N.D. | 122.7 | 70.4 | N.D. | N.D. | 76 |
| PECAM1 | 5175 | Hs.514412 | platelet/endothelial cell adhesion molecule (CD31 antigen) | N.D. | N.D. | 40 | 49.1 | N.D. | N.D. | 77 |
| FABP2 | 2169 | Hs.282265 | fatty acid binding protein 2, intestinal | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 78 |
| HNF4A | 3172 | Hs.116462 | hepatocyte nuclear factor 4, alpha | N.D. | N.D. | 16 | 10 | N.D. | N.D. | 79 |
| FGF8 | 2253 | Hs.57710 | fibroblast growth factor 8 (androgen-induced) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 80 |
| HGF | 3082 | Hs.396530 | Hepatocyte growth factor | N.D. | N.D. | 32.2 | 39.4 | 0 | 1 | 81 |
| FOXA2 | 3170 | Hs.155651 | Forkhead box A2 | N.D. | N.D. | 94.2 | 83.2 | 2 | 0 | 82 |
| Mesoderm marker | ||||||||||
| COL1A1 | 1277 | Hs.172928 | collagen, type I, alpha 1 | 3889 | 68430 | 208.7 | 2424.2 | 67 | 658 | 83 |
| HAND1 | 9421 | Hs.152531 | heart and neural crest derivatives expressed 1 | 0 | 500 | 389.8 | 9466.3 | 0 | 8 | 84 |
| MSX1 | 4487 | Hs.424414 | msh homeo box homolog 1 (Drosophila) | 15 | 218 | 467.3 | 2198.1 | 1 | 3 | 85 |
| ACTC | 70 | Hs.118127 | actin, alpha, cardiac muscle | 112 | 141 | 952.2 | 2963.6 | 1 | 4 | 86 |
| GATA6 | 2627 | Hs.514746 | GATA binding protein 6 | 4 | 53 | 206.7 | 964.2 | N.D. | N.D. | 87 |
| COL2A1 | 1280 | Hs.408182 | collagen, type II, alpha 1 (primary osteoarthritis, spondyloepiphyseal dysplasia, congenital) | 70 | 49 | 144.5 | 632.6 | 1 | 19 | 88 |
| HBZ | 3050 | Hs.449632 | hemoglobin, zeta | 0 | 34 | 30 | 408.7 | N.D. | N.D. | 89 |
| T | 6862 | Hs.389457 | T, brachyury homolog (mouse) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 90 |
| WT1 | 7490 | Hs.408453 | Wilms tumor 1 | N.D. | N.D. | 40.6 | 0.9 | N.D. | N.D. | 91 |
| MYF5 | 4617 | Hs.178023 | myogenic factor 5 | N.D. | N.D. | 20.2 | 12.1 | N.D. | N.D. | 92 |
| DES | 1674 | Hs.471419 | desmin | N.D. | N.D. | 6.6 | 4.5 | N.D. | N.D. | 93 |
| NPPA | 4878 | Hs.75640 | natriuretic peptide precursor A | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 94 |
| HBB | 3043 | Hs.523443 | hemoglobin, beta | N.D. | N.D. | 33.1 | 46 | N.D. | N.D. | 95 |
| RUNX2 | 860 | Hs.535845 | runt-related transcription factor 2 | N.D. | N.D. | 21.9 | 5.3 | 0 | 3 | 96 |
| IGF2 | 3481 | Hs.523414 | insulin-like growth factor 2 (somatomedin A) | N.D. | N.D. | 115.8 | 5369.7 | 1 | 18 | 97 |
| Trophoblast | ||||||||||
| EOMES | 8320 | Hs.147279 | eomesodermin homolog (Xenopus laevis) | 10 | 0 | 247.2 | 63.4 | N.D. | N.D. | 98 |
| CDX2 | 1045 | Hs.174249 | caudal type homeo box transcription factor 2 | 0 | 5 | 13.4 | 343.9 | 0 | 1 | 99 |
| GCM1 | 8521 | Hs.28346 | glial cells missing homolog 1 (Drosophila) | N.D. | N.D. | 16.5 | 9.4 | N.D. | N.D. | 100 |
| KRT1 | 3848 | Hs.80828 | keratin 1 (epidermolytic hyperkeratosis) | N.D. | N.D. | 13.4 | 17.1 | N.D. | N.D. | 101 |
| Germ Cells | ||||||||||
| SYCP3 | 50511 | Hs.506504 | synaptonemal complex protein 3 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 102 |
| DDX4 | 54514 | Hs.223581 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | N.D. | N.D. | 10 | 5.5 | N.D. | N.D. | 103 |
| IFITM1 | 8519 | Hs.458414 | Fragilis-1 | N.D. | N.D. | 8844 | 2188.5 | 4 | 1 | 104 |
| IFITM2 | 10581 | Hs.174195 | Fragilis-2 | N.D. | N.D. | 129.2 | 204 | 2 | 0 | 105 |
Markers of the ESC state
Forty-four genes highly expressed in undifferentiated hESCs but down-regulated in EBs were selected as markers of stem cells to represent the undifferentiated ESC state (Table 1). Several of these genes such as Oct4, Nanog and TERT are generally accepted markers of pluripotency. Many genes in the list including UTF1, Sox2, Lin28, Lin 41, PODXL, LeftyB, GJA1, FoxD3 and Rex1 (ZFP42) have recently been reported to be expressed in undifferentiated hESCs by several research groups [4; 17-19; 34]. Other undifferentiated markers included genes that encoded transcriptional factors, growth factors, signal transducers, cell surface antigens and receptors. In addition, 11 genes were included as ESC markers although they did not meet our selection criteria. Of them, eight genes, NOG, TFCP2L1, CommD3, TERT, NR5A2, DPPA5, NODAL, ITGB1BP3, were selected by convention. Two genes, GJA1 and IMP2, were selected because they expressed at significant higher levels in hESCs when compared to human universal RNA by microarray analysis, although their expression were equally high in EBs. Finally, despite higher level expression in EBs, ITGB1 was included because it is a cell surface receptor which may bind fibronectin that has been reported to be a substrate capable of supporting hESC growth [35; 36].
Markers of the EB state
To represent the complexity of EBs, we included as many types of early markers of differentiation as possible, and selected the following: 1) 12 ectoderm markers including markers for neural precursors such as nestin and Sox1, and for terminal differentiated neural cells like Tuj1 (TUBB3), TH and GFAP, 2) 19 endoderm markers including pancreatic marker insulin, imprinted maternally expressed gene H19, HNF and AFP, 3) 15 mesoderm markers including collagen, Brachyury and ACTC, 4) Four trophoblast markers, KRT1, EOMES, GCM1 and CDX2, and 5) Four germ cell markers SYCP3, DDX4, IFITM1, IFITM2 (Table 2).
Expression of candidate markers by RT-PCR
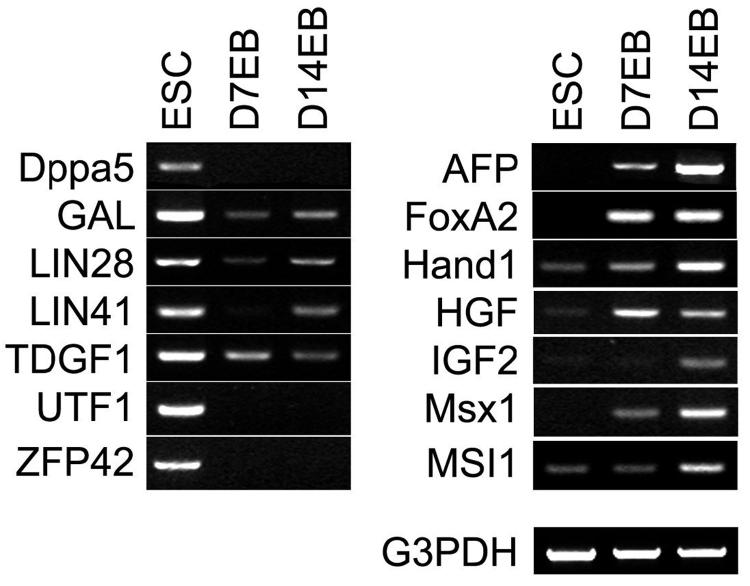
Our selection criteria indicated that candidate markers will be expressed in the appropriate stage of development and should be detected by RT-PCR. We therefore generated a PCR primer list for all 109 genes (Supplementary Table1) and tested 35 genes (Fig. 1 and data not shown, highlighted in Supplementary Table 1) using three different cell lines. Representative results for a subset of markers for the I6 line are shown. Of the 7 ESC markers shown, all were expressed in undifferentiated hESCs. A subset (Dppa5, UTF1 and ZFP42) were undetectable in 14-day EBs while the others were down-regulated (GAL, Lin28, Lin41 and TDGF1) in EBs (Fig. 1, left panel). Likewise, the 7 EB specific markers (AFP, FoxA2, Hand1, HGF, IGF2, Msx1 and MSI1) were expressed in EBs but absent (AFP, HNF3b, IGF2 and Msx1) in undifferentiated hESCs or only slightly expressed (Hand1, HGF and MSI1) in undifferentiated hESCs (Fig. 1, right panel). These results showed the relatively specific pattern of expression of candidate ESC and EB markers and indicate the suitability of using some or all of these to assess the overall state of cultured cells.
Fig 1.
Differential expression of ESC or EB specific genes by RT-PCR. Expression of selected markers of ESC (Dppa5, GAL, LIN28, LIN41, TDGF1, UTF1, and ZFP42) and EB (AFP, FoxA2, HAND1, HGF, IGF2, Msx1, MSI1) was examined by RT-PCR in undifferentiated hESCs, 7-day and 14-day EBs. Consistent with other independent analyses, all the ESC markers are highly expressed in hESCs but quickly down regulated in the two EB populations, whereas all the EB markers are detected in EB samples, but not or slightly expressed in undifferentiated hESCs.
Antibodies to test the expression of candidate markers
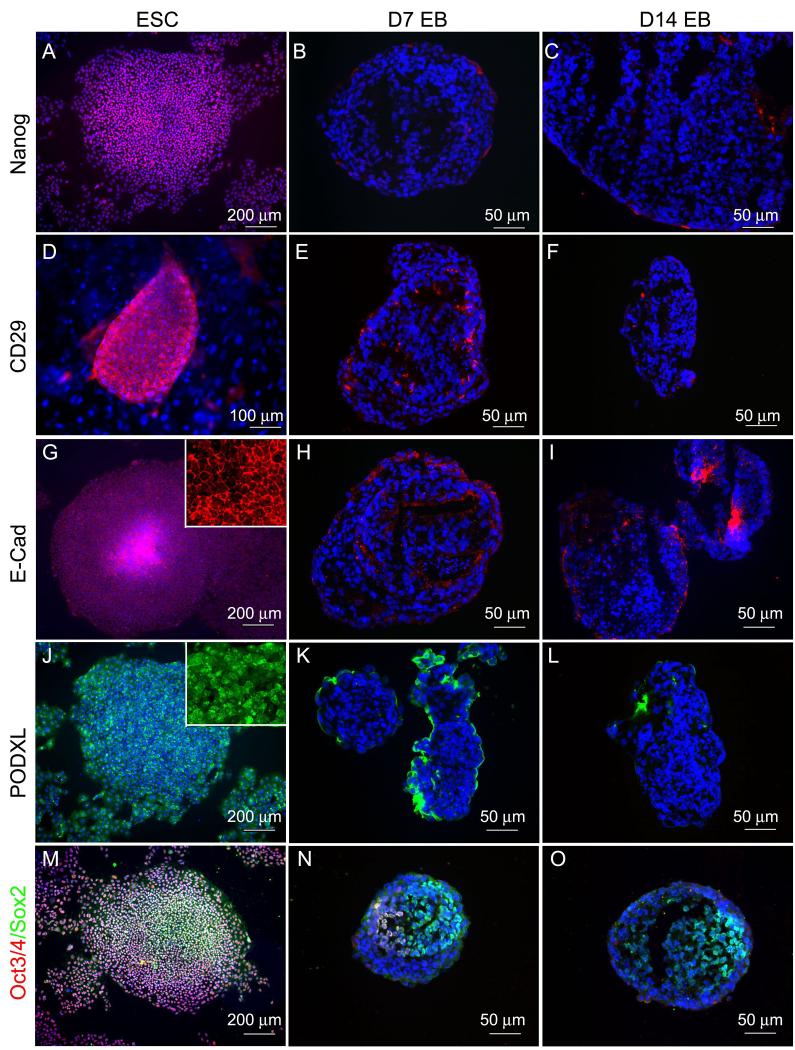
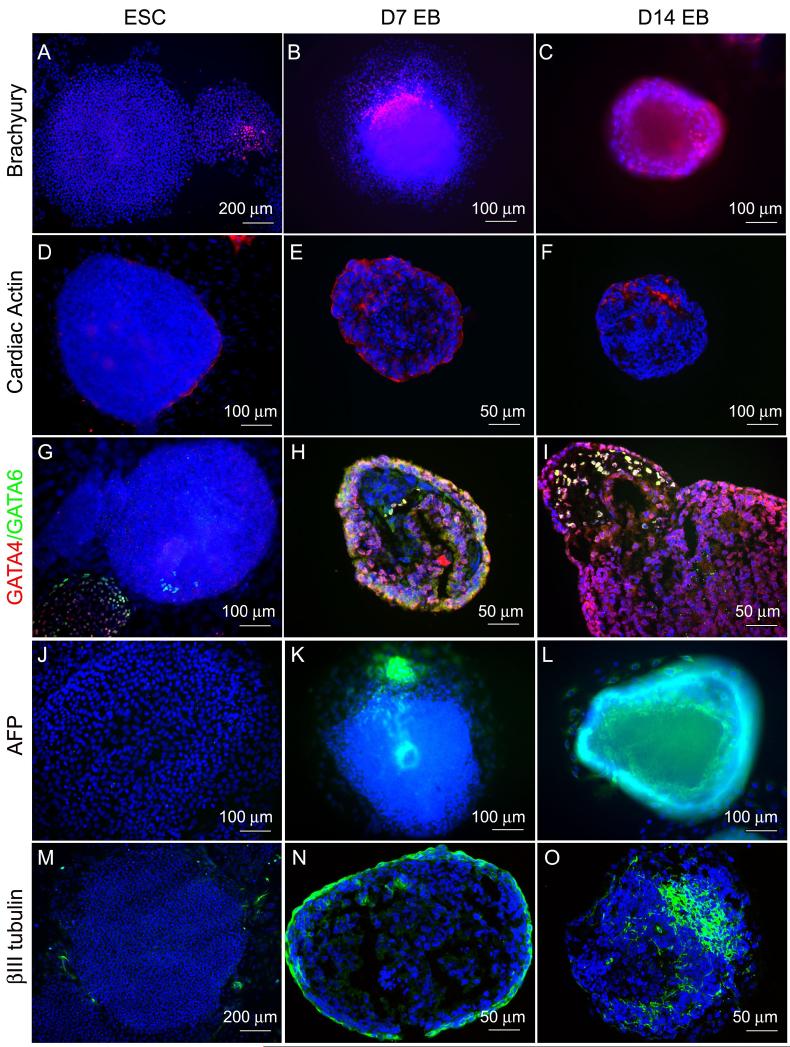
Of the 109 genes, we were able to locate commercially available antibodies for about two thirds (76) with reactivity against human antigens and these antibodies and their source are provided in Supplementary Table 1. We tested a subset of these commercial available antibodies by immunocytochemistry and at least 15 of them worked well (highlighted in Supplementary Table 1). Representative staining of ESC or EB specific genes in 7-day and 14-day differentiated EBs and undifferentiated hESCs are shown in Figs. 2 and 3. In general, all the ESC markers (Nanog, Oct4, ITGB1, CDH1 and PODXL) were strongly positive in undifferentiated hESCs but weakly expressed in 7-day EBs, and not expressed in 14-day EBs (Fig.2, panel A-L). The one exception was Sox2, which was expressed in both undifferentiated hESCs and the two stages of EBs (Fig. 2, panel M-O). All the markers of differentiation examined (Brachyury, ACTC, AFP, GATA4, GATA6, TUBB3) were strongly expressed in both 7-day and 14-day EBs but negative or only weakly expressed in undifferentiated hESCs (Fig. 3). In addition to these markers, known pluripotency markers SSEA (SSEA3 and 4) and TRA (TRA-1-60, 1-81) were down-regulated in differentiated EBs and Table 3 summarizes the results of immunocytochemistry. Thus antibodies to most of the candidate markers exist and a significant subset can be used for immunocytochemistry. We notice, however, that ITGB1 was strongly positive stained in undifferentiated hESC but down-regulated in EBs which is in conflict with the MPSS and array data. This suggests that not all genes could be used in all methodologies.
Fig 2.
Staining of ESC markers on undifferentiated hESCs, 7-day and 14-day EBs. ESC markers Nanog, ITGB1, CDH1, PODXL, Oct4, and Sox2 are expressed by most of undifferentiated hESCs (A, D, G, J, M), while their expression were downregulated in both 7-day and 14-day EBs (B-C, E-F, H-I, K-L, red in N-O) except Sox2 (green in N-O), which is also a neural stem cell marker.
Fig 3.
Staining of differentiated markers on 7-day and 14-day EBs and undifferentiated hESCs. Mesoderm markers Brachyury and ACTC, endoderm markers AFP, GATA4, and GATA6, and ectoderm marker TUBB3 were detectable in both 7-day and 14-day EBs (A-B, D-E, G-H, J-K, M-N), while their expression were not detected in undifferentiated hESCs (C, F, I, L, O). Spontaneously differentiated hESCs also expressed Brachyury (C), GATA4 and GATA6 (I).
Table 3.
Expression levels of ESC and EB markers in hESCs.
| Markers | ESCs | EB | |||
|---|---|---|---|---|---|
| IC | PCR | IC | PCR | ||
| ESC | LIN28 | N.A | ++ | N.A | -/+ |
| POU5F1 (Oct4) | +++ | +++ | - | -/+ | |
| GJA1 | N.A | +++ | N.A | ++ | |
| SOX2 | +++ | +++ | + | -/+ | |
| GAL | N.A | +++ | N.A | -/+ | |
| LIN41 | N.A | ++ | N.A | -/+ | |
| UTF1 | N.A | +++ | N.A | - | |
| TDGF1 | N.A | +++ | N.A | -/+ | |
| FOXD3 | N.A | ++ | N.A | N.A | |
| CDH1 | +++ | N.A | - | N.A | |
| NANOG | +++ | +++ | - | - | |
| ZFP42 (Rex1) | N.A | ++ | N.A | - | |
| TERT | N.A | ++ | N.A | - | |
| PODXL | +++ | N.A | - | N.A | |
| FGF4 | N.A | +++ | N.A | N.A | |
| DPPA5 | N.A | + | N.A | - | |
| Tra-1-60 | ++ | N.A. | - | N.A. | |
| Tra-1-81 | ++ | N.A. | - | N.A. | |
| SSEA-1 | +++ | N.A. | - | N.A. | |
| SSEA-4 | +++ | N.A. | - | N.A. | |
| ITGB1 (CD29) | +++ | N.A. | - | N.A. | |
| Ectoderm | MSI1 | N.A. | -/+ | N.A. | ++ |
| MAP2 | N.A. | -/+ | N.A. | ++ | |
| SOX1 | N.A. | - | N.A. | ++ | |
| NES | -/+ | N.A | +++ | N.A | |
| TH | - | - | ++ | ++ | |
| TUBB3 | - | N.A | +++ | N.A | |
| Endoderm | DCN | N.A | - | N.A | +++ |
| H19 | N.A | - | N.A | +++ | |
| AFP | - | - | ++ | +++ | |
| GATA4 | - | - | +++ | +++ | |
| HGF | N.A. | - | N.A. | ++ | |
| FoxA2 | N.A. | - | N.A. | ++ | |
| Mesoderm | HAND1 | N.A. | -/+ | N.A. | +++ |
| MSX1 | N.A. | - | N.A. | ++ | |
| ACTC | - | N.A. | +++ | N.A. | |
| GATA6 | - | N.A. | + | N.A. | |
| T (Brachyury) | - | - | ++ | ++ | |
| IGF2 | N.A. | - | N.A. | + | |
Monitor differentiation by microarray
Although offering sufficient resolution, RT-PCR and immunocytochemistry are difficult to perform for a large number of genes and cannot be easily automated. To test whether differences in gene expression were of a sufficient magnitude that they could be detected by a more global and less quantitative measurement, we assessed the expression of candidate ESC and EB markers by analyzing their expression in three hESC lines derived by BresaGen (three undifferentiated hESC samples of BG01, 02 and 03 and EBs derived from them) using the Illumina BeadArray containing about 48,000 unique features. All samples were examined in duplicate and only data from duplicates samples that showed 99% or greater correlation was used. The present results were focused on expression of the genes that were selected as candidate markers of the ESC and EB state.
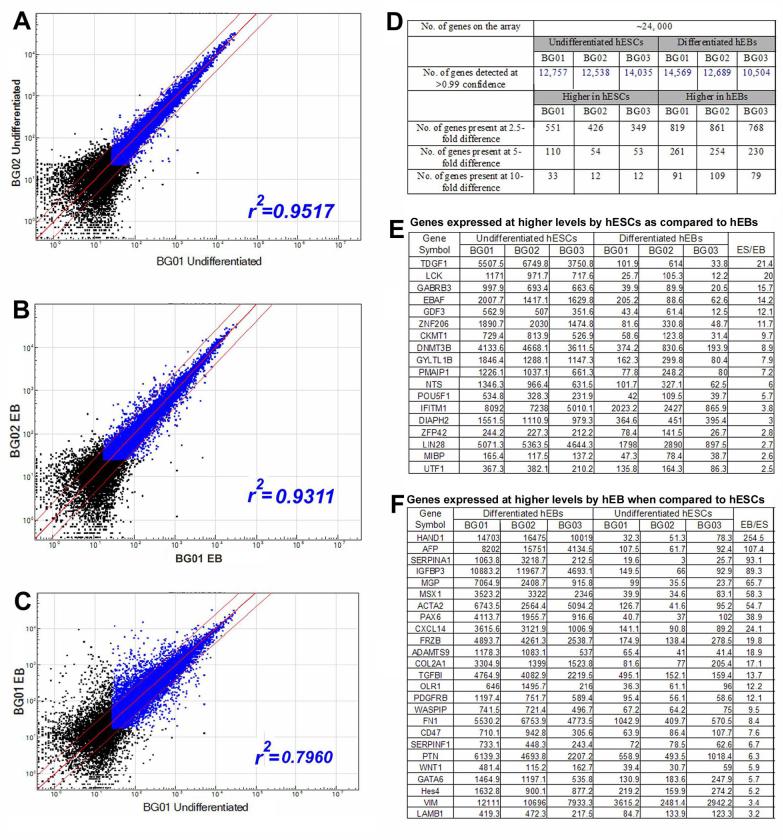
Global pairwise comparisons between different hESC lines (Fig. 4, panel A, and Supplementary Table 2) or different EBs (Fig. 4, panel B), showed similar levels of gene expression and around 90% of the genes detected at greater than 99% confidence limit were expressed at approximately similar levels (within the 2.5-fold range) (Fig. 4F). Pairwise comparisons of hESCs with hESCs or EBs with EBs showed a high degree of similarity of samples (correlation coefficient greater than 0.90). Most of the differential expression seen in Fig.4 A and B is the result of biological differences between the cultures; technical replicates have correlation coefficients greater than 0.90 (Loring, et al., in preparation). Comparisons of hESCs to EBs showed a much lower degree of similarity (correlation coefficient <0.8, Fig. 4, panel C). This suggests that different hESCs are similar to each other and that this similarity is greater than that between hESC and EB derived from the same line. The entire comparison is presented in Supplementary Table 2 and a restricted list of genes which were selected as ESC or EB markers that showed a greater than 2.5-fold difference in expression is shown in Fig. 4 (Panel G and H). The large difference between hESCs and EBs detected by this global comparison indicates that arrays can readily distinguish hESCs from EBs derived from them.
Fig 4.
Assessment of potential hESC and EB markers using microarrays. Three different ESC lines (BG01, 02 and 03) and 14-day EBs that differentiated from them were compared using an Illumina Bead array. (A-B) Comparisons of gene expression among three hESC lines or three EBs show similar levels with a correlation coefficient (r2) greater than 0.93. (C) Pairwise comparisons of undifferentiated hESCs with their matched EBs reveal that about 50% of the genes are expressed with at least a 2.5-fold difference. (D) Summary of the numbers of genes detected in this array. (E-F) Selected genes that are differentially expressed in three hESC lines (BG01, 02 and 03) and in their matched EBs. Note that only the genes that are detected at >0.99 confidence (blue dots) are considered valid for further analysis. Dots that fall between the thin red lines represent genes that are commonly expressed in hESCs and EBs, while dots outside red lines correspond to differentially expressed genes at >2.5 fold.
To further test whether the similarity in gene expression between hESC lines can be generalized, we analyzed an additional hESC lines H9, obtained from WiCell Institute (Madison, WI, http://www.wicell.org) rather than from BresaGen. As shown in Fig. 4D, gene expression profiles of H9 was remarkably similar to the BG lines, with a correlation coefficient of 0.93 when compared to BG01. This suggests that gene expression profiles in hESC lines derived from different laboratories are similar.
qPCR to monitor differentiation
Our results suggested that a global assessment by a relatively non-quantitative method such as RT-PCR or microarray could be used to detect differentiation. Given the dramatic differences in gene expression, we reasoned that assessing a smaller number of markers using a more quantitative measurement could be sufficient in monitoring the overall state of hESCs. To test this hypothesis, we selected a small number of genes from the 109-list and tested their expression in undifferentiated hESCs and in two differentiating stages of EBs (7-day and 14-day) using BG03. These included 6 undifferentiated ESC markers (Oct4, Nanog, UTF1, DPPA5, Lin 41 and Sox2) and 6 markers of differentiation with at least one gene from each germ layer (Sox1, DCN, H19, IGF2, GATA4 and Hand1). The expression level of these genes were determined as the ratio to the level of 18S RNA, and differential expression of these genes in undifferentiated hESCs and EBs were shown as the ratio of expression in hESC to EB (ESC markers) or EB to hESC (EB markers).
As expected, the expression of ESC markers Oct4, Nanog, UTF1, DPPA5 and Lin41 were higher in undifferentiated hESCs than in EBs, whereas the expression of EB markers Sox1, DCN, H19, IGF2, GATA4 and Hand1 were up-regulated in EBs compared to in undifferentiated hESCs (Fig. 5). In particular, expression of UTF1 and Nanog was rapidly down-regulated upon differentiation with more than a 10-fold decrease in 7-day EBs and 200-fold lower in 14-day EBs. Down-regulation of Oct4, an important gene for the maintenance of pluripotency in both hESCs and mESCs, was less marked with only 5-fold decrease in 14-day EBs. Sox2 was expressed in both undifferentiated hESCs and differentiated EBs (3-fold higher in 14-day EB and 5-fold higher in 7-day EB) which is expected as Sox2 is known to express in neural stem cells which are present in EBs. Interestingly, expression of Lin41 was rapidly decreased in 7-day EBs but the expression level increased in 14-day EBs (Fig. 5).
Fig 5.
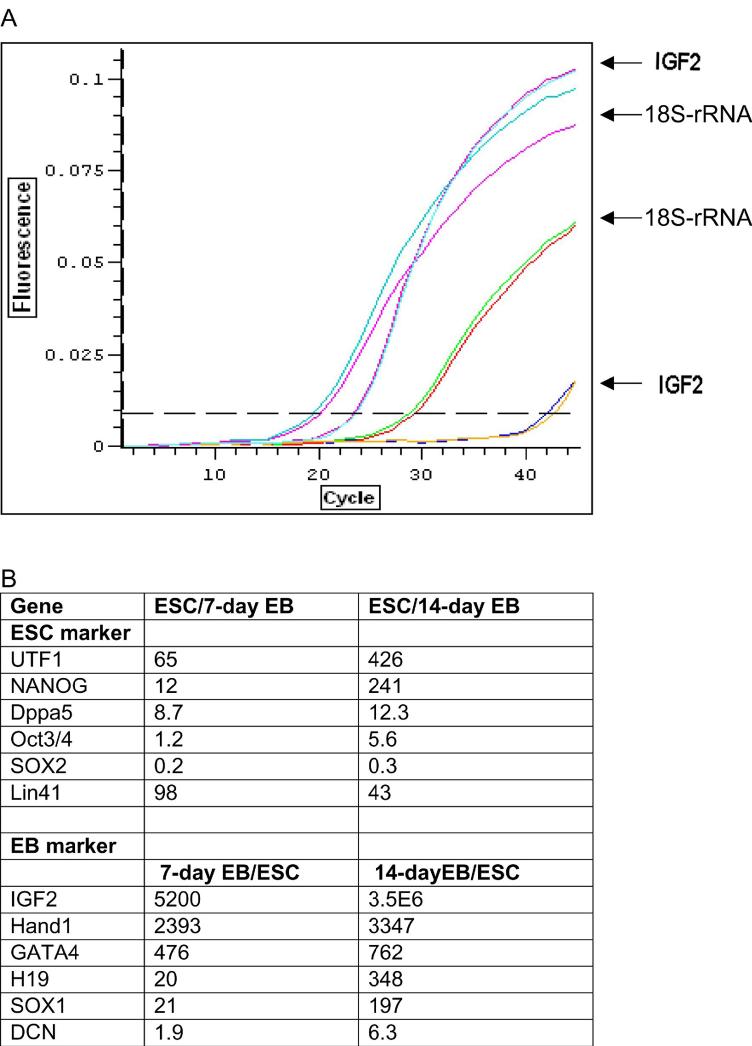
qPCR analysis of 12 genes during hESC differentiation. Expression of 6 markers of each ESC and EB were quantified by qPCR. (A) Amplification curves corresponding to IGF2 and 18S RNA (standard curve) are shown from left to right: IGF2 of 14-day EB (blue and red); 18S RNA of 14-day EB (red and blue); 18S RNA of undifferentiated hESCs (green and red); and IGF2 of undifferentiated hESCs (blue and yellow). (B) The expression level of these genes were determined as the ratio to the level of 18S RNA, and differential expression of these genes in undifferentiated hESCs and EBs were shown as the ratio of expression in hESC to EB (ESC markers) or EB to hESC (EB markers).
All of the differentiation markers, except for Decorin, were rapidly induced as the cells differentiated. The most dramatic changes were seen for an imprinted gene IGF2 whose levels were several thousand fold higher in EBs than in undifferentiated hESCs. Expression of the imprinted gene H19 as well as Hand1 and GATA4 was also rapidly increased as the cells underwent differentiation.
qPCR detects changes that may be missed by immunocytochemistry
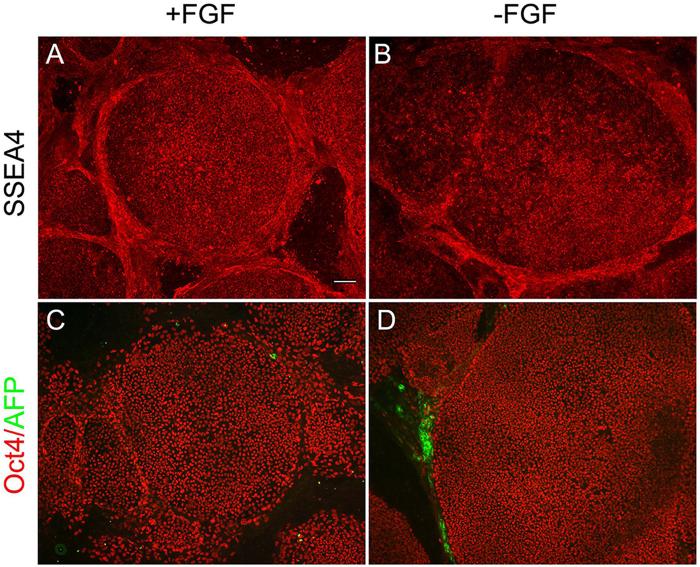
To test whether our qPCR assay can detect more subtle changes in hESC cultures that affect the undifferentiated state, we examined hESC cultures maintained with bFGF and cultures where bFGF was withdrawn for a period of 72 hours by qPCR and immunocytochemistry. For qPCR assay, we chose to analyze two markers of the ESC (UTF1 and Nanog) and EB (IGF2 and Hand1) state as expression of these four genes changed most significantly upon differentiation in our qPCR analysis (Fig. 4). No change in expression of either ESC (SSEA4 and Oct4) or EB markers (AFP) could be detected in this time period by immunocytochemistry (Fig. 6). However, qPCR readily detected a significant change in cultures maintained without bFGF for 72 hours: IGF2 and Hand1 were expressed 3.6-fold and 2.3-fold higher in hESC cultures without bFGF, whereas no significant changes were observed for the two most differentially expressed ESC markers (UTF: 1.4-fold and Nanog: 1.5-fold) detected by our qPCR analysis. These changes, despite smaller than those seen in 7-day and 14-day EBs, were similar in profile to changes when cells undergo differentiation.
Fig 6.
No difference in ESC and EB marker expression by immunocytochemistry between hESC cultures maintained with bFGF and cultures where bFGF was withdrawn for a period of 72 hours. (A-B) Immunostaining of SSEA4 (red) shows similar expression levels in these two cell populations (with and without supplement bFGF). (C-D) Immunostaining of Oct4 (red) shows most of the cells are positive, while only occasional AFP positive cells (green) are seen outside the colonies in both hESC populations. Scale bar=100 μm.
Discussion
Since the first derivation of hESC lines in 1998, information on gene expression in hESCs and other human stem cells has been accumulated rapidly using a variety of techniques such as microarray, SAGE, EST scan and MPSS [17-21; 24]. A large number of genes have been identified that are expressed at high levels in undifferentiated hESCs and “stemness” genes that define a stem cell state have been proposed [18; 37]. Expression of many of these genes is down-regulated as hESCs differentiate, and parallel to this, many genes are induced during differentiation. Nevertheless, there are no defined set of markers that can be routinely used for assessing the different states of hESCs, i.e., the undifferentiated ESC stage and different stages of differentiating EBs. In the present study, we compared published reports on gene expression in ESCs and EBs and selected a set of 109 known genes including 55 stem cell, 12 ectoderm, 19 endoderm, 15 mesoderm, 4 trophoblast and 4 germ cell markers as potential ESC or EB specific markers. We show that this set of genes can serve as indicators of the states of hESCs using four independent methods, qPCR, immunocytochemistry, RT-PCR and microarray, using at least three different hESC lines for each method.
It is clear that no single marker is sufficient to define the state of hESCs. Several surface antigens including SSEA and TRA are useful markers for undifferentiated hESCs as their level of expression is down-regulated as hESCs differentiate. The genes encoding them however, have yet to be identified. Other pluripotency genes including two well-characterized transcription factors, Oct4 and Nanog, are good markers to assess the presence of undifferentiated hESCs. Oct4 and Nanog are essential for the maintenance of pluripotency in both hESCs and mESCs and knock-out or knock-down of either gene causes differentiation [38; 39]. Oct4 and Nanog are, however, not uniquely expressed in undifferentiated hESCs: Oct4 is expressed in germ cells and Nanog has recently been reported to be expressed in mature tissues [40; 41]. Moreover, the expression of Oct4 declines slowly as cells differentiate and the change in levels is small. For example, we have detected Oct4 expression in hESCs that underwent differentiation for a week or underwent neuronal differentiation on PA6 cells for 2 weeks (unpublished results). Our qPCR results likewise showed only a moderate decrease in Oct4 expression in 7-day EBs and only a 5-fold decrease in 14-day EBs. Taken together, we believe that while SSEA, TRA, Oct4 and Nanog are useful for distinguishing undifferentiated hESCs from their differentiated progeny, expression of these markers alone or in combination is not enough to define the undifferentiated hESC populations. Likewise, expression of a single marker is not a definitive indicator of the differentiated EB state. Indeed, some of the early differentiation genes including keratin, actin and tubulin were expressed at low levels in undifferentiated hESCs; however, their expression was strongly up-regulated as hESCs formed EBs [19]. Since the differentiated progeny of hESCs include a number of cell types, it is important to assess EBs using a combination of endoderm, mesoderm and ectoderm markers.
Our assessment of RT-PCR as a method of examining hESC cultures showed that even though it is not quantitative, it is quite robust provided appropriate genes are selected for assessment. Primers were designed to all 109 genes and the expression of 35 candidate ESC and EB genes was confirmed by RT-PCR. Our data showed that undifferentiated hESCs could be readily distinguished from differentiating EBs by assessing 10-20 markers using semi-quantitative RT-PCR. The relatively specific expression of ESC and EB markers in undifferentiated hESCs and in EBs provides a simple method to assess the quality of RNA samples for different purposes and to estimate the level of differentiation in hESC culture.
In addition to the confirmation of differential expression in hESCs and EBs by RT-PCR, we examined the expression of many of these genes by immunocytochemistry. These included markers that have not previously been analyzed by antibody staining in hESCs such as PODXL, ITGB1 and Nanog. Nanog, PODXL, ITGB1 and CDH1 were down-regulated in 7-day EBs, and further decreased in 14-day EBs. Similarly, expression of differentiation markers such as AFP, GATA4 and GATA6 were strongly up-regulated in 7-day EBs. These markers, together with the SSEA and TRA surface markers can reliably detect the differentiation of hESCs and can be used for routine examination of differentiation in hESC cultures. Although immunostaining is relatively more time consuming, it offers unprecedented resolution allowing one to rapidly assess the degree of contamination or the extent of differentiation.
RT-PCR and immunocytochemistry, however, are not suitable for scaling up or processing of a large number of markers. We therefore examined if the genes identified as candidate markers can be used to assess differentiation using a microarray platform. Our results showed that many though not all genes show detectable changes in gene expression even in a relatively poor quantitative method such as microarray. For example, Oct4, Lin28, TDGF1 and GDF3 were present at significantly higher levels in hESCs than in EBs; while Col2A1, Col1A1 and SerpinA1 were present higher in EBs using the Illumina BeadArray. The difference in gene expression were present in five hESC lines tested in this study and in six other cell lines evaluated (Jeanne Loring, Burnham Institute, personal communication), indicating that these genes may serve as a standard measure of changes irrespective of the cell line being used. Other genes that were expected to serve as useful markers and showed utility in immunocytochemistry and RT-PCR were not as useful in this microarray format. Such genes included Nanog, Sox2 and Sox1 (see Supplementary Table 2 for a complete list), indicating that candidate genes will have to be assessed in each individual platform to determine if they are adequate within the limitations of that particular technology.
Among the 109 genes, six of each undifferentiated and differentiated markers (at least one marker of each germ layer) were further examined by qPCR in undifferentiated hESCs and two stages of EBs (7-day and 14-day). We reasoned that careful quantitation may allow one to use only a small subset of markers. Indeed our results showed that as few as six markers may be sufficient provided both positive and negative markers were used. Whereas down-regulation of Oct4 was gradual during differentiation, expression of Nanog and UTF1 declined more than 200-fold in 14-day EBs, suggesting that these markers are good indicators of the undifferentiated ESC state. Dramatic up-regulation of expression in EBs was also found for an imprinted gene IGF2, and for Hand1. Expression of IGF2 and Hand1 were quickly up-regulated in 7-day EBs by several thousand folds and by day 14, the expression levels were 3 million-fold higher for IGF2. It therefore seems that undifferentiated hESCs and their derivatives can be discriminated by examining a few genes using a quantitative method, if the genes are appropriately selected. UTF1 and Nanog are excellent candidates for markers of the undifferentiated state, whereas IGF2 and Hand1 are good markers for differentiated EBs. The dramatic changes in expression level of these four genes upon differentiation can be reliably used for assessing the undifferentiated ESC and differentiated states. Moreover, negative markers (differentiated EB markers) are more sensitive than positive markers (undifferentiated ESC markers) in detecting differentiation in hESCs. These conclusions are supported by experiments designed to detect smaller changes of differentiation in hESC cultures where bFGF was removed for a period of three days. Whereas immunocytochemistry could not detect any difference in bFGF treated and bFGF withdrawn cultures using ESC or EB markers, bFGF withdrawn hESC cultures could be readily distinguished from their sister culture maintained with bFGF by qPCR using negative markers (IGF2 and Hand1).
As more data on hESCs are collected, additional markers for undifferentiated and differentiated cells will undoubtedly be identified. For example, a large number of novel genes or genes of unknown function that show a similar robust alteration in expression levels as hESCs differentiate [18] may be included in future arrays or qPCR sets to provide an additional level of sensitivity to allow a finer dissection of the state of differentiation. The present lists, however, provides useful information for evaluating the states of hESC populations, extent of differentiation or for quality control of hESC cultures. Our results suggest that any of the four methods we describe here can be used to monitor the transition of undifferentiated hESCs to differentiated EBs when a combination of ESC and EB markers from our list were tested.
Conclusion
Our strategy of including a combination of genes that are down-regulated and up-regulated during differentiation which includes genes that represent different cell types (undifferentiated cells and cell types of the three germ layers, as well as trophoblast and germ cells) allows one to identify a set of markers that can be readily assessed by routine molecular or cellular biology methods. We believe that any of the methods we tested is sufficient to monitor the state of hESC but each method has its advantages and disadvantages. If qPCR is used, a small number of genes are sufficient provided both positive and negative markers are used [see present results and 42]. However, the most cost-effective method for the wealth of information obtained may be a focused array that includes many markers such as the genes we have described. Efforts to generate such an array are in progress [43, Ian Lyons, Invitrogen, personal communication]. Alternatively, microfluidic plates allow to custom design markers and have the advantages of being able to be adapted to very small number of cells.
Acknowledgements
We acknowledge the support of CNS foundation, NIA, NIDA and the Packard Center. We thank Drs. Tim McDaniel and David L Barker for sharing the Illumina BeadArray analysis and Rose Amable for technical assistance.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter MK, Rosler E, Rao MS. Characterization and differentiation of human embryonic stem cells. Cloning Stem Cells. 2003;5:79–88. doi: 10.1089/153623003321512193. [DOI] [PubMed] [Google Scholar]
- 3.Mitalipova M, Calhoun J, Shin S, et al. Human embryonic stem cell lines derived from discarded embryos. Stem Cells. 2003;21:521–526. doi: 10.1634/stemcells.21-5-521. [DOI] [PubMed] [Google Scholar]
- 4.Zeng X, Miura T, Luo Y, et al. Properties of pluripotent human embryonic stem cells BG01 and BG02. Stem Cells. 2004;22:292–312. doi: 10.1634/stemcells.22-3-292. [DOI] [PubMed] [Google Scholar]
- 5.Amit M, Itskovitz-Eldor J. Derivation and spontaneous differentiation of human embryonic stem cells. J Anat. 2002;200:225–232. doi: 10.1046/j.1469-7580.2002.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heins N, Englund MC, Sjoblom C, et al. Derivation, characterization, and differentiation of human embryonic stem cells. Stem Cells. 2004;22:367–376. doi: 10.1634/stemcells.22-3-367. [DOI] [PubMed] [Google Scholar]
- 7.Park JH, Kim SJ, Oh EJ, et al. Establishment and maintenance of human embryonic stem cells on STO, a permanently growing cell line. Biol Reprod. 2003;69:2007–2014. doi: 10.1095/biolreprod.103.017467. [DOI] [PubMed] [Google Scholar]
- 8.Ginis I, Luo Y, Miura T, et al. Differences between human and mouse embryonic stem cells. Dev Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter MK, Inokuma MS, Denham J, Mujtaba T, Chiu CP, Rao MS. Enrichment of neurons and neural precursors from human embryonic stem cells. Exp Neurol. 2001;172:383–397. doi: 10.1006/exnr.2001.7832. [DOI] [PubMed] [Google Scholar]
- 10.Kaufman DS, Hanson ET, Lewis RL, Auerbach R, Thomson JA. Hematopoietic colony-forming cells derived from human embryonic stem cells. Proc Natl Acad Sci U S A. 2001;98:10716–10721. doi: 10.1073/pnas.191362598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kehat I, Kenyagin-Karsenti D, Snir M, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 13.Rambhatla L, Chiu CP, Kundu P, Peng Y, Carpenter MK. Generation of hepatocyte-like cells from human embryonic stem cells. Cell Transplant. 2003;12:1–11. doi: 10.3727/000000003783985179. [DOI] [PubMed] [Google Scholar]
- 14.Reubinoff BE, Itsykson P, Turetsky T, et al. Neural progenitors from human embryonic stem cells. Nat Biotechnol. 2001;19:1134–1140. doi: 10.1038/nbt1201-1134. [DOI] [PubMed] [Google Scholar]
- 15.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 16.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 17.Brandenberger R, Khrebtukova I, Thies RS, et al. MPSS profiling of human embryonic stem cells. BMC Dev Biol. 2004;4:10. doi: 10.1186/1471-213X-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharya B, Miura T, Brandenberger R, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 19.Miura T, Luo Y, Khrebtukova I, et al. Monitoring early differentiation events in human embryonic stem cells by massively parallel signature sequencing and expressed sequence tag scan. Stem Cells Dev. 2004;13:694–715. doi: 10.1089/scd.2004.13.694. [DOI] [PubMed] [Google Scholar]
- 20.Sperger JM, Chen X, Draper JS, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abeyta MJ, Clark AT, Rodriguez RT, Bodnar MS, Pera RA, Firpo MT. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet. 2004;13:601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- 22.Robson P. The maturing of the human embryonic stem cell transcriptome profile. Trends Biotechnol. 2004;22:609–612. doi: 10.1016/j.tibtech.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 24.Richards M, Tan SP, Tan JH, Chan WK, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 25.Brimble SN, Zeng X, Weiler DA, et al. Karyotypic stability, genotyping, differentiation, feeder-free maintenance, and gene expression sampling in three human embryonic stem cell lines derived prior to August 9, 2001. Stem Cells Dev. 2004;13:585–597. doi: 10.1089/scd.2004.13.585. [DOI] [PubMed] [Google Scholar]
- 26.Wei CL, Miura T, Robson P, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharya B, Luo Y, Miura T, et al. Comparison of the gene expression profile of undifferentiated human embryonic stem cell lines and embryoid bodies differentiated from them. BMC Dev Biol. 2005 doi: 10.1186/1471-213X-5-22. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humpherys D, Eggan K, Akutsu H, et al. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 29.Dean W, Bowden L, Aitchison A, et al. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development. 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- 30.Xu C, Inokuma MS, Denham J, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 31.Rosler ES, Fisk GJ, Ares X, et al. Long-term culture of human embryonic stem cells in feeder-free conditions. Dev Dyn. 2004;229:259–274. doi: 10.1002/dvdy.10430. [DOI] [PubMed] [Google Scholar]
- 32.Zeng X, Chen J, Sanchez JF, et al. Stable expression of hrGFP by mouse embryonic stem cells: promoter activity in the undifferentiated state and during dopaminergic neural differentiation. Stem Cells. 2003;21:647–653. doi: 10.1634/stemcells.21-6-647. [DOI] [PubMed] [Google Scholar]
- 33.Kuhn K, Baker SC, Chudin E, et al. A novel, high-performance random array platform for quantitative gene expression profiling. Genome Res. 2004;14:2347–2356. doi: 10.1101/gr.2739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong RC, Pebay A, Nguyen LT, Koh KL, Pera MF. Presence of functional gap junctions in human embryonic stem cells. Stem Cells. 2004;22:883–889. doi: 10.1634/stemcells.22-6-883. [DOI] [PubMed] [Google Scholar]
- 35.Amit M, Shariki C, Margulets V, Itskovitz-Eldor J. Feeder layer- and serum-free culture of human embryonic stem cells. Biol Reprod. 2004;70:837–845. doi: 10.1095/biolreprod.103.021147. [DOI] [PubMed] [Google Scholar]
- 36.Draper JS, Moore HD, Ruban LN, Gokhale PJ, Andrews PW. Culture and characterization of human embryonic stem cells. Stem Cells Dev. 2004;13:325–336. doi: 10.1089/scd.2004.13.325. [DOI] [PubMed] [Google Scholar]
- 37.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 38.Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz-Eldor J, Daley GQ. High-efficiency RNA interference in human embryonic stem cells. Stem Cells. 2005;23:299–305. doi: 10.1634/stemcells.2004-0252. [DOI] [PubMed] [Google Scholar]
- 39.Mitsui K, Tokuzawa Y, Itoh H, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 40.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 41.Hart AH, Hartley L, Ibrahim M, Robb L. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev Dyn. 2004;230:187–198. doi: 10.1002/dvdy.20034. [DOI] [PubMed] [Google Scholar]
- 42.Noaksson K, Zoric N, Zeng X, et al. Monitoring differentiation of human embryonic stem cells using real-time PCR. Stem Cells. 2005 doi: 10.1634/stemcells.2005-0093. In Press. [DOI] [PubMed] [Google Scholar]
- 43.Yang AX, Mejido J, Luo Y, et al. Development of a focused microarray to assess human embryonic stem cell differentiation. Stem Cells Dev. 2005 doi: 10.1089/scd.2005.14.270. In Press. [DOI] [PubMed] [Google Scholar]
- 44.Rosner MH, Vigano MA, Ozato K, et al. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- 45.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 46.Cui L, Johkura K, Yue F, et al. Spatial distribution and initial changes of SSEA-1 and other cell adhesion-related molecules on mouse embryonic stem cells before and during differentiation. J Histochem Cytochem. 2004;52:1447–1457. doi: 10.1369/jhc.3A6241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anisimov SV, Tarasov KV, Tweedie D, Stern MD, Wobus AM, Boheler KR. SAGE identification of gene transcripts with profiles unique to pluripotent mouse R1 embryonic stem cells. Genomics. 2002;79:169–176. doi: 10.1006/geno.2002.6687. [DOI] [PubMed] [Google Scholar]
- 48.Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 49.Okuda A, Fukushima A, Nishimoto M, et al. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. Embo J. 1998;17:2019–2032. doi: 10.1093/emboj/17.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lei W, Hirose T, Zhang LX, et al. Cloning of the human orphan receptor germ cell nuclear factor/retinoid receptor-related testis-associated receptor and its differential regulation during embryonal carcinoma cell differentiation. J Mol Endocrinol. 1997;18:167–176. doi: 10.1677/jme.0.0180167. [DOI] [PubMed] [Google Scholar]
- 51.Hanna LA, Foreman RK, Tarasenko IA, Kessler DS, Labosky PA. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 2002;16:2650–2661. doi: 10.1101/gad.1020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 53.Rogers MB, Hosler BA, Gudas LJ. Specific expression of a retinoic acid-regulated, zinc finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development. 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- 54.Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- 55.Kim SK, Suh MR, Yoon HS, et al. Identification of developmental pluripotency associated 5 expression in human pluripotent stem cells. Stem Cells. 2005;23:458–462. doi: 10.1634/stemcells.2004-0245. [DOI] [PubMed] [Google Scholar]
- 56.Schmid E, Tapscott S, Bennett GS, et al. Differential location of different types of intermediate-sized filaments in various tissues of the chicken embryo. Differentiation. 1979;15:27–40. doi: 10.1111/j.1432-0436.1979.tb01031.x. [DOI] [PubMed] [Google Scholar]
- 57.Sharma MK, Saxena V, Liu RZ, et al. Differential expression of the duplicated cellular retinoic acid-binding protein 2 genes (crabp2a and crabp2b) during zebrafish embryonic development. Gene Expr Patterns. 2005;5:371–379. doi: 10.1016/j.modgep.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 58.Kolodkin AL, Matthes DJ, Goodman CS. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75:1389–1399. doi: 10.1016/0092-8674(93)90625-z. [DOI] [PubMed] [Google Scholar]
- 59.Good P, Yoda A, Sakakibara S, et al. The human Musashi homolog 1 (MSI1) gene encoding the homologue of Musashi/Nrp-1, a neural RNA-binding protein putatively expressed in CNS stem cells and neural progenitor cells. Genomics. 1998;52:382–384. doi: 10.1006/geno.1998.5456. [DOI] [PubMed] [Google Scholar]
- 60.Garner CC, Tucker RP, Matus A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 61.Reeves SA, Helman LJ, Allison A, Israel MA. Molecular cloning and primary structure of human glial fibrillary acidic protein. Proc Natl Acad Sci U S A. 1989;86:5178–5182. doi: 10.1073/pnas.86.13.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 63.Malas S, Duthie SM, Mohri F, Lovell-Badge R, Episkopou V. Cloning and mapping of the human SOX1: a highly conserved gene expressed in the developing brain. Mamm Genome. 1997;8:866–868. doi: 10.1007/s003359900597. [DOI] [PubMed] [Google Scholar]
- 64.Hockfield S, McKay RD. Identification of major cell classes in the developing mammalian nervous system. J Neurosci. 1985;5:3310–3328. doi: 10.1523/JNEUROSCI.05-12-03310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 66.Masserano JM, Weiner N. Tyrosine hydroxylase regulation in the central nervous system. Mol Cell Biochem. 1983;53:54–129. doi: 10.1007/BF00225250. [DOI] [PubMed] [Google Scholar]
- 67.Burgoyne RD, Cambray-Deakin MA, Lewis SA, Sarkar S, Cowan NJ. Differential distribution of beta-tubulin isotypes in cerebellum. Embo J. 1988;7:2311–2319. doi: 10.1002/j.1460-2075.1988.tb03074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kornblihtt AR, Vibe-Pedersen K, Baralle FE. Isolation and characterization of cDNA clones for human and bovine fibronectins. Proc Natl Acad Sci U S A. 1983;80:3218–3222. doi: 10.1073/pnas.80.11.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fisher LW, Hawkins GR, Tuross N, Termine JD. Purification and partial characterization of small proteoglycans I and II, bone sialoproteins I and II, and osteonectin from the mineral compartment of developing human bone. J Biol Chem. 1987;262:9702–9708. [PubMed] [Google Scholar]
- 70.Ruoslahti E, Pihko H, Seppala M. Alpha-fetoprotein: immunochemical purification and chemical properties. Expression in normal state and in malignant and non-malignant liver disease. Transplant Rev. 1974;20:38–60. doi: 10.1111/j.1600-065x.1974.tb00140.x. [DOI] [PubMed] [Google Scholar]
- 71.Lohi J, Leivo I, Franssila K, Virtanen I. Changes in the distribution of integrins and their basement membrane ligands during development of human thyroid follicular epithelium. Histochem J. 1997;29:337–345. doi: 10.1023/a:1026482700109. [DOI] [PubMed] [Google Scholar]
- 72.Smyth N, Vatansever HS, Murray P, et al. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to failure of endoderm differentiation. J Cell Biol. 1999;144:151–160. doi: 10.1083/jcb.144.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wozney JM, Rosen V, Celeste AJ, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 74.Kurachi K, Chandra T, Degen SJ, et al. Cloning and sequence of cDNA coding for alpha 1-antitrypsin. Proc Natl Acad Sci U S A. 1981;78:6826–6830. doi: 10.1073/pnas.78.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kendall RL, Thomas KA. Inhibition of vascular endothelial cell growth factor activity by an endogenously encoded soluble receptor. Proc Natl Acad Sci U S A. 1993;90:10705–10709. doi: 10.1073/pnas.90.22.10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.ten Dijke P, Ichijo H, Franzen P, et al. Activin receptor-like kinases: a novel subclass of cell-surface receptors with predicted serine/threonine kinase activity. Oncogene. 1993;8:2879–2887. [PubMed] [Google Scholar]
- 77.Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Novak U, Wilks A, Buell G, McEwen S. Identical mRNA for preproglucagon in pancreas and gut. Eur J Biochem. 1987;164:553–558. doi: 10.1111/j.1432-1033.1987.tb11162.x. [DOI] [PubMed] [Google Scholar]
- 79.Andik I, Donhoffer S. The effect of insulin on food intake and selection of mice. Z Vitam Horm Fermentforsch. 1949;3:208–212. [PubMed] [Google Scholar]
- 80.Newman PJ, Berndt MC, Gorski J, et al. PECAM-1 (CD31) cloning and relation to adhesion molecules of the immunoglobulin gene superfamily. Science. 1990;247:1219–1222. doi: 10.1126/science.1690453. [DOI] [PubMed] [Google Scholar]
- 81.Lowe JB, Boguski MS, Sweetser DA, Elshourbagy NA, Taylor JM, Gordon JI. Human liver fatty acid binding protein. Isolation of a full length cDNA and comparative sequence analyses of orthologous and paralogous proteins. J Biol Chem. 1985;260:3413–3417. [PubMed] [Google Scholar]
- 82.Chartier FL, Bossu JP, Laudet V, Fruchart JC, Laine B. Cloning and sequencing of cDNAs encoding the human hepatocyte nuclear factor 4 indicate the presence of two isoforms in human liver. Gene. 1994;147:269–272. doi: 10.1016/0378-1119(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 83.Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 84.Zarnegar R, Michalopoulos G. Purification and biological characterization of human hepatopoietin A, a polypeptide growth factor for hepatocytes. Cancer Res. 1989;49:3314–3320. [PubMed] [Google Scholar]
- 85.Yamada S, Zhu Q, Aihara Y, et al. Cloning of cDNA and the gene encoding human hepatocyte nuclear factor (HNF)-3 beta and mutation screening in Japanese subjects with maturity-onset diabetes of the young. Diabetologia. 2000;43:121–124. doi: 10.1007/s001250050016. [DOI] [PubMed] [Google Scholar]
- 86.Termine JD, Kleinman HK, Whitson SW, Conn KM, McGarvey ML, Martin GR. Osteonectin, a bone-specific protein linking mineral to collagen. Cell. 1981;26:99–105. doi: 10.1016/0092-8674(81)90037-4. [DOI] [PubMed] [Google Scholar]
- 87.Russell MW, Baker P, Izumo S. Cloning, chromosomal mapping, and expression of the human eHAND gene. Mamm Genome. 1997;8:863–865. doi: 10.1007/s003359900596. [DOI] [PubMed] [Google Scholar]
- 88.Robert B, Sassoon D, Jacq B, Gehring W, Buckingham M. Hox-7, a mouse homeobox gene with a novel pattern of expression during embryogenesis. Embo J. 1989;8:91–100. doi: 10.1002/j.1460-2075.1989.tb03352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gunning P, Ponte P, Blau H, Kedes L. alpha-skeletal and alpha-cardiac actin genes are coexpressed in adult human skeletal muscle and heart. Mol Cell Biol. 1983;3:1985–1995. doi: 10.1128/mcb.3.11.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suzuki E, Evans T, Lowry J, et al. The human GATA-6 gene: structure, chromosomal location, and regulation of expression by tissue-specific and mitogen-responsive signals. Genomics. 1996;38:283–290. doi: 10.1006/geno.1996.0630. [DOI] [PubMed] [Google Scholar]
- 91.Glant TT, Hadhazy C, Mikecz K, Sipos A. Appearance and persistence of fibronectin in cartilage. Specific interaction of fibronectin with collagen type II. Histochemistry. 1985;82:149–158. doi: 10.1007/BF00708199. [DOI] [PubMed] [Google Scholar]
- 92.Lau ET, Kwok YK, Chui DH, Wong HS, Luo HY, Tang MH. Embryonic and fetal globins are expressed in adult erythroid progenitor cells and in erythroid cell cultures. Prenat Diagn. 2001;21:529–539. doi: 10.1002/pd.81. [DOI] [PubMed] [Google Scholar]
- 93.Herrmann BG. Action of the Brachyury gene in mouse embryogenesis. Ciba Found Symp. 1992;165:78–86. doi: 10.1002/9780470514221.ch5. discussion 86-91. [DOI] [PubMed] [Google Scholar]
- 94.Call KM, Glaser T, Ito CY, et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms′ tumor locus. Cell. 1990;60:509–520. doi: 10.1016/0092-8674(90)90601-a. [DOI] [PubMed] [Google Scholar]
- 95.Braun T, Buschhausen-Denker G, Bober E, Tannich E, Arnold HH. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. Embo J. 1989;8:701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li ZL, Lilienbaum A, Butler-Browne G, Paulin D. Human desmin-coding gene: complete nucleotide sequence, characterization and regulation of expression during myogenesis and development. Gene. 1989;78:243–254. doi: 10.1016/0378-1119(89)90227-8. [DOI] [PubMed] [Google Scholar]
- 97.Kangawa K, Matsuo H. Purification and complete amino acid sequence of alpha-human atrial natriuretic polypeptide (alpha-hANP) Biochem Biophys Res Commun. 1984;118:131–139. doi: 10.1016/0006-291x(84)91077-5. [DOI] [PubMed] [Google Scholar]
- 98.Marotta CA, Forget BG, Cohen-Solal M, Weissman SM. Nucleotide sequence analysis of coding and noncoding regions of human beta-globin mRNA. Prog Nucleic Acid Res Mol Biol. 1976;19:165–175. doi: 10.1016/s0079-6603(08)60917-4. [DOI] [PubMed] [Google Scholar]
- 99.Ogawa E, Maruyama M, Kagoshima H, et al. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bell GI, Merryweather JP, Sanchez-Pescador R, et al. Sequence of a cDNA clone encoding human preproinsulin-like growth factor II. Nature. 1984;310:775–777. doi: 10.1038/310775a0. [DOI] [PubMed] [Google Scholar]
- 101.Ryan K, Garrett N, Mitchell A, Gurdon JB. Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell. 1996;87:989–1000. doi: 10.1016/s0092-8674(00)81794-8. [DOI] [PubMed] [Google Scholar]
- 102.Suh E, Chen L, Taylor J, Traber PG. A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol Cell Biol. 1994;14:7340–7351. doi: 10.1128/mcb.14.11.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamada K, Ogawa H, Honda S, Harada N, Okazaki T. A GCM motif protein is involved in placenta-specific expression of human aromatase gene. J Biol Chem. 1999;274:32279–32286. doi: 10.1074/jbc.274.45.32279. [DOI] [PubMed] [Google Scholar]
- 104.Cserhalmi-Friedman PB, Squeo R, Gordon D, et al. Epidermolytic hyperkeratosis in a Hispanic family resulting from a mutation in the keratin 1 gene. Clin Exp Dermatol. 2000;25:241–243. doi: 10.1046/j.1365-2230.2000.00625.x. [DOI] [PubMed] [Google Scholar]
- 105.Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell. 2000;5:73–83. doi: 10.1016/s1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
- 106.Castrillon DH, Quade BJ, Wang TY, Quigley C, Crum CP. The human VASA gene is specifically expressed in the germ cell lineage. Proc Natl Acad Sci U S A. 2000;97:9585–9590. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deblandre GA, Marinx OP, Evans SS, et al. Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. J Biol Chem. 1995;270:23860–23866. doi: 10.1074/jbc.270.40.23860. [DOI] [PubMed] [Google Scholar]
- 108.Tanaka SS, Matsui Y. Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Gene Expr Patterns. 2002;2:297–303. doi: 10.1016/s0925-4773(02)00384-2. [DOI] [PubMed] [Google Scholar]