Abstract
Resistances to tetracycline and mercury were identified in an environmental strain of Serratia marcescens isolated from a stream highly contaminated with heavy metals. As a step toward addressing the mechanisms of coselection of heavy metal and antibiotic resistances, the tetracycline resistance determinant was cloned in Escherichia coli. Within the cloned 13-kb segment, the tetracycline resistance locus was localized by deletion analysis and transposon mutagenesis. DNA sequence analysis of an 8.0-kb region revealed a novel gene [tetA(41)] that was predicted to encode a tetracycline efflux pump. Phylogenetic analysis showed that the TetA(41) protein was most closely related to the Tet(39) efflux protein of Acinetobacter spp. yet had less than 80% amino acid identity with known tetracycline efflux pumps. Adjacent to the tetA(41) gene was a divergently transcribed gene [tetR(41)] predicted to encode a tetracycline-responsive repressor protein. The tetA(41)-tetR(41) intergenic region contained putative operators for TetR(41) binding. The tetA(41) and tetR(41) promoters were analyzed using lacZ fusions, which showed that the expression of both the tetA(41) and tetR(41) genes exhibited TetR(41)-dependent regulation by subinhibitory concentrations of tetracycline. The apparent lack of plasmids in this S. marcescens strain, as well as the presence of metabolic genes adjacent to the tetracycline resistance locus, suggested that the genes were located on the S. marcescens chromosome and may have been acquired by transduction. The cloned Tet 41 determinant did not confer mercury resistance to E. coli, confirming that Tet 41 is a tetracycline-specific efflux pump rather than a multidrug transporter.
Antibiotic resistance has become one of the greatest obstacles to the successful treatment of bacterial diseases. Resistance to antimicrobial agents is widespread and occurs in numerous bacterial genera (7). Abundant evidence shows that resistance also occurs in many commensal or environmental organisms (5, 7, 20, 31, 37), which may serve as reservoirs of antibiotic resistance genes that can subsequently be transferred to pathogens by horizontal gene transfer. Consequently, elucidating sources and mechanisms that select for antibiotic resistance in the environment is critical to a complete understanding of the problem of bacterial antibiotic resistance.
Tetracycline and its many derivatives have been used extensively for nearly 6 decades (7) to treat many types of bacterial illnesses in humans and in animals and have been included as growth promoters in animal feeds or sprayed onto fruit trees for disease prevention (7, 30). Despite its widespread use, the effectiveness of tetracycline therapy has been limited due to antibiotic resistance, which began to arise shortly after the discovery of tetracycline (30). Resistance to tetracycline can be made manifest by ribosomal protection, enzymatic inactivation, or drug efflux. Of the nearly 40 known classes of tetracycline resistance determinants, the majority belong to the drug efflux group (7, 22). Membrane-bound tetracycline efflux proteins actively transport the drug from the cytoplasm, preventing the accumulation of tetracycline within the cell. Some classes of efflux proteins (multidrug transporters) can transport tetracycline as well as other drugs or toxic compounds, such as heavy metals (7).
The synthesis of some (but not all) gram-negative tetracycline pumps is tightly regulated by the presence of tetracycline (7, 16). In the absence of tetracycline, expression of the efflux protein is negatively regulated by a repressor protein (TetR) that is encoded by a gene divergently transcribed from that encoding the resistance protein (16). When tetracycline is present in as little as nanomolar concentrations, it is bound by TetR, which then loses affinity for the operator of the resistance gene. Expression of the efflux protein is then rapidly and dramatically upregulated (16).
Several factors influencing the direct selection of tetracycline resistance have been proposed, including misuse (e.g., overprescription) during therapy of humans, the use of subtherapeutic doses in animals, and the stability of tetracycline in the environment (7, 21). Direct selection of antibiotic resistance by exposure to antibiotics such as tetracycline, however, is not the only mechanism by which resistance can be selected or maintained. It is well known that resistances to antibiotics and other compounds such as antibacterial compounds and heavy metals can be found on the same genetic elements. Few experimental studies have been conducted to investigate the role of various selective agents in co- or cross-resistance. One study examined the effects of Ni, Cd, tetracycline, or ampicillin on the frequency of resistance traits found in aquatic bacteria (36). When microcosms containing river water were exposed to various concentrations of these selective agents, increases in both metal and antibiotic resistance were observed, suggesting the potential for environmentally mediated increases in unrelated resistance traits.
Serratia marcescens is an opportunistic gram-negative pathogen that is a significant cause of nosocomial infections, such as pneumonia, urinary tract infections, bacteremia, meningitis, and myocarditis (1, 18, 28, 33), including troublesome and recurrent hospital outbreaks, for example, in neonatal intensive care units (11). S. marcescens infections (septicemia, pneumonia, cellulitis) are also problematic in human immunodeficiency virus-infected persons (24). S. marcescens is ubiquitous in nature and can be found in water, soil, plants, insects, and animals (1). While tetracycline resistance is reportedly widespread in S. marcescens (6, 12, 15, 38), the only characterized S. marcescens tetracycline resistance determinant is the Tet(B) protein expressed by the conjugative plasmid R478 (13).
This study was initiated in an effort to further our understanding of the possible indirect selection of antibiotic resistance in bacteria living in environments contaminated with heavy metals. While characterizing the mechanism of antibiotic resistance genes from this environment, we isolated a strain of S. marcescens that contained a previously unreported class of the tetracycline resistance gene tetA(41).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in these studies are listed in Table 1. Strain FMC 1-23-O, identified as S. marcescens and resistant to tetracycline and mercury, is an environmental isolate obtained from Four Mile Creek at the Savannah River site (near Aiken, SC).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Resistance | Source or reference |
|---|---|---|---|
| Strains | |||
| FMC 1-23-O | S. marcescens environmental isolate | Tetracycline | Savannah River Site |
| JM109 | E. coli cloning host | Promega | |
| Plasmids | |||
| pBluescript II SK(−) | Cloning vector | Ampicillin | Stratagene |
| pEVM1 | 13-kb EcoRI fragment of FMC 1-23-O in pBluescript II SK(−) | Tetracycline | This study |
| pEVM2 | 4.4-kb BamHI fragment from pEVM1 in pBluescript II SK(−) | Tetracycline | This study |
| pEVM3 | tetA(41)-lacZ transcriptional fusion in pMW10 | Kanamycin | This study |
| pEVM4 | tetR(41)-lacZ transcriptional fusion in pMW10 | Kanamycin | This study |
| pMOD-Kan | EZ::TN transposon containing Kmr cassette (aphA gene from pUC4K) | Kanamycin | This study |
| pMW10 | Promoterless lacZ shuttle vector | Kanamycin | 41 |
Samples were collected from the top 10 cm of sediment from select locations in Four Mile Creek, placed in sterile bags, and transported on ice to the laboratory. Each sample was homogenized by kneading, and ∼10 g of sediment was removed and diluted with 10 ml sterile saline and sonicated for 5 minutes (Fisher Bransonic ultrasonic cleaner, model B2200R-1). The resulting slurry was used to isolate and enumerate Escherichia coli organisms using USEPA approved method no. 10029. Strain FMC 1-23-O was putatively identified as E. coli based on colony color and morphology but later identified as S. marcescens as described below. The isolate was screened for antibiotic resistance by plating it onto nutrient agar amended with gentamicin (10 mg/liter), streptomycin (10 mg/liter), kanamycin (30 mg/liter), tetracycline (30 mg/liter), chloramphenicol (30 mg/liter), or HgCl2 (10 mg/liter).
Competent E. coli JM109 (Promega, Madison, WI) was used in all transformations, and pBluescript II SK(−) was used as the cloning vector (Stratagene, La Jolla, CA). When appropriate, E. coli were selected in Luria-Bertani (LB) medium using the antibiotics listed above or ampicillin (100 mg/liter).
DNA techniques.
Restriction enzymes and DNA ligase (Promega), Taq DNA polymerase and DNA extraction kits (QIAGEN, Chatsworth, CA), and PfuTurbo DNA polymerase (Stratagene) were each used according to manufacturers' recommendations. PCR and sequencing primers (Table 2) were synthesized by MWG Biotech (High Point, NC). DNA sequencing was performed by the MCG Genomics Core Facility using an ABI Prism 377 XL DNA sequencer (Applied Biosystems, Foster City, CA).
TABLE 2.
PCR primers
| Primer | Sequence (5′→3′) (reference) |
|---|---|
| 519fta | GTT TCA GCM GCC GCG GTA ATW C (27) |
| 1390rta | GTT TGA CGG GCG GTG TGT RCA A (27) |
| EC-LAC-F1100 | AAA TCT AGA GTG AGC TGA TAC CGC TCG CC |
| EC-LAC-R817 | TTT TCT AGA AGC TGT TTC CTG TGT GAA ATT GTT |
| Tet-F2 | CCC TCT AGA GTT AGC ATA GGT CAG CGG ATC CCG |
| Tet-R2 | GGT CTA GAC CTA TAG ATA AGC CTG GTT TTA TGA TT |
| Tet-Prom-F1 | CCC TCT AGA ATG ATC AGA CCG ATG CCC ACC GCA |
| Tet-Prom-R1 | GGG TCT AGA GAT GTC ATC CAG TAA CGC CAG CGC |
Identification of FMC 1-23-O.
Two methods were used to identify FMC 1-23-O. Initial identification was achieved from the partial sequence of the 16S rRNA gene as described previously (27), following PCR with universal primers 519fta and 1390rta (Table 2). The DNA sequence of the FMC 1-23-O 16S rRNA gene was compared to sequences in the National Center for Biotechnology Information (NCBI) sequence database using the BLAST program (3). Conclusive identification of FMC 1-23-O as S. marcescens was provided by the MCG Clinical Microbiology Laboratory using biochemical assays contained in a MicroScan broth microdilution kit (Dade-Behring, West Sacramento, CA).
Construction of genomic library and cloning procedures.
To prepare a genomic library, FMC 1-23-O chromosomal DNA was purified, digested to completion with EcoRI, and ligated with EcoRI-digested pBluescript II SK(−). The ligation was used to transform E. coli JM109 (32), which was plated on LB agar containing ampicillin and tetracycline to select for transformants containing the cloned tetracycline resistance determinant.
Restriction mapping and deletion mapping of and transposon insertion into pEVM1.
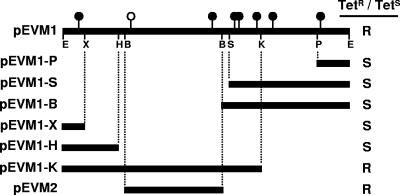
The plasmid from a single tetracycline-resistant transformant (pEVM1) was digested with six different restriction endonucleases (XhoI, HindIII, BamHI, SacI, KpnI, and PstI) (Fig. 1). The vector-containing fragments were purified, self-ligated, and used to transform E. coli JM109. The plasmids of the resulting subclones (i.e., the religated restriction fragment containing the vector) were purified, digested with the appropriate restriction enzyme, and analyzed to create a restriction map. The subclones were also tested for their abilities to confer tetracycline resistance and compared to the restriction map to localize the tetracycline resistance determinant.
FIG. 1.
A restriction map of the ∼13-kb pEVM1 insert containing the tetracycline resistance locus was created using the following enzymes: EcoRI (E), XhoI (X), HindIII (H), BamHI (B), SacI (S), KpnI (K), and PstI (P). Shaded bars below the map indicate DNA that is present in various deletion subclones of pEVM1, and the effect of the deletions on tetracycline resistance is shown in the right margin (R, resistance; S, susceptibility). Filled circles represent the sites of transposon insertions that did not disrupt tetracycline resistance; the single open circle represents a transposon insertion that disrupted tetracycline resistance. pEVM2 contains a subcloned BamHI fragment identified as containing the tetracycline resistance determinant.
In vitro transposon insertions into pEVM1 were accomplished with the EZ::TN transposon construction vector pMOD-2 (Epicenter, Madison, WI). A selectable transposon was created by subcloning the E. coli kanamycin resistance (aphA) cassette from pUC4K as a BamHI fragment, to yield the kanamycin-resistant transposon donor plasmid pMOD-Kan. Transposon insertions into pEVM1 were recovered by selection with ampicillin and kanamycin and subsequently tested for resistance or sensitivity to tetracycline.
To subclone the BamHI restriction fragment containing the tetracycline resistance determinant, pEVM1 was digested to completion with BamHI. The 4.2-kb pEVM1 BamHI fragment of interest was gel purified and ligated with BamHI-digested pBluescript II SK(−), and transformants of E. coli JM109 were selected with ampicillin and tetracycline. Subclone pEVM2 was subjected to restriction analysis and DNA sequencing to verify the presence of the expected insert.
DNA sequencing and protein and phylogenetic analyses.
The DNA sequences of portions of pEVM1, deletion plasmids thereof, and pEVM2 (Fig. 1) were determined on both strands using vector primers and primer walking (32). The sequence was analyzed for open reading frames (ORFs) using VectorNTI 7.0 (Invitrogen, Carlsbad, CA). Sequences of known tetracycline resistance proteins were obtained from the NCBI database and were aligned using ClustalW (40) of VectorNTI. A phylogenetic comparison of representative tetracycline resistance proteins was performed using the parsimony algorithm, and the statistical significance of the tree nodes was determined by bootstrap analysis (1,000 bootstrap replicates), both done using PAUP 4.0b10 (39). Prediction of transmembrane segments was achieved using the TMpred algorithm (17) of VectorNTI. The presence of helix-turn-helix motifs was evaluated by the method of Dodd and Egan (10) using the NPS@ server (8).
Regulation of tetA(41) and tetR(41) promoters by tetracycline.
The tetA(41)-tetR(41) intergenic region along with the tetR(41) repressor gene was amplified using PCR with high-fidelity PfuTurbo DNA polymerase, primers Tet-Prom-F1 and Tet-R2 (Table 2), and the following PCR conditions: 95°C for 2 min, followed by 30 cycles of 30 seconds of denaturation at 95°C, 30 seconds of annealing at 58.3°C, and a 2-min extension at 72°C, with a final extension reaction of 10 min at 72°C. The resulting 0.8-kb fragment was purified and digested with XbaI and then ligated with XbaI-digested pMW10 (41), a promoterless lacZ shuttle vector, which allows the screening of promoter activity by β-galactosidase activity (26). Transformants were recovered as blue colonies on LB-kanamycin plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and a subinhibitory concentration of tetracycline (0.5 μg/ml) reported to induce the expression of tetracycline resistance determinants (7).
To determine the β-galactosidase activities of lacZ fusions, all samples were grown in LB-kanamycin, with and without tetracycline (0.5 μg/ml). β-Galactosidase assays were performed as described previously (26). The optimal tetracycline concentration for induction of the tetA(41) promoter was determined by growing E. coli containing pEVM3 in various concentrations of tetracycline, ranging from 0 to 10 μg/ml, and measuring β-galactosidase activities. β-Galactosidase assays were repeated twice on different days, with each experiment being performed using samples in triplicate.
Nucleotide sequence accession numbers.
The S. marcescens tetA(41) tetracycline resistance locus and partial 16S rRNA gene sequences were deposited in GenBank under accession numbers AY264780 and EF210775, respectively.
RESULTS
Isolation of an S. marcescens tetracycline resistance gene.
In the course of studies investigating the role of indirect selection of antibiotic-resistant environmental organisms growing in contaminated streams at the U.S. Department of Energy Savannah River site near Aiken, SC, we isolated FMC 1-23-O, an environmental strain resistant to mercury and tetracycline. FMC 1-23-O was identified as Serratia marcescens using both 16S rRNA gene sequencing and diagnostic biochemical assays performed by the MCG clinical microbiology laboratory (data not shown). A single tetracycline-resistant clone, pEVM1, was isolated from a genomic library of FMC 1-23-O. Restriction mapping of pEVM1 revealed the presence of a 13-kb EcoRI insert, and several restriction sites were localized within this fragment (Fig. 1). The approximate location of the tetracycline resistance within the pEVM1 insert was determined using in vitro transposon mutagenesis. Eight independent transposon insertions were chosen, and the locations of the transposons were mapped using restriction analysis. The plasmids containing transposon insertions were also screened for resistance or sensitivity to tetracycline. Seven of the eight transposon insertions did not disrupt the pEVM1-encoded tetracycline resistance (Fig. 1). However, a single transposon insertion inactivated the tetracycline resistance phenotype, and this insertion mapped within a 4.4-kb BamHI-HindIII fragment (Fig. 1).
To further localize the region responsible for tetracycline resistance, we digested pEVM1 with each of the restriction enzymes mapped previously, each of which has a single site in pBluescript II. The vector-containing restriction fragments were self-ligated, and E. coli bacteria containing these deleted plasmids were screened for sensitivity or resistance to tetracycline (Fig. 1). Results from this experiment showed that the tetracycline resistance determinant was located on the same 4.4-kb BamHI-HindIII fragment implicated by transposon mutagenesis (Fig. 1).
To confirm the localization of the tetracycline resistance gene to the indicated 4.4-kb BamHI-HindIII fragment, a 4.2-kb BamHI fragment of pEVM1 (an additional BamHI site is present just to the right of the HindIII site implicated by deletion analysis as a boundary of the resistance-containing fragment) was subcloned to yield pEVM2. Resistance to tetracycline was conferred by pEVM2, confirming that the entire tetracycline resistance determinant was contained within this region.
Features of the tetracycline resistance locus.
The DNA sequence of the pEVM2 insert was determined using vector primers. Immediately adjacent to the leftmost BamHI site of pEVM2 (Fig. 1 and 2) was an ORF with substantial homology to bacterial tetracycline efflux pumps. Within this region were two divergently transcribed ORFs (Fig. 2). The larger ORF was 1,179 nucleotides in length, preceded by an intergenic region of 64 nucleotides, and followed by a second ORF of 639 nucleotides. The larger ORF predicted a basic 41-kDa protein (393 amino acids, pI 10.1) that was most similar to the tetracycline efflux protein Tet(39) from Acinetobacter sp. strain LUH5605 (GenBank accession no. AAW66497; 63% amino acid identity/78% similarity) (2). The protein was 43% identical/62% similar to the only previously known tetracycline resistance protein of S. marcescens, the Tet(B) protein of conjugative plasmid R478 (13). Because this predicted tetracycline resistance protein shared less than 80% amino acid identity with known tetracycline resistance alleles, it was given the allele number TetA(41) in accordance with current nomenclature of tetracycline resistance determinants (22). DNA sequence analysis of the single transposon insertion into pEVM1 that disrupted tetracycline resistance (Fig. 1) verified that the transposon had inserted within codon 351 of the larger ORF and that the interruption of this ORF resulted in tetracycline sensitivity. Together, these results indicated that the expression of TetA(41) was necessary and sufficient for the tetracycline resistance phenotype of pEVM1 and pEVM2.
FIG. 2.
Genetic organization of the tetA(41)-tetR(41) tetracycline resistance locus and its flanking regions. (Top) DNA sequence of the 64-bp tetA(41)-tetR(41) intergenic region. Putative start codons are underlined. Repeated DNA sequences suggestive of putative operator regions (OL and OR) for TetR(41) binding are designated by gray shading and arrows. (Middle and bottom) Diagrams showing the region flanking the tetA(41) locus in S. marcescens FMC 1-23-O, including orthologs of a putative P. aeruginosa carbon monoxide dehydrogenase (coxS, coxL, cccA), and Yersinia bglA and rpiR genes (middle). In sequenced Yersinia genomes, the location corresponding to tetA(41) is occupied by Mu-like phage genes (bottom) (only the Y. pestis CO92 genome is shown). The shaded box indicates the extent of homology between DNA cloned in pEVM1 and the Y. pestis CO92 genome.
The TetA(41) protein was compared to other representative tetracycline alleles in a phylogenetic analysis (data not shown). As predicted from BLAST searches, TetA(41) was most closely related to the Tet(39) allele of Acinetobacter, as well as to Tet(30) from Agrobacterium tumefaciens. Furthermore, TetA(41) was found within a cluster of proteins that comprise the group 1 family of tetracycline efflux proteins (7). Consistent with this observation, TetA(41) was predicted to have 12 transmembrane α-helices (located at amino acids 7 to 30, 43 to 65, 72 to 100, 94 to 117, 131 to 149, 159 to 179, 209 to 236, 239 to 259, 277 to 294, 300 to 321, 341 to 359, and 363 to 384)—properties also characteristic of group 1 efflux proteins (7).
The smaller ORF adjacent to and divergently transcribed from tetA(41) predicted a protein of 23.7 kDa (213 amino acids, pI 6.1). As with TetA(41) homologies, TetR(41) was most closely related to the TetR(39) protein of Acinetobacter (57% amino acid identity/73% similarity), as well as to numerous TetR orthologs in other bacteria. These proteins are involved in the regulation of the tetracycline efflux proteins such that the pump becomes highly upregulated in the presence of tetracycline. The putative S. marcescens TetA(41) repressor protein had a helix-turn-helix motif (amino acids 31 to 52) as well as several specific amino acid residues that are signatures of TetR family members (29), supporting its proposed role as a regulator of tetA(41) transcription (7).
The 64-nucleotide intergenic region between tetA(41) and tetR(41) contained two regions of imperfect dyad symmetry that are candidate operator sequences for TetR(41) binding (designated OL and OR for the left and right operators, respectively) (Fig. 2, top). These sequences had the consensus sequence CtCTAtCA(G/C)TGATAGAG (lowercase letters indicate nucleotides that do not match perfect dyad symmetry) and were separated by 20 nucleotides. These sequences closely matched the TetR binding sites that have been well studied in other TetR-regulated systems (29). Thus, the tetA(41) locus in S. marcescens resembled those in other bacteria encoding group 1 tetracycline efflux pumps, and its expression was inducible by the presence of tetracycline.
DNA sequence flanking the tetA(41) locus.
Because gram-negative, group 1 tetracycline efflux pumps are usually found on plasmids (7), we made numerous attempts to isolate plasmids from S. marcescens FMC 1-23-O. However, plasmids were not detected in this strain (data not shown). To gain further insight into the context of the tetracycline resistance determinant (i.e., chromosomal or plasmid), additional DNA sequencing was performed on the region of pEVM1 containing the S. marcescens FMC 1-23-O tetA(41) locus (Fig. 2). DNA sequencing of a total of 8.0 kb containing and adjacent to the S. marcescens tetA(41) locus revealed that the tetA(41) and tetR(41) genes were flanked by predicted regulatory and metabolic genes (Fig. 2, middle). Downstream of tetR(41) was a cluster of three genes orthologous to those comprising a putative Pseudomonas aeruginosa carbon monoxide dehydrogenase (coxS, coxL, and cccA). The DNA downstream of tetA(41) encoded two genes most closely related to genes from Yersinia pestis. Separated by 256 bp from tetA(41) was a predicted lacI family regulatory gene (rpiR), followed by the bglA gene, predicted to encode the catabolic enzyme 6-phospho-β-glucosidase. Interestingly, comparison of this region of pEVM1 with the corresponding regions from three sequenced Y. pestis genomes and one Y. pseudotuberculosis genome revealed that the tetA(41) locus was present in a position occupied in all four sequenced Yersinia genomes by clusters of genes constituting Mu-like bacteriophages (Fig. 2, bottom). Analysis of the intergenic regions flanking the tetA(41) tetracycline resistance locus [i.e., between bglA and rpiR, rpiR and tetA(41), and tetR(41) and coxS] did not reveal direct or inverted repeat sequences indicative of remnants of bacteriophages or transposons. Due to our inability to isolate plasmids, the presence of several metabolic genes, and the apparent lack of transmissible elements flanking the tetracycline resistance gene, we concluded that the tetA(41) locus is most likely found on the S. marcescens chromosome.
Regulation of tetA(41) and tetR(41) promoters by tetracycline.
Because the S. marcescens tetA(41) locus was similar to efflux pumps inducible by tetracycline, we examined the regulation of tetA(41) and tetR(41) expression in E. coli. The area of the tetA(41) locus encompassing the tetA(41)-tetR(41) intergenic region and the entirety of the tetR(41) gene was amplified and cloned in both orientations into the promoter probe vector pMW10 (41). This resulted in two plasmids, pEVM3 and pEVM4, containing transcriptional fusions of lacZ to the tetA(41) and tetR(41) promoters, respectively (Fig. 3A). These plasmids were used in β-galactosidase assays to determine the activities of these promoters in the presence or absence of a subinhibitory concentration (0.5 μg/ml) of tetracycline. In the absence of tetracycline, the tetA(41) promoter (in pEVM3) yielded minimal β-galactosidase activity (Fig. 3B), indicating nearly complete repression of this promoter. Without tetracycline, the tetR(41) promoter (in pEVM4) gave β-galactosidase activity approximately six times higher than that of the tetA(41) promoter. In the presence of tetracycline, however, the activities of both promoters were equivalent and substantially higher than in the absence of tetracycline (Fig. 3B), with the tetA(41) and tetR(41) promoters induced 27-fold and 4-fold, respectively. Therefore, like many other gram-negative efflux pumps, the tetA(41) system of S. marcescens was inducible by tetracycline.
FIG. 3.
Regulation of the tetA(41) and tetR(41) promoters by tetracycline. (A) Diagram of the lacZ fusions used to assay tetracycline regulation of the tetA(41) and tetR(41) promoters; (B) β-galactosidase activity of E. coli containing the tetA(41)- and tetR(41)-lacZ fusions [in the presence of the TetR(41) repressor protein] either in the presence (+) or absence (−) of 0.5 μg/ml tetracycline.
To determine the amount of tetracycline needed to induce the tetA(41) promoter, E. coli containing pEVM3 was incubated with various concentrations of tetracycline. Tetracycline concentrations less than 0.063 μg/ml did not induce the expression of the tetA(41) promoter (data not shown). Antibiotic concentrations of 0.125 μg/ml or higher caused the induction of β-galactosidase activity, with a maximum at 0.5 μg/ml tetracycline. Concentrations of tetracycline higher than 0.5 μg/ml did not result in greater expression of the tetA(41) promoter.
Does tetA(41) confer resistance to mercury?
S. marcescens FMC 1-23-O was originally isolated based on its resistance to mercury and subsequently was shown to be resistant to tetracycline as well. To determine whether both the resistance to mercury and that to tetracycline were conferred by the tetA(41) efflux pump, we plated E. coli containing either pEVM1, pEVM2, or pBluescript (control) on various inhibitory concentrations of mercury. Resistance to mercury was the same for each strain, indicating that tetA(41) was not responsible for the mercury resistance of strain FMC 1-23-O (data not shown).
DISCUSSION
Resistance to antimicrobials is a tremendous public health problem. While the misuse and overuse of antibiotics certainly selects for resistance in both pathogenic and commensal bacteria, there may be underappreciated environmental sources of indirect selection for antibiotic resistance. One possible source of indirect selection is exposure to heavy metal contamination, which is abundant throughout the world (5). Simultaneous selection of multiple resistances can be due to distinct resistances present on the same genetic element (coresistance) or to the same protein(s) mediating resistance to multiple compounds (cross-resistance). As a step in characterizing the possible indirect selection of antibiotic resistance by heavy metal contamination at the Savannah River site, we selected an environmental bacterium that was resistant to mercury. It was screened for resistance to several antibiotics and found to also be resistant to tetracycline.
We first showed by biochemical assays and by 16S rRNA gene sequencing that the environmental organism was S. marcescens. Tetracycline resistance is reportedly abundant in S. marcescens (6, 12, 15, 38), although with the exception of the tet(B) allele found on the broad-host-range conjugative plasmid R478 (13), the resistance determinants have not been characterized. In addition to tet(B), S. marcescens also contains tet(A), tet(C), and tet(E) alleles (7), including tet(E) found in S. marcescens isolated from a polluted aquatic environment (4).
The selection of pEVM1 showed that the S. marcescens determinant was expressed and able to confer tetracycline resistance to E. coli. Using a combination of deletion analyses, transposon mutagenesis, and subcloning, we identified the region of the cloned DNA that was responsible for tetracycline resistance. This region contained two genes, tetA(41) and tetR(41), that were orthologous to numerous genetic elements encoding tetracycline-inducible drug efflux pumps. The two components of such systems include the structural genes for the single protein efflux pump itself [tetA(41)] and for a repressor protein responsible for the tetracycline inducibility of the efflux pump [tetR(41)].
The TetA(41) protein is most similar to proteins comprising group 1 tetracycline efflux pumps and most closely related to the recently identified Tet(39) protein of Acinetobacter (2) and to Tet(30) of Agrobacterium tumefaciens (23). TetA(41) is clearly different from the only previously sequenced tetracycline determinant from Serratia [Tet(B) from plasmid R478], sharing only 43% amino acid identity. Consistent with the identification of this determinant as a group 1 member, the G+C content of the tetA(41) genes is 65.4%. This further shows that tetA(41) is a “gram-negative-like” resistance gene, as these typically have >40% G+C (7).
Despite the divergence of the TetA(41) and TetR(41) proteins being significant enough for them to constitute a novel tet allele, each protein has features known to be present in orthologous proteins. First, the size of the TetA(41) protein (41 kDa) is consistent with the sizes of known group 1 efflux proteins, which are ∼46 kDa (7). Second, as with group 1 efflux proteins, TetA(41) is predicted to have 12 α-helical transmembrane segments separated by alternating cytoplasmic and periplasmic loops. These transmembrane segments allow the formation of a pore through which tetracycline-cation complexes are extruded (7). Third, mutational studies of the TetA(C) efflux pump defined eight amino acid residues in cytoplasmic loops 2 to 3 and 10 to 11 that are involved in coupling drug transport to the bacterial proton motive force (25). Seven of eight of these are conserved in the TetA(41) protein [serine-65, aspartate-66, glycine-69, arginine-70, alanine-320, glycine-332, and glycine-336 of TetA(41)]. The eighth residue (alanine-323) is a conservative substitution from the serine at the analogous position in TetA(C). Furthermore, residues arginine-70 and aspartate-120 of Tet(B) were shown to be required for proper insertion of the transmembrane regions into the cytoplasmic membrane (34). These amino acids were also present at the corresponding positions in TetA(41), suggesting that these residues serve similar functions.
TetR and related proteins constitute a large family of bacterial transcriptional regulators (29). E. coli TetR contains a helix-turn-helix motif located at the amino terminus of the protein, and this has been shown by mutational and crystallographic studies to constitute the DNA binding region of this protein family (29). The DNA binding region consists of four α-helical segments (α1 to α4), with turns between α1 to α2 and α3 to α4 (29). Within α2 and α3, specific amino acid residues that are involved in DNA contact have been identified. Likewise, S. marcescens TetR(41) possesses a similar helix-turn-helix structure at its amino terminus (data not shown). Of the eight amino acids identified in DNA binding by other TetR family proteins, seven were conserved in TetR(41). Therefore, the mechanistic paradigm for DNA binding and repression by TetR proteins appears to occur with TetR(41) as well, and we have no reason to suggest that transcriptional regulation by TetR(41) is atypical.
The tetA(41) and tetR(41) genes are divergently transcribed, sharing a 64-nucleotide regulatory region. Within this region are two predicted operator regions, designated OL and OR (Fig. 2). In other tetracycline-regulated systems, similar operator sequences are bound by TetR proteins when tetracycline is absent. Mutational, biochemical, and crystallographic studies have identified points of interaction between E. coli TetR and its cognate operators (29) and defined a consensus TetR operator consisting of an imperfect palindromic 15-bp sequence (TCTATCATTGATAGG). Amino acid residues in the helix-turn-helix binding motif of TetR (described above) interact with the majority of the nucleotides within these operators. The putative palindromic OL and OR operators in the tetA(41)-tetR(41) intergenic region are slightly longer (17 bp) than those in E. coli but have 12 bp (OL) or 13 bp (OR) in common with the E. coli counterparts. The S. marcescens TetR(41) protein appears to have differing affinities for the OL and OR operators, demonstrated by the finding that the tetA(41) gene is more stringently repressed in the absence of tetracycline than is tetR(41). The putative tetA(41) OL and OR operators are not identical but have three nucleotide differences from each other, and these differences may account for variation in the levels of binding of TetR(41) to these operators.
The majority of antibiotic resistance genes are found on mobile genetic elements such as plasmids and transposons, facilitating the dissemination of resistances among diverse bacteria (7). However, certain tetracycline genes are not found on readily mobilizable elements. For example, tet(B) genes found in some Haemophilus, Moraxella catarrhalis, and Treponema denticola strains are chromosomal (7). Many tet(E) genes are found on nonconjugative and nonmobile plasmids (9, 35), and in numerous E. coli isolates the tet(E) gene was found to be chromosomal (19). These tet(E) alleles are predicted to be of more limited distribution (7) and, interestingly, are prevalent in aquatic environments and polluted marine sediments (4). In S. marcescens strain FMC 1-23-O, the tetA(41)-tetR(41) genes are flanked by predicted metabolic genes, including bglA (6-phospho-β-glucosidase), rpiR (lacI family repressor protein), and coxSL-cccA (carbon monoxide dehydrogenase). Combined with our inability to isolate or detect plasmids in this strain, this strongly suggests that the tetA(41) locus is found on the S. marcescens chromosome. However, we cannot rule out the possibility that both the predicted metabolic genes and the tetracycline resistance determinant are carried on a hypothetical low-copy-number plasmid that was undetected in our studies. The G+C content of each of the genes in the S. marcescens tetA(41) region [bglA, 60.0% G+C; rpiR, 54.0% G+C; tetA(41)-tetR(41), 65.4% G+C; and coxSL-cccA, 65.9% G+C] is not significantly different from that of the S. marcescens chromosome as a whole (58% G+C) (14). This suggests that these genes were not recently acquired from a bacterium with a different G+C content but rather have been long-term constituents of S. marcescens. It is interesting to note that the closest similarities of the bglA and rpiR genes flanking the left side of the tetA(41) locus are to orthologs in Yersinia. Immediately adjacent to the bglA and rpiR genes in all four sequenced Yersinia genomes are genes comprising predicted prophage genomes of Mu-like bacteriophages (Fig. 2). It is tempting to speculate, therefore, that the tetA(41) locus entered S. marcescens via transduction by a Mu-like bacteriophage. We did not detect sequence remnants of such a hypothetical transduction event.
One of the original questions pertaining to this project was whether exposure to heavy metals can indirectly select for antibiotic resistance in this strain. For example, both mercury and tetracycline resistances are found on the conjugative plasmid R478 (13), which is found in S. marcescens, and selection for either resistance can select for maintenance of the plasmid. Furthermore, some multidrug efflux pumps can mediate the removal of both tetracycline and toxic metals from the cell (7). Because S. marcescens FMC 1-23-O was resistant to both mercury and tetracycline, we cloned the tetracycline resistance gene in E. coli on pEVM1. However, pEVM1 did not confer mercury resistance to E. coli, showing that the mercury resistance was not due to the tetA(41) determinant, nor was the mercury resistance gene located on the ∼13-kb segment of DNA cloned in pEVM1. In consideration of these findings together with the probable chromosomal location of tetA(41), we therefore conclude that the mercury and tetracycline resistances of S. marcescens FMC 1-23-O do not represent either coresistance (two different resistances on the same mobile genetic element) or cross-resistance (a single determinant conferring both resistances) but rather independent selection of two separate resistances.
Acknowledgments
We thank Dawn Israel, Vanderbilt University, for the kind gift of pUC4K and Stuart Levy and Laura McMurry, Tufts University, for assistance in assigning proper nomenclature for the new tetracycline resistance protein. Craig Baker-Austin provided comments on a draft version of the manuscript.
This work was supported by a grant from the Medical College of Georgia Research Institute (to S.A.T.) and by financial award no. DE-FC09-96-SR18546 from the U.S. Department of Energy to the University of Georgia Research Foundation (to J.V.M.).
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Abbott, S. 1999. Klebsiella, Enterobacter, Citrobacter, and Serratia, p. 475-482. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, DC.
- 2.Agersø, Y., and L. Guardabassi. 2005. Identification of Tet 39, a novel class of tetracycline resistance determinant in Acinetobacter spp. of environmental and clinical origin. J. Antimicrob. Chemother. 55:566-569. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, S. R., and R. A. Sandaa. 1994. Distribution of tetracycline resistance determinants among gram-negative bacteria isolated from polluted and unpolluted marine sediments. Appl. Environ. Microbiol. 60:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker-Austin, C., M. S. Wright, R. Stepanauskas, and J. V. McArthur. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176-182. [DOI] [PubMed] [Google Scholar]
- 6.Carbonell, G. V., H. H. Della Colleta, T. Yano, A. L. Darini, C. E. Levy, and B. A. Fonseca. 2000. Clinical relevance and virulence factors of pigmented Serratia marcescens. FEMS Immunol. Med. Microbiol. 28:143-149. [DOI] [PubMed] [Google Scholar]
- 7.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 9.DePaola, A., and M. C. Roberts. 1995. Class D and E tetracycline resistance determinants in gram-negative bacteria from catfish ponds. Mol. Cell. Probes 9:311-313. [DOI] [PubMed] [Google Scholar]
- 10.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleisch, F., U. Zimmermann-Baer, R. Zbinden, G. Bischoff, R. Arlettaz, K. Waldvogel, D. Nadal, and C. Ruef. 2002. Three consecutive outbreaks of Serratia marcescens in a neonatal intensive care unit. Clin. Infect. Dis. 34:767-773. [DOI] [PubMed] [Google Scholar]
- 12.Fontana, R., G. Lo Cascio, M. Ligozzi, O. Friscia, and T. Oldoni. 2001. Antimicrobial susceptibility of respiratory isolates of Enterobacteriaceae and Staphylococcus aureus in Italy: incidence and trends over the period 1997-1999. Eur. J. Clin. Microbiol. Infect. Dis. 20:854-863. [DOI] [PubMed] [Google Scholar]
- 13.Gilmour, M. W., N. R. Thomson, M. Sanders, J. Parkhill, and D. E. Taylor. 2004. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52:182-202. [DOI] [PubMed] [Google Scholar]
- 14.Grimont, F., and P. A. D. Grimont. 1992. The genus Serratia, p. 2822-2848. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 3. Springer-Verlag, New York, NY. [Google Scholar]
- 15.Haddy, R. I., B. L. Mann, D. D. Nadkarni, R. F. Cruz, D. J. Elshoff, F. C. Buendia, T. A. Domers, and A. M. Oberheu. 1996. Nosocomial infection in the community hospital: severe infection due to Serratia species. J. Fam. Pract. 42:273-277. [PubMed] [Google Scholar]
- 16.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10-encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 18.Johnson, J. S., J. Croall, J. S. Power, and G. R. Armstrong. 1998. Fatal Serratia marcescens meningitis and myocarditis in a patient with an indwelling urinary catheter. J. Clin. Pathol. 51:789-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, C., B. E. Langlois, and K. A. Dawson. 1993. Detection of tetracycline resistance determinants in pig isolates from three herds with different histories of antimicrobial agent exposure. Appl. Environ. Microbiol. 59:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy, S. B. 2002. The 2000 Garrod lecture. Factors impacting on the problem of antibiotic resistance. J. Antimicrob. Chemother. 49:25-30. [DOI] [PubMed] [Google Scholar]
- 21.Levy, S. B. 2001. Antibiotic resistance: consequences of inaction. Clin. Infect. Dis. 33(Suppl. 3):S124-S129. [DOI] [PubMed] [Google Scholar]
- 22.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, Z. Q., and S. K. Farrand. 1999. Cloning and characterization of a tetracycline resistance determinant present in Agrobacterium tumefaciens C58. J. Bacteriol. 181:618-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manfredi, R., A. Nanetti, M. Ferri, and F. Chiodo. 2000. Clinical and microbiological survey of Serratia marcescens infection during HIV disease. Eur. J. Clin. Microbiol. Infect. Dis. 19:248-253. [DOI] [PubMed] [Google Scholar]
- 25.McNicholas, P., I. Chopra, and D. M. Rothstein. 1992. Genetic analysis of the tetA(C) gene on plasmid pBR322. J. Bacteriol. 174:7926-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Orphan, V. J., L. T. Taylor, D. Hafenbradl, and E. F. Delong. 2000. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 66:700-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostrowsky, B. E., C. Whitener, H. K. Bredenberg, L. A. Carson, S. Holt, L. Hutwagner, M. J. Arduino, and W. R. Jarvis. 2002. Serratia marcescens bacteremia traced to an infused narcotic. N. Engl. J. Med. 346:1529-1537. [DOI] [PubMed] [Google Scholar]
- 29.Ramos, J. L., M. Martinez-Bueno, A. J. Molina-Henares, W. Teran, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 31.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412-416. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Schaberg, D. R., D. H. Culver, and R. P. Gaynes. 1991. Major trends in the microbial etiology of nosocomial infection. Am. J. Med. 91:72S-75S. [DOI] [PubMed] [Google Scholar]
- 34.Someya, Y., T. Kimura-Someya, and A. Yamaguchi. 2000. Role of the charge interaction between Arg(70) and Asp(120) in the Tn10-encoded metal-tetracycline/H(+) antiporter of Escherichia coli. J. Biol. Chem. 275:210-214. [DOI] [PubMed] [Google Scholar]
- 35.Sorum, H., M. C. Roberts, and J. H. Crosa. 1992. Identification and cloning of a tetracycline resistance gene from the fish pathogen Vibrio salmonicida. Antimicrob. Agents Chemother. 36:611-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stepanauskas, R., T. C. Glenn, C. H. Jagoe, R. C. Tuckfield, A. H. Lindell, C. J. King, and J. V. McArthur. 2006. Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 8:1510-1514. [DOI] [PubMed] [Google Scholar]
- 37.Stepanauskas, R., T. C. Glenn, C. H. Jagoe, R. C. Tuckfield, A. H. Lindell, and J. V. McArthur. 2005. Elevated microbial tolerance to metals and antibiotics in metal-contaminated industrial environments. Environ. Sci. Technol. 39:3671-3678. [DOI] [PubMed] [Google Scholar]
- 38.Stock, I., T. Grueger, and B. Wiedemann. 2003. Natural antibiotic susceptibility of strains of Serratia marcescens and the S. liquefaciens complex: S. liquefaciens sensu stricto, S. proteamaculans and S. grimesii. Int. J. Antimicrob. Agents 22:35-47. [DOI] [PubMed] [Google Scholar]
- 39.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), ed. 4.0. Sinauer Associates, Sunderland, MA.
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wösten, M. M., M. Boeve, M. G. Koot, A. C. van Nuene, and B. A. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]