Abstract
Although the nuclear envelope is a dynamic structure that disassembles and reforms during mitosis, the formation of membranous vesicles derived from the nuclear envelope has not yet been described in noninfected cells. However, during herpesvirus maturation, intranuclear capsids initiate transit to the cytosol for final maturation by budding at the inner nuclear membrane. Two conserved herpesvirus proteins are required for this primary envelopment, designated in the alphaherpesviruses as pUL31 and pUL34. Here, we show that simultaneous expression of pUL31 and pUL34 of the alphaherpesvirus pseudorabies virus in stably transfected rabbit kidney cells resulted in the formation of vesicles in the perinuclear space that resemble primary envelopes without a nucleocapsid. They contain pUL31 and pUL34 as shown by immunolabeling and are derived from the nuclear envelope. Thus, coexpression of only two conserved herpesvirus proteins without any other viral factor is sufficient to induce the formation of vesicles from the nuclear membrane. This argues for the contribution of cellular factors in this process either recruited from their natural cytoplasmic location or not yet identified as components of the nuclear compartment.
Keywords: nuclear envelope, primary envelopment, pseudorabies virus, herpesvirus egress
Herpesvirus particles are complex macromolecular assemblies consisting of >30 virally encoded proteins, which make up four morphologically distinct structures, core, capsid, tegument, and envelope. Herpesvirus morphogenesis proceeds in two different cellular compartments. While capsid assembly and DNA packaging take place in the nuclei of infected cells, acquisition of the majority of tegument and final envelopment occur in the cytoplasm. To gain access to the cytoplasm the nucleocapsid has to traverse the nuclear lamina and the inner and outer nuclear membranes, because the diameter of the nuclear pores is too small to allow exit of the ≈110-nm particle. Although alternative mechanisms for nuclear egress have been proposed, most data support a model that entails primary envelopment by budding of capsids at the inner nuclear membrane resulting in the formation of primary virions in the perinuclear space whose envelope then fuses with the outer nuclear membrane to release the capsid into the cytoplasm (reviewed in ref. 1).
The molecular mechanism of this budding, scission, and fusion reaction is unknown. Glycoproteins gB and gH, which are essential for fusion of the viral envelope with the plasma membrane during entry (2) as well as for cell–cell spread, are not required for virion formation indicating that a different molecular mechanism is responsible for fusion during nuclear egress (3). Cellular fusion processes involving the nuclear membranes occur during mitosis. However, the fusion machinery involved is unknown. Soluble N-ethylmaleimide sensitive factor attachment protein receptors (SNAREs) involved in intracytoplasmic vesicle targeting and fusion (reviewed in refs. 4 and 5) are suspected to be located in the nuclear envelope but have not yet been detected (reviewed in ref. 6).
Two conserved herpesvirus proteins, in the alphaherpesviruses designated as pUL31 and pUL34, are required for nuclear egress of herpesviruses from all three subfamilies (7–13). pUL34 is a predicted type II membrane protein, which is targeted to the nuclear envelope even in the absence of other viral proteins. In contrast, pUL31 is found diffusely distributed in the nucleus in transfected cells but is relocated to the nuclear rim in the presence of pUL34. Both proteins interact, and formation of the pUL34/pUL31 complex is required for proper positioning of both complex partners, local dissolution of the nuclear lamina, modification of the host cell chromatin, and efficient release of nucleocapsids from the nucleus (14–21). pUL31 and pUL34 bind nuclear lamins A/C and B (15, 18, 20) and are required for recruitment of cellular protein kinase C (PKC). Phosphorylation of lamins presumably leads to local dissolution of the lamina (12, 21). Both proteins are also components of the primary virion but are absent from mature virus particles (8, 10, 11, 22, 23). In contrast, major tegument proteins of mature virions such as pUL36, pUL37, pUL46, pUL47, or pUL49 are not detectable in primary virions (24). In addition to pUL31 and pUL34, other herpesviral proteins are also involved in nuclear virus egress. The alphaherpesvirus pUS3 kinase is required for efficient fusion of primary envelopes because, in the absence of pUS3, primary enveloped virions accumulate in the perinuclear space (25). The conserved pUL25 capsid-associated protein apparently plays a role in triggering primary envelopment of DNA-containing mature capsids at the inner nuclear membrane (26).
Even in the absence of virus capsids, secondary envelopment in the cytoplasm proceeds, resulting in the formation of so-called “light” (L-) particles (27, 28). Capsidless enveloped particles (“primary L-particles”) have also been detected in the perinuclear space in electron microscopic analyses, although they are rare in wild-type virus-infected cells. This indicates that nucleocapsids may also be dispensable for primary envelopment and vesicle formation at the inner nuclear membrane (3, 28, 29). To investigate whether pUL34 and pUL31 are sufficient for vesicle formation from the nuclear envelope, we generated cell lines expressing both proteins of the alphaherpesvirus pseudorabies virus (PrV). We show here that in pUL31/pUL34 doubly expressing cells membranous vesicles are formed from the nuclear envelope that are located in the perinuclear space and contain pUL31 and pUL34. Thus, coexpression of pUL31 and pUL34 is sufficient for the formation of perinuclear vesicles that resemble primary enveloped particles without a nucleocapsid.
Results
Generation of RK13-UL31/34 Cells.
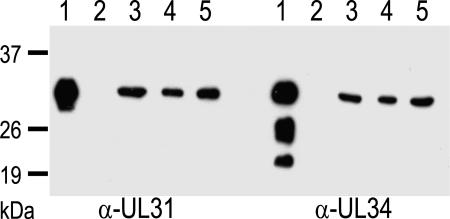
To generate a cell line that coexpresses pUL31 and pUL34, the corresponding ORFs were inserted into modified plasmid pcDNA3 (Invitrogen, Karlsruhe, Germany) under control of the human or murine cytomegalovirus immediate-early 1 promotor/enhancer, respectively [see supporting information (SI) Fig. 6]. Rabbit kidney cells (RK13) were transfected and selected for G418 resistance. Cell clones were screened for pUL31 and pUL34 expression by indirect immunofluorescence and for complementation of respective virus mutants (8, 11). Three cell clones yielding the highest virus titers in the complementation assays expressed both proteins in comparable amounts, as tested by Western blot (Fig. 1). Electrophoretic mobility also correlated with the corresponding proteins in wild-type PrV strain Kaplan- (PrV-Ka; Fig. 1, lanes 1; ref. 30) infected cell lysates, although truncated forms of pUL34 of unknown function and origin seen in infected cells were not observed in the cell lines. Clone RK13-UL31/34–2 (Fig. 1, lanes 3) was randomly selected for further analysis.
Fig. 1.
Identification of pUL31 and pUL34 in RK13-UL31/34 cells. Three different geneticin-resistant cell clones (lanes 3–5) were probed with antisera against pUL31 or pUL34 (8, 11). For control, wild-type PrV-Ka (lanes 1) and mock-infected RK13 cell lysates (lanes 2) were analyzed. Location of molecular mass markers is indicated on the left.
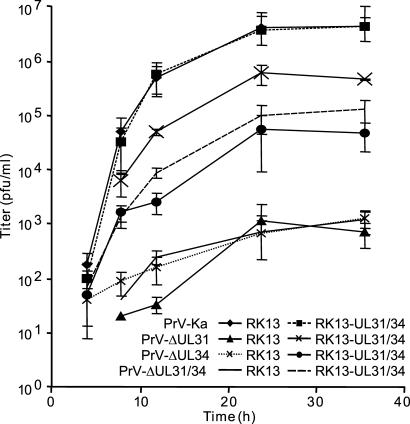
To test for functionality of the proteins, a PrV mutant lacking the UL31 and UL34 genes was isolated. Southern and Western blot analyses verified absence of UL31 and UL34 in this mutant (data not shown). One-step growth kinetics of mutants lacking pUL31 (PrV-ΔUL31) or pUL34 (PrV-ΔUL34) or both (PrV-ΔUL31/34) was analyzed on either RK13 or RK13-UL31/34 cells. Propagation on RK13 cells resulted in a maximum titer of only ≈1 × 103 pfu/ml, with no significant difference between the single or double mutants (Fig. 2). After propagation on RK13-UL31/34 cells, titers reached 5 × 104−1 × 105 for PrV-ΔUL34 and PrV-ΔUL31/34, and 5 × 105 pfu/ml for PrV-ΔUL31 indicating that both proteins were functional, although titers did not reach the level of wild-type PrV-Ka. This is most likely due to the fact that not all cells coexpress both proteins at detectable levels at any given time point. Titers for PrV-Ka were similar after propagation on RK13 or RK13-UL31/34 cells, demonstrating that simultaneous ectopic expression of pUL31 and pUL34 had no gross negative effect on PrV replication. Diameters of plaques induced by PrV-ΔUL31/34 on RK13-UL31/34 cells were similar to those formed by wild-type PrV-Ka (data not shown).
Fig. 2.
One-step growth kinetics of wild-type and mutant viruses. RK13 and RK13-UL31/34 cells were infected with PrV-Ka or the respective mutants and analyzed at the indicated times after low-pH treatment.
Colocalization of pUL31 and pUL34 in Speckles Associated with the Nuclear Membrane.
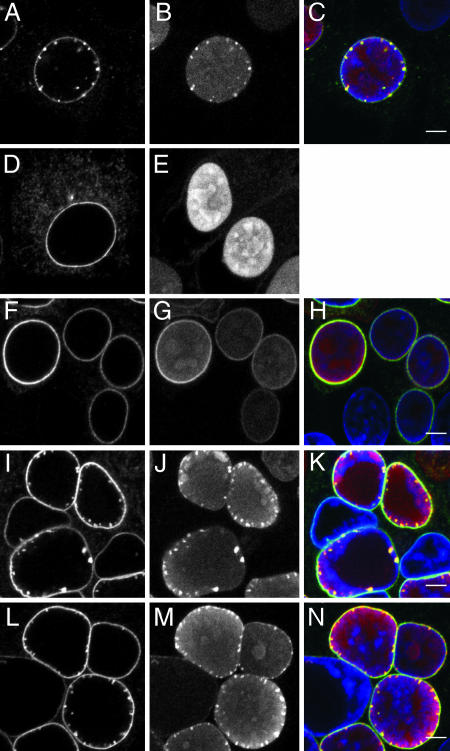
To investigate whether simultaneous expression altered localization of pUL31 and pUL34, cells on glass cover slips were fixed and analyzed by immunofluorescence with a rabbit anti-pUL31 (11) and a murine anti-pUL34 serum. In RK13-UL31/34 cells, pUL34 fluorescence appeared as a strong nuclear rim staining accompanied with a speckled pattern (Fig. 3A), which contrasts the smoother nuclear rim and cytoplasmic staining in cells expressing only pUL34 (Fig. 3D; ref. 8) or the exclusive nucleoplasmic staining in pUL31-expressing cells (Fig. 3E). The speckles were also labeled with the anti-pUL31 serum (Fig. 3B). The merged image (Fig. 3C) demonstrates that both proteins colocalize in these punctae. In wild-type PrV-Ka-infected cells, no apparent alterations of the nuclear envelope were observed (Fig. 3 F–H). In contrast, absence of pUS3, which is involved in fusion of primary virions with the outer nuclear membrane and whose absence results in accumulation of primary enveloped virions (Fig. 4E) (25), yielded a picture similar to that observed after coexpression of pUL31 and pUL34 (Fig. 3 I–K). Because coexpression of pUL31 and pUL34 is sufficient for their formation, speckles at the nuclear envelope should also be formed during wild-type virus infection. However, they were never observed, which might be due to a more efficient envelopment of capsids and/or rapid fusion of primary envelopes with the outer nuclear envelope. To address this question, cells were infected with PrV-ΔUL25/US3 lacking pUL25, required for primary envelopment, as well as pUS3 promoting deenvelopment. As shown in Fig. 3 L–N, in the absence of nuclear egress of capsids and the fusion-promotion function of pUS3, extensive punctate staining lining the inner nuclear membrane was indeed observed.
Fig. 3.
Laser-scanning confocal microscopy. Noninfected RK13-UL31/34 (A–C), RK13-UL34 (D), or RK13-UL31 cells (E), or RK13 cells infected with PrV-Ka (F–H), PrV-ΔUS3 (I–K), or PrV-ΔUL25/US3 (L–N) were fixed and immunostained with mouse anti-pUL34 (Left) and rabbit anti-pUL31 serum (Center). Right shows merged images (green: anti-pUL34; red: anti-pUL31). Nuclei were visualized by chromatin stain with TO-PRO-3 (blue). [Scale bars (green: anti-pUL34; red: anti-pUL31), 5 μm.]
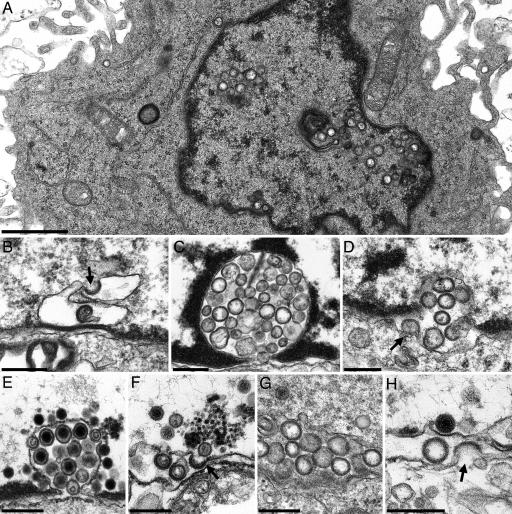
Fig. 4.
Electron microscopy. (A) Overview of an RK13-UL31/34 cell exhibiting intranuclear vesicles. The intranuclear appearance with no apparent connection to the inner nuclear membrane of most vesicles is due to the level of section (see SI Fig. 7). (B) Vesicle formation at the inner nuclear membrane resulting in invaginations of the inner nuclear membrane filled with membranous vesicles (C). (D) Budding/fusion stage at the outer nuclear membrane. (E) Invagination of inner nuclear membrane filled with primary virions after infection of RK13 cells with PrV-ΔUS3. (F–H) The same stages were observed after infection of RK13 cells with PrV-ΔUL25/US3. Arrows in B, D, F, and H show budding/fusion of vesicles. [Scale bars, 1.0 μm (A) and 300 nm (B–H).]
Simultaneous Expression of pUL31 and pUL34 Leads to Formation of Nuclear Membrane-Derived Vesicles.
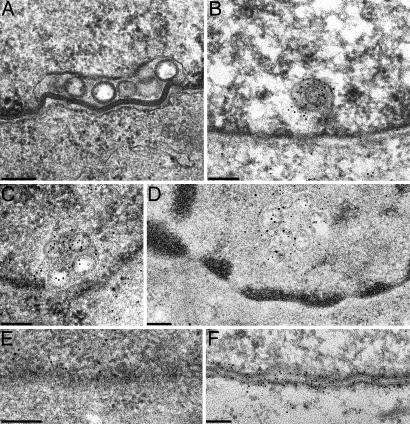
To investigate whether the speckled fluorescence corresponded to alterations of the nuclear membrane, RK13-UL31/34 cells were fixed and processed for electron microscopy either for ultrathin sectioning (Fig. 4) or for immunogold labeling (Fig. 5). Although expression of either pUL31 (Fig. 5E) or pUL34 (Fig. 5F) had no obvious effect on the nuclear membranes (8, 11), RK13-UL31/UL34 cells showed local proliferations of the inner nuclear membrane (Fig. 4 A, C, and D), which contained membranous vesicles of on average ≈130- to 160-nm diameter, resembling in size and appearance primary virions but lacking nucleocapsids. This is particularly evident in a comparison between noninfected RK13-UL31/34 cells (Fig. 4C) and RK13 cells infected with PrV-ΔUS3, which accumulate primary virions (Fig. 4E). Concerning the origin of these vesicles, budding (or fusion) stages were observed at the inner (Fig. 4B) and the outer nuclear membrane (Fig. 4D). To test whether the vesicles contain both proteins, RK13-UL31/34 cells (Fig. 5A) were incubated either separately with anti-UL34 or anti-UL31 rabbit sera or simultaneously with anti-UL34 murine and -UL31 rabbit serum followed by incubation with gold-conjugated secondary antibodies. A pUL31- and pUL34-specific label was found associated with these vesicles either by using the rabbit sera (Fig. 5 B and C) or in colabeling experiments with gold particles of 15- and 10-nm diameter for the detection of pUL31 or pUL34 (Fig. 5D). Cells expressing only pUL31 or pUL34 served as controls. Without the respective complex partner, pUL31 exhibited a diffuse intranuclear label (Fig. 5E), whereas pUL34 was associated with both leaflets of the nuclear envelope (Fig. 5F). Infection with PrV-ΔUL25/US3 also resulted in vesicle formation from the nuclear membrane (Fig. 4G), and budding/fusion stages could be observed at either leaflet of the nuclear envelope (Fig. 4 F and H).
Fig. 5.
Immunoelectron microscopy. Ultrathin sections of RK13-UL31/34 cells. (A) were incubated with rabbit anti-UL34 serum (B) or rabbit anti-UL31 serum (C) or simultaneously with murine anti-UL34 serum and rabbit anti-UL31 serum (D). Bound antibodies were visualized with gold-conjugated secondary anti-rabbit sera containing 10-nm gold for B and C and 15-nm gold for anti-rabbit and 10-nm gold for anti-mouse sera for D. For control, RK13-UL31 cells were labeled with the anti-UL31 serum (E) and RK13-UL34 cells with the anti-UL34 serum (F). (Scale bars, 150 nm.)
Discussion
Viruses have often been used to elucidate fundamental cell biological processes, because they divert or alter them for their replication. During herpesvirus infection, the morphology of the cell nucleus changes drastically by enlargement, proliferation of the nuclear membranes, and marginalization of host cell chromatin. Although these effects are well known, their molecular basis is largely unclear. Here we demonstrate that vesicle formation from the nuclear envelope is induced by coexpression of two herpesviral proteins, pUL31 and pUL34, a process that resembles primary envelopment during herpesvirus infection.
Intracellular formation and trafficking of vesicles are highly specific and coordinated. All known fusion events require Rab-GTPases, which organize the fusion site; SNARE proteins, which act during fusion; and N-ethylmaleimide sensitive factor with its cofactor SNAP, which are apparently required for recycling of the cellular fusion machinery (reviewed in ref. 5). The nuclear envelope is a dynamic structure that disassembles through mitosis and reforms on chromatin during the telophase by the fusion of multiple membrane vesicles controlled by the small GTPase Ran (reviewed in ref. 6).
However, so far, vesicle formation from the nuclear envelope without complete membrane disassembly and fusion with the outer nuclear membrane have not been described in noninfected cells indicating that vesicle budding processes at the nuclear membrane normally do not occur. Moreover, nuclear membrane-specific SNARE proteins, tethers, or cofactors that may be required for subsequent fusion of perinuclear membrane vesicles with the nuclear envelope have not been detected. Thus, our finding that vesicle formation from and probably vesicle fusion with the nuclear membrane can be induced by ectopic expression of only two viral proteins, pUL31 and pUL34, is intriguing. The absence of any further viral protein in the cells expressing pUL31 and pUL34 indicates that no other viral proteins are essential for this process or mimic components of the cellular machinery required for vesicle formation. Thus, the requirement of only two viral proteins for vesicle formation in fact argues in favor of subversion of coat proteins, SNAREs, and tethers from cellular compartments to induce vesicle formation at the nuclear envelope. However, further experiments are required to demonstrate whether pUL31 and pUL34 are able to execute functions of the cellular vesicle formation machinery themselves, or whether relevant cellular proteins required for vesicle formation (and subsequent fusion) are recruited by the two viral proteins similar to the recruitment of PKC for dissolution of the nuclear lamina (12, 21).
It remains unclear whether the perinuclear vesicles in RK13-UL31/34 cells indeed are fusion competent. Although budding and/or fusion stages at either nuclear membrane have been observed in ultrastructural studies (Fig. 5), these cannot be differentiated unequivocally by electron microscopy. pUL34 is a predicted type II membrane protein, and it is highly likely that it locates to the vesicle membrane. However, in its predicted topology, only few amino acids extend outside of the vesicle, which appear insufficient for mediating the complex processes resulting in membrane fusion. Therefore, if fusion occurs, it would have to be mediated by other cellular proteins incorporated into the vesicle membrane. In herpesvirus infection, the budding of capsids at the inner nuclear membrane and subsequent fusion of the primary envelope with the outer nuclear membrane are part of the replicative cycle. This fusion appears to be a rapid process, because primary enveloped particles as well as fusion stages at the outer nuclear membrane are observed only infrequently in cells infected by wild-type viruses. However, nuclear egress is impaired and primary virions accumulate in the perinuclear cleft when the alphaherpesviral pUS3 kinase is deleted or inactivated (10, 25, 31, 32), indicating that phosphorylation might regulate fusion. In RK13-UL31/34 cells pUS3 is absent, which may account for the abundance of vesicles within the perinuclear space. To induce fusion of these vesicles, RK13-UL31/34 cells were transfected with a pUS3 expression plasmid. However, ectopic pUS3 expression leads to irreversible morphological changes, including cell rounding and formation of filopodia, frequently resulting in cell death (33–35). This was also true for RK13-UL31/34 cells (data not shown) preventing the analysis of fusion induction by pUS3.
Alterations of the nuclear envelope have also been observed after coexpression of pUL31 and pUL34 homologs of Epstein–Barr virus (18). However, they did not include formation of perinuclear vesicles but rather presented as reduplications of the nuclear membrane. The formation of intranuclear membrane structures has also been described in noninfected cells after overexpression of proteins containing a CaaX (36) or CxxM (37) motif, which is recognized as a substrate by farnesyl- and geranylgeranyltransferases. Neither PrV pUL31 nor pUL34 contains one of these motifs, indicating a different mechanism for the formation of pUL31/pUL34-induced perinuclear vesicles.
The observed nuclear envelope-derived vesicles contain pUL31 and pUL34, a property shared with primary enveloped virions during herpesvirus infection (8, 11). Although these vesicles vary in size, most of them display a diameter of between 130 and 160 nm and resemble in size and morphology primary virions without a capsid. Apparently, both proteins are necessary for formation of these structures, because the expression of either PrV pUL31 or pUL34 did not result in detectable alterations of the nuclear membranes (refs. 8 and 11; this study). Similar effects were also observed after infection with a PrV mutant deficient not only in pUS3 but also in pUL25, a capsid-associated protein required for primary envelopment which indicates that, given the proper conditions, capsidless nuclear envelope-derived vesicles are also formed during PrV infection. Concerning their origin, accumulations of these vesicles were found in invaginations of the inner nuclear membrane, whereas budding (or fusion) stages were observed at the inner as well as the outer nuclear membrane. Because electron microscopy does not provide directionality, it remains unresolved whether these indeed represent budding at the inner and fusion at the outer nuclear membrane, as would be congruent with the envelopment–deenvelopment model of nuclear egress. However, that pUL31, one of the two proteins required for vesicle formation, is predominantly located in the nucleus and thus is available primarily in this compartment strongly argues for a budding mechanism at the inner nuclear membrane.
The formation of capsidless enveloped viral structures is a hallmark of herpesvirus replication. These L-particles (27) or dense bodies (38) are formed in the cytoplasm and released from infected cells. They consist of tegument, which apparently initiates and completes secondary envelopment by budding into trans-Golgi vesicles without the need for a capsid. Formation of these capsidless particles, however, requires tegument and envelope proteins, because L-particle formation is blocked when specific viral tegument proteins, such as pUL11, and envelope proteins, e.g., gM, are absent (39). L-particles and dense bodies are heterogeneous in size but clearly resemble complete viral particles. So far, formation of perinuclear capsidless enveloped viral particles, i.e., vesicular capsidless structures containing noncapsid viral proteins that originate from invagination of the inner nuclear membrane and are located in the perinuclear cleft, has only rarely been observed (28, 29). pUL31 has been hypothesized to represent a component of the primary tegument, and pUL34, a type II transmembrane protein, is considered a component of the primary envelope. This assumed localization of the two proteins within the primary virion is supported by our results, because the interaction of a tegument component (pUL31) with an envelope protein (pUL34) is presumably required to initiate and drive the budding process analogous to the formation of secondary L-particles in the cytoplasm. Both proteins are components of the vesicles in the perinuclear cleft, which is congruent with the envelopment–deenvelopment–reenvelopment model of herpesvirus maturation.
In summary, we demonstrate that coexpression of pUL31 and pUL34 of the alphaherpesvirus pseudorabies virus is sufficient to induce the formation of membranous vesicles from the nuclear envelope, which resemble primary envelopes. Although the mechanism of the formation of primary herpes virions and subsequent fusion of the primary envelope with the outer nuclear membrane is still largely unclear, our data strongly support the envelopment–deenvelopment–reenvelopment model for herpesvirus morphogenesis (reviewed in ref. 1). Most interestingly, the finding that only two herpesviral proteins, pUL31 and pUL34, are sufficient for vesicle formation at the nuclear envelope strongly argues in favor of the contribution of cellular factors either recruited from their natural cytoplasmic location or not yet identified as components of the nuclear compartment. Identification of these factors will further add to the understanding of herpesvirus nuclear egress and may provide novel insights into the cellular mechanisms involved in regulating membrane vesicle formation and fusion.
Materials and Methods
Cells and Viruses.
Mutants PrV-ΔUL34 (8), PrV-ΔUL31 (11), and PrV-ΔUS3 (25) have been derived from PrV-Ka (30). Mutant PrV-ΔUL25/US3 was isolated after cotransfection of PrV-ΔUL25 genomic DNA (26) and plasmid pΔUS3gfp (25) into RK13-UL25 cells (26). Mutant viruses were propagated on RK13 or RK13-UL31, RK13-UL34, or RK13-UL25 cells (8, 11, 26). A cell line complementing simultaneous lack of UL31 and UL34 was generated after cotransfection of a plasmid containing a genomic 5.2-kb DrdI-fragment [nucleotides 28099–33282; GenBank accession no. BK001744 (40); SI Fig. 6] and pSV2neo (41). G418-resistant cell clones were isolated and tested with PrV-ΔUL31 and PrV-ΔUL34 for complementation in plaque assays. One cell clone that allowed plaque formation of both virus mutants was named RK13-DrdI and was used to isolate PrV-ΔUL31/34. RK13-UL31/34 cells were selected after transfection with plasmid p3ie-UL31/34 (see below).
Construction of PrV-ΔUL31/34.
For generation of a mutant simultaneously lacking UL31 and UL34 (SI Fig. 6 A and B) genomic DNA of mutant PrV-ΔUL31F (11) was cotransfected with plasmid pΔUL34gfp (8) into RK13-DrdI cells. The progeny virus was isolated on RK13-DrdI cells, and one single plaque isolate, designated PrV-ΔUL31/34, was analyzed further. Restriction fragment length and Southern blot analyses verified deletion of UL31 and UL34 sequences (data not shown).
Construction of pUL31/pUL34 Double Expression Vector.
A DNA fragment encompassing the murine cytomegalovirus immediate-early 1 enhancer/promoter followed by a polylinker sequence and the polyadenylation signal of the bovine herpesvirus 1 glycoprotein D gene was isolated from plasmid promI (42) with PflMI and HindIII and blunt-ended with Klenow polymerase. The purified fragment was cloned into pcDNA3 (Invitrogen) after cleavage with BglII and blunt-ending with Klenow polymerase, resulting in plasmid p3ie in which the immediate-early enhancer/promoter elements of human and murine cytomegalovirus direct transcription in opposite directions (SI Fig. 6 C). The UL31 ORF was excised as a 0.85-kb EcoRI fragment from plasmid pcDNA-UL31 (11) and cloned into the EcoRI site of p3ie. The UL34 ORF was excised from plasmid pcDNA-UL34 (8) as a 0.8-kb BamHI/XhoI fragment and subsequently cloned into BglII-digested p3ie-UL31 after blunt-ending by Klenow polymerase, generating plasmid p3ie-UL31/34 (SI Fig. 6 C).
Generation of Monospecific Antisera.
Rabbit sera against PrV pUL31 and pUL34 have been described (8, 11). To generate monospecific anti-PrV pUL34 mouse serum, two mice were immunized with 20 μg of GST-UL34 fusion protein (8). Sera were collected after the fourth boost.
One-Step Growth Analysis.
RK13 or RK13-UL31/34 cells were infected at a multiplicity of infection of 5 with PrV-Ka or the different mutants and incubated on ice for 1 h. The inoculum was removed, prewarmed medium was added, and cells were incubated for 1 additional hour at 37°C. Nonpenetrated virus was inactivated by low-pH treatment, and cells and supernatant were harvested either immediately (0 h) or after 4, 8, 12, 24, and 36 h. Progeny virus titers were determined by plaque assays on RK13-DrdI and RK13 cells. Mean values of three independent experiments and the corresponding standard deviations were calculated and plotted.
Western Blot Analysis and Immunofluorescence.
Cell lysates of infected (18 h postinfection), mock-infected RK13, or noninfected RK13-UL31/34 cells were harvested as described (11). Cell lysates were separated on 12% polyacrylamide gels, blotted, and incubated with rabbit anti-pUL31 (dilution 1:50.000; ref. 11) or anti-pUL34 sera (dilution 1:50,000; ref. 8). Bound antibody was detected with peroxidase-conjugated anti-rabbit antibodies (Dianova, Hamburg, Germany) and visualized by chemiluminescence (Super Signal, Pierce, Bonn, Germany) recorded on x-ray films.
For immunofluorescence, RK13-UL31/34, RK13-UL31, RK13-UL34, or RK13 cells on coverslips were infected with PrV-Ka, PrV-ΔUS3, or PrV-ΔUL25/US3 and fixed with 3% paraformaldehyde for 30 min and subsequently with 3% paraformaldehyde and 0.5% Triton X-100 for 20 min at room temperature. Cells were incubated with the murine anti-pUL34 (1:300) and rabbit anti-pUL31 (1:500; ref. 11) sera and overlaid with Alexa Fluor 488-conjugated anti-mouse and Alexa Fluor 555-conjugated anti-rabbit secondary antibodies (Molecular Probes, Invitrogen, Karlsruhe, Germany). TO-PRO-3 (1 μl/ml; Molecular Probes) was added for staining of cell chromatin. Fluorescence was preserved with a 9:1 mixture of glycerol in PBS containing 25 mg of 1,4-diazabicyclooctane per milliliter and analyzed in a confocal laser-scanning microscope (LSM510; Zeiss, Oberkochen, Germany).
Electron Microscopy and Immunolabeling.
RK13 cells expressing pUL31, pUL34, or both proteins or RK13 cells infected with PrV-ΔUS3 or PrV-ΔUL25/US3 were processed as described (8). For immunogold labeling, antisera against pUL31 and pUL34 were decorated with gold-tagged goat-antispecies antibodies (GAR10 and GFAF10, for single labeling and GAR15/GFAF10 for double labeling; British BioCell International, Cambridge, U.K.). Specificity was controlled on uninfected and infected cells by using gold-conjugate without primary antibody and nonherpesvirus protein-specific antibodies (data not shown). Counterstained ultrathin sections were analyzed with a transmission electron microscope (Philips EM 400 T, Tecnai 12, Eindhoven, The Netherlands).
Supplementary Material
Acknowledgments
We thank Dr. Egbert Mundt for preparation of the mouse antiserum; Petra Meyer, Diana Werner, Kathrin Müller, and Katrin Giesow for expert technical assistance; and Mandy Jörn for photographic help. This study was supported by Deutsche Forschungsgemeinschaft Grant DFG Me 854/8-1.
Abbreviations
- SNARE
soluble N-ethylmaleimide sensitive factor attachment protein receptor
- PrV
pseudorabies virus
- PrV-Ka
PrV strain Kaplan
- RK13
rabbit kidney cells
- L-
light.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701757104/DC1.
References
- 1.Mettenleiter TC, Klupp BG, Granzow H. Curr Opin Microbiol. 2006;9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Spear PG, Longnecker R. J Virol. 2003;77:10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granzow H, Klupp BG, Fuchs W, Veits J, Osterrieder N, Mettenleiter TC. J Virol. 2001;75:3675–3684. doi: 10.1128/JVI.75.8.3675-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jahn R, Scheller RH. Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 5.Ungermann C, Langosch D. J Cell Sci. 2005;118:3819–3828. doi: 10.1242/jcs.02561. [DOI] [PubMed] [Google Scholar]
- 6.Prunuske AJ, Ullman KS. Curr Opin Cell Biol. 2006;18:1–9. doi: 10.1016/j.ceb.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roller RJ, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. J Virol. 2000;74:117–129. doi: 10.1128/jvi.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klupp BG, Granzow H, Mettenleiter TC. J Virol. 2000;74:10063–10073. doi: 10.1128/jvi.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. J Virol. 2001;75:8803–8817. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. J Virol. 2002;76:8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs W, Klupp BG, Granzow H, Osterrieder N, Mettenleiter TC. J Virol. 2002;76:364–378. doi: 10.1128/JVI.76.1.364-378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH. Science. 2002;297:854–857. doi: 10.1126/science.1071506. [DOI] [PubMed] [Google Scholar]
- 13.Farina A, Feederle R, Raffa S, Gonnella R, Santarelli R, Frati L, Angeloni A, Torrisi MR, Faggioni A, Delecluse H-J. J Virol. 2005;79:3703–3712. doi: 10.1128/JVI.79.6.3703-3712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott ES, O'Hare P. J Virol. 2001;75:8818–8830. doi: 10.1128/JVI.75.18.8818-8830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds AE, Liang L, Baines JD. J Virol. 2004;78:5564–5575. doi: 10.1128/JVI.78.11.5564-5575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lake CM, Hutt-Fletcher LM. Virology. 2004;320:99–106. doi: 10.1016/j.virol.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 17.Simpson-Holley M, Colgrove RC, Nalepa G, Harper JW, Knipe DM. J Virol. 2005;79:12840–12851. doi: 10.1128/JVI.79.20.12840-12851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonnella R, Farina A, Santarelli R, Raffa S, Feederle R, Bei R, Granato M, Modesti A, Frati L, Delecluse H-J, et al. J Virol. 2005;79:3713–3727. doi: 10.1128/JVI.79.6.3713-3727.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang L, Baines JD. J Virol. 2005;79:3797–3806. doi: 10.1128/JVI.79.6.3797-3806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjerke S, Roller R. Virology. 2006;347:261–276. doi: 10.1016/j.virol.2005.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park R, Baines JD. J Virol. 2006;80:494–504. doi: 10.1128/JVI.80.1.494-504.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johannsen E, Luftig M, Chase MR, Weicksel S, Chair-McFarland E, Illanes D, Sarracino D, Kieff E. Proc Natl Acad Sci USA. 2004;101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michael K, Klupp BG, Mettenleiter TC, Karger A. J Virol. 2006;80:1332–1339. doi: 10.1128/JVI.80.3.1332-1339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granzow H, Klupp BG, Mettenleiter TC. J Virol. 2004;78:1314–1323. doi: 10.1128/JVI.78.3.1314-1323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp BG, Granzow H, Mettenleiter TC. J Gen Virol. 2001;82:2363–2371. doi: 10.1099/0022-1317-82-10-2363. [DOI] [PubMed] [Google Scholar]
- 26.Klupp BG, Granzow H, Keil GM, Mettenleiter TC. J Virol. 2006;80:6235–6246. doi: 10.1128/JVI.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLauchlan J, Rixon FJ. J Gen Virol. 1992;73:269–276. doi: 10.1099/0022-1317-73-2-269. [DOI] [PubMed] [Google Scholar]
- 28.Alemañ N, Quiroga MI, López-Peña M, Vázquez S, Guerrero FH, Nieto JM. J Virol. 2003;77:5657–5667. doi: 10.1128/JVI.77.10.5657-5667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Granzow H, Weiland F, Jöns A, Klupp BG, Karger A, Mettenleiter TC. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan AS, Vatter AE. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 31.Wagenaar F, Pol JM, Peeters B, Gielkens AL, de Wind N, Kimman TG. J Gen Virol. 1995;76:1851–1859. doi: 10.1099/0022-1317-76-7-1851. [DOI] [PubMed] [Google Scholar]
- 32.Schumacher D, Tischer BK, Trapp S, Osterrieder N. J Virol. 2005;79:3987–3997. doi: 10.1128/JVI.79.7.3987-3997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Minnebruggen G, Favoreel HW, Jacobs L, Nauwynck HJ. J Virol. 2003;77:9074–9080. doi: 10.1128/JVI.77.16.9074-9080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calton CM, Randall JA, Adkins MW, Banfield BW. Virus Genes. 2004;29:131–145. doi: 10.1023/B:VIRU.0000032796.27878.7f. [DOI] [PubMed] [Google Scholar]
- 35.Favoreel HW, van Minnebruggen G, Adriaensen D, Nauwynck HJ. Proc Natl Acad Sci USA. 2005;102:8990–8995. doi: 10.1073/pnas.0409099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ralle T, Grund C, Franke WW, Stick R. J Cell Sci. 2004;117:6095–6104. doi: 10.1242/jcs.01528. [DOI] [PubMed] [Google Scholar]
- 37.Prüfert K, Vogel A, Krohne G. J Cell Sci. 2004;117:6105–6116. doi: 10.1242/jcs.01532. [DOI] [PubMed] [Google Scholar]
- 38.Irmiere A, Gibson W. Virology. 1983;130:118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- 39.Kopp M, Granzow H, Fuchs W, Klupp BG, Mettenleiter TC. J Virol. 2004;78:3024–3034. doi: 10.1128/JVI.78.6.3024-3034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klupp BG, Hengartner CJ, Mettenleiter TC, Enquist LW. J Virol. 2004;78:424–440. doi: 10.1128/JVI.78.1.424-440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Southern PJ, Berg P. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 42.Kühnle G, Collins RA, Scott JE, Keil GM. J Gen Virol. 1996;77:2231–2240. doi: 10.1099/0022-1317-77-9-2231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.