Abstract
Anoxygenic photosynthesis based on Fe(II) is thought to be one of the most ancient forms of metabolism and is hypothesized to represent a transition step in the evolution of oxygenic photosynthesis. However, little is known about the molecular basis of this process because, until recently (Y. Jiao and D. K. Newman, J. Bacteriol. 189:1765-1773, 2007), most phototrophic Fe(II)-oxidizing bacteria have been genetically intractable. In this study, we circumvented this problem by taking a heterologous-complementation approach to identify a three-gene operon (the foxEYZ operon) from Rhodobacter sp. strain SW2 that confers enhanced light-dependent Fe(II) oxidation activity when expressed in its genetically tractable relative Rhodobacter capsulatus SB1003. The first gene in this operon, foxE, encodes a c-type cytochrome with no significant similarity to other known proteins. Expression of foxE alone confers significant light-dependent Fe(II) oxidation activity on SB1003, but maximal activity is achieved when foxE is expressed with the two downstream genes foxY and foxZ. In SW2, the foxE and foxY genes are cotranscribed in the presence of Fe(II) and/or hydrogen, with foxZ being transcribed only in the presence of Fe(II). Sequence analysis predicts that foxY encodes a protein containing the redox cofactor pyrroloquinoline quinone and that foxZ encodes a protein with a transport function. Future biochemical studies will permit the localization and function of the Fox proteins in SW2 to be determined.
Oxygenic photosynthesis, the biological process by which water is oxidized to molecular oxygen (O2) using solar energy with the concomitant fixation of inorganic carbon, has had a profound impact on the biology and chemistry of the Earth; however, its origin remains enigmatic (7, 21, 31). Lipid biomarker and stratigraphic geochemical analyses of stromatolite assemblages may date the existence of the first cyanobacteria to ∼2.7 billion years ago (Ga) (9, 11); however, whether this is indeed the case is debatable (12, 18). Recent phylogenetic studies using several photosynthesis-related genes suggest that the anoxygenic form of photosynthesis evolved before the oxygenic form, with the purple photosynthetic group representing the most ancient taxon (53). In addition, the structural and biophysical similarities between the reaction centers of purple phototrophs and the oxygen-evolving reaction center (PSII) of cyanobacteria are hypothesized to reflect a common ancestry (41, 43). Thus, it has been proposed that PSII evolved from the reaction center of purple bacteria via transitional stages involving reaction centers able to accept electrons from compounds such as Fe(II), Mn-bicarbonate clusters or H2O2 (5, 8, 19, 40). Fe(II) is thought to have been abundant in the ancient ocean, making it likely that Fe(II)-oxidizing organisms evolved early on (15, 50). Further, today Fe(II) oxidation is performed by phylogenetically diverse organisms including purple and green phototrophic bacteria—a phylogenetic distribution that may reflect the antiquity of this form of photosynthesis (14, 15). In this context, understanding the molecular mechanisms by which anoxygenic bacteria oxidize Fe(II) phototrophically is of evolutionary interest.
Although first discovered over a decade ago, the majority of Fe(II)-oxidizing phototrophs are not currently amenable to genetic analyses and little is known about how they oxidize Fe(II) (14, 52). Recent studies of the newly isolated strain Rhodopseudomonas palustris TIE-1, where the ability to generate mutants has been developed, revealed that a homolog of a cobalt chelatase and an integral membrane protein that is likely a component of an ABC transport system are required for photoautotrophic Fe(II) oxidation (26). The roles of these proteins in Fe(II) oxidation by R. palustris TIE-1 remain unclear, but given their lack of redox cofactor binding motifs, it seems unlikely that they are involved directly in the transfer of electrons from Fe(II). In the companion paper to this article, we report the discovery of a three-gene operon from TIE-1, designated the pio operon; deletion of this operon leads to a specific growth defect on Fe(II). One of the genes in this operon, pioA, encodes a c-type cytochrome that is predicted to function in the periplasm of this strain as the Fe(II) oxidoreductase (27). Several years ago, at least two c-type cytochromes were found to have increased expression in Rhodomicrobium vannielii strain BS-1 during phototrophic growth on Fe(II), but whether these cytochromes were specific to or required for Fe(II) oxidation was not determined (25).
Beyond serving as an electron donor for photosynthesis, Fe(II) can support the growth of both aerobic and anaerobic chemolithoautotrophs (reference 14 and references therein). Yet, very little is understood about how Fe(II) oxidation works at the molecular level for these organisms either. To date, our deepest understanding of microbial Fe(II) oxidation comes from genetic and biochemical studies of Acidithiobacillus ferrooxidans, a bacterium that couples Fe(II) oxidation to the reduction of O2 at low pH. Several proteins, including the blue copper protein rusticyanin (13), a high-redox potential Fe-S protein (34), an outer membrane porin (37), and several types of cytochromes (4, 36, 48, 54, 55) have been implicated in the enzymatic oxidation of Fe(II) by A. ferrooxidans. How these proteins work together, however, is uncertain and may differ strain by strain (4, 34, 55). Nevertheless, in A. ferrooxidans strain ATCC 33020, the protein proposed to be the primary acceptor for electrons from Fe(II), Cyc2, is a c-type cytochrome. In A. ferrooxidans strain ATCC 33020, this cytochrome is known to be localized to the outer membrane (4, 55), which stands in contrast to the predicted periplasmic localization of PioA in TIE-1 (27). Thus, while knowledge of the mechanism of Fe(II) oxidation by aerobic acidophiles can inform our studies of anoxygenic phototrophs, the mechanism of Fe(II) oxidation between these organisms is likely to be different given that anoxygenic phototrophs oxidize Fe(II) under anaerobic conditions at neutral pH, where the ferric iron product of this metabolism is a mineral, whereas Acidithiobacillus grows in aerobic acidic environments, where the ferric iron product is soluble.
Here, we used a heterologous complementation approach to identify genes involved in phototrophic Fe(II) oxidation in the genetically intractable Fe(II)-oxidizing photosynthetic bacterium Rhodobacter sp. strain SW2. We identified the foxEYZ operon—a three-gene operon from SW2 that confers enhanced light-dependent Fe(II) oxidation activity to Rhodobacter capsulatus SB1003.
MATERIALS AND METHODS
Strains, vectors, and growth conditions.
The bacterial strains, plasmids, and cosmids used or constructed in this study are described in Table 1. For aerobic growth of R. capsulatus SB1003 (SB1003), YP medium was used (51). For phototrophic growth of SB1003 and Rhodobacter sp. strain SW2 (SW2), a previously described anoxic minimal-salts medium for freshwater cultures (pH 6.8) was used (20). As substrates for phototrophic growth, H2 was provided as a headspace of 80% H2/20% CO2, and acetate was added to a final concentration of 10 mM.
TABLE 1.
Strains, cosmids, and plasmids used in this study
| Strain or plasmid | Genotype, markers, characteristics, and uses | Source and/or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli WM3064 | Donor strain for conjugation; thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::[erm pir(wt)] | W. Metcalf (University of Illinois, Urbana-Champaign) |
| E. coli DH10β | Host for E. coli cloning; F−mcrA Δ(mrr-hsdRMS-mcrBC) Δ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK rpsL nupG (Strr) | Invitrogen (Carlsbad, CA) |
| R. capsulatus SB1003 | rif-10 | R. Haselkorn (University of Chicago); 56 |
| Rhodobacter sp. strain SW2 | Wild type | F. Widdel (MPI, Bremen, Germany); 20 |
| Cosmids and plasmids | ||
| pLAFR5 | 21.5-kb broad-host-range cosmid cloning vector derivative of pLAFR3; ori RK2 (TcrlacZα) | 30 |
| 9E12 | Contains SW2 genomic DNA cloned into BamHI site of pLAFR5 that confers Fe(II) oxidation activity to SB1003 | This work |
| pBBR1MCS5 | Gmr derivative of pBBR1 | 33 |
| pP3-gm1 | Contains ∼9.4-kb PstI fragment from 9E12 in pBBR1MCS5 (Gmr) | This work |
| pP3-gm2 | Contains ∼9.4-kb PstI fragment from 9E12 in pBBR1MCS5 (Gmr); insert is cloned in opposite orientation to that of pP3-gm1 | This work |
| pBBR1MCS2 | Knr derivative of pBBR1 | 33 |
| pAK20 | Derivative of pBBR1MCS2 (Knr); inserts can be expressed from tac promoter fused to GFPa | A. Komeili (University of California Berkeley); 32 |
| pfoxEYZ | Contains 3,235-bp PCR product derived from 9E12 containing foxE, foxY, and foxZ genes cloned into EcoRI/SpeI sites of pAK20 | This work |
| pfoxE | Contains 1,273-bp PCR product derived from 9E12 containing foxE gene cloned into EcoRI/SpeI sites of pAK20 | This work |
| pfoxY | Contains 1,351-bp PCR product derived from 9E12 containing foxY gene cloned into EcoRI/SpeI sites of pAK20 | This work |
| pfoxZ | Contains 1,290-bp PCR product derived from 9E12 containing foxZ gene cloned into EcoRI/SpeI sites of pAK20 | This work |
GFP, green fluorescent protein.
To test SB1003 for the ability to grow phototrophically on insoluble Fe(II) alone or in the presence of added cosubstrates, this strain was first grown phototrophically on H2 and then transferred to fresh phototrophic medium containing the following substrates for growth: (i) 9 mM Fe(II)Cl2·H2O, (ii) 9 mM Fe(II)Cl2·H2O plus 0.1% yeast extract, and (iii) 9 mM Fe(II)Cl2·H2O plus H2. To test SB1003 for the ability to grow phototrophically on Fe(II) under a condition where both the Fe(II) substrate and the Fe(III) product of this metabolism remained dissolved, SB1003 was grown phototrophically on H2 and transferred to fresh phototrophic medium containing 3 to 4 mM Fe(II)Cl2·H2O plus 5 mM nitrilotriacetic acid (NTA). These concentrations of Fe(II) and NTA were used because higher concentrations of NTA were toxic to SB1003 and at this concentration of NTA, higher concentrations of Fe(II) resulted in Fe(III) mineral precipitation as bacterial Fe(II) oxidation progressed. To test SB1003 carrying the Fe(II) oxidation activity-conferring cosmids for the ability to grow phototrophically on Fe(II), these strains were first grown phototrophically on H2 and then transferred to fresh phototrophic medium containing Fe(II)Cl2·H2O as the sole electron donor. Concentrations of both 4 mM and 9 mM Fe(II)Cl2·H2O were tested for these cosmid-carrying stains. In all of these cultures, Fe(II) oxidation was used as a proxy for growth and was monitored via the ferrozine assay (46).
Phototrophically grown cultures of SB1003 and SW2 were incubated under continuous illumination ∼30 cm from a 34-W incandescent light at 30°C and 16°C, respectively. Luria-Bertani (LB) medium was used for routine culturing of Escherichia coli strains DH10β and WM3064 at 37°C. A 0.2 mM concentration of diaminopimelic acid was added to permit growth of WM3064 cultures. Antibiotic concentrations were as follows. For SB1003, 1 μg/ml tetracycline (TC), 5 μg/ml kanamycin, and 3 μg/ml gentamicin were used. For E. coli strains, 15 μg/ml TC, 50 μg/ml kanamycin, and 20 μg/ml gentamicin were used. Phototrophically grown cultures supplemented with TC were incubated behind UV light filters to minimize light-mediated degradation of this drug (17).
Rhodobacter sp. strain SW2 genomic cosmid library construction.
Genomic DNA was isolated from SW2 according to standard protocols (16). Plasmid and cosmid DNA was purified with QIAGEN Mini and Maxi Kits, respectively. After purification, the cosmid vector pLAFR5 (30) was digested sequentially with ScaI and BamHI. SW2 genomic DNA was partially digested with Sau3AI, dephosphorylated, and ligated with digested pLAFR5 at a 9:1 molar ratio of insert to vector in the presence of 5 mM ATP. The ligation was packaged into recombinant λ phage with the Stratagene Gigapack III XL packaging extract, and E. coli WM3064 was infected with the resultant phage lysate. Cosmids from WM3064 were transferred to SB1003 via conjugation. Selection against the donor strain was achieved by omitting diaminopimelic acid from the medium. The resulting library contained 1,536 clones with an average insert size of 23.5 kb. On the basis of comparison to the genome sizes of R. sphaeroides and R. capsulatus, which are ∼4.5 and 3.6 Mb, respectively, we estimate that our library represents five- to six-times coverage of the SW2 genome (24, 35).
Cell suspension assay for Fe(II) oxidation activity.
All cell suspension assay mixtures were prepared and conducted at room temperature in an anaerobic chamber containing an atmosphere of 5% H2/80% N2/15% CO2. A light intensity of ∼500 lx from an incandescent bulb was used for light-incubated assay mixtures. Our initial screening to identify cosmids able to enhance Fe(II) oxidation was an endpoint assay. Here, SB1003 transconjugants were grown photoautotrophically on H2 in 96-well plates, washed once with an anoxic buffer containing 50 mM HEPES and 20 mM NaCl at pH 7 (assay buffer) and resuspended in 100 μl of assay buffer containing ∼0.2 mM Fe(II)Cl2·H2O and 20 mM NaHCO3. After an ∼20-h incubation in the light, the concentration of Fe(II) remaining in each clone-containing well was measured by adding 100 μl of ferrozine solution (0.1% [wt/vol] ferrozine in 50% [wt/vol] ammonium acetate solution) (46). Cosmids from clones that showed less purple color than the negative control, SB1003/pLAFR5, were purified and moved into a clean SB1003 genetic background, and the resultant strains were retested quantitatively for their Fe(II) oxidation activity through a time course cell suspension assay.
All time course cell suspension assay mixtures contained the same number of cells. To prepare these assay mixtures, ∼50 ml of an early-exponential-phase culture was harvested by centrifugation, washed once with an equal volume of assay buffer, resuspended to a final optical density at 570 nm (OD570) of ∼0.3 in assay buffer containing 0.5 mM Fe(II)Cl2·H2O and 20 mM NaHCO3 and dispensed in 300-μl aliquots to a 96-well plate. For each Fe(II) measurement, 10 μl of cell suspension was transferred to 90 μl of 1 M HCl. A 100-μl volume of ferrozine solution was added, and the OD570 was read after 10 min. Samples for total Fe measurements were diluted 1/10 with a solution of hydroxylamine hydrochloride (10% [wt/vol] in 1 M HCl) and incubated at 65°C overnight to facilitate the reductive dissolution of Fe(III) precipitates. One-hundred-microliter volumes of these samples were combined with 100 μl of ferrozine solution, and the OD570 was read after 10 min. Fe concentration measurements were corrected for cell interference, and rates of Fe(II) oxidation were calculated through the initial linear portion of the Fe(II) oxidation curves.
Cloning, sequencing, annotation, and reverse transcriptase (RT)-PCR.
Cosmid 9E12 was digested with PstI, and the subsequent restriction fragments were gel purified, ligated with PstI-digested pBBR1MCS5 (33), and transformed into E. coli DH10β. SB1003 strains with representative plasmids containing the correct-size insert in both transcriptional orientations were constructed and tested for light-dependent Fe(II) oxidation activity.
Shotgun cloning and sequencing of P3 were performed by Laragen (Los Angeles, CA). To annotate the sequence, an initial set of putative open reading frames (ORFs) was identified with ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Initial functional assignments and homology identifications were made by comparison of the translated ORFs to proteins in the BLAST database with BlastP (http://www.ncbi.nlm.nih.gov/BLAST/). Additional protein analyses (e.g., membrane-spanning domains, subcellular localization, and motif identification) were performed with the tools on the ExPASy proteomics server (http://us.expasy.org/). Predicted operons, promoters, and terminators were identified with the tools at Softberry (http://www.softberry.com/berry.phtml). The R. capsulatus genome was accessed at the Integrated Genomics website (http://www.integratedgenomics.com/).
The insert fragments of pfoxE, pfoxY, pfoxZ, and pfoxEYZ were generated by PCR amplification from 9E12 with Mix D from the FailSafe PCR System by Epicenter and the primers listed in Table 2. Primers were designed with SpeI or MfeI and PstI restriction sites to facilitate cloning. PCR products were digested with SpeI and MfeI and ligated into EcoRI- and SpeI-digested pAK20 (32) to create pfoxE, pfoxY, pfoxZ, and pfoxEYZ. After insert sequence verification, versions of strain SB1003 carrying these plasmids were constructed and tested for Fe(II) oxidation activity.
TABLE 2.
Oligonucleotides used in this studya
| Oligonucleotide name | Description | Sequence (5′-3′) |
|---|---|---|
| 9 | Forward primer used to amplify pfoxE insert | cgcccaattgctgcagTTATTGCGCCCTTTTTCATC |
| 10 | Reverse primer used to amplify pfoxE insert | ggcgactagtAGGCGATCCTCACCATAGG |
| 13 | Forward primer used to amplify pfoxY insert | cgcccaattgctgcagTCCACGAGCTGAACCTGAC |
| 14 | Reverse primer used to amplify pfoxY insert | ggcgactagtGTTATAGGTGGCGGTTGCTG |
| 17 | Forward primer used to amplify pfoxZ insert | cgcccaattgctgcagTCAGACCACCGATTACGACA |
| 18 | Reverse primer used to amplify pfoxZ insert | ggcgactagtGGTTTGAGTTTGAGGCAGGA |
| 21 | Forward primer used to amplify pfoxEYZ insert | cgcccaattgctgcagAGGAAGTGCTGACCGACATC |
| 18 | Reverse primer used to amplify pfoxEYZ insert | ggcgactagtGGTTTGAGTTTGAGGCAGGA |
| 1 | Used for RT-PCR | AAGGTGTTCCAGCACCTGAC |
| 2 | Used for RT-PCR | GGCATAGGCGATGATGGTAT |
| 3 | Used for RT-PCR | CCTTTGACGGCAAGCTTTAT |
| 4 | Used for RT-PCR | GTGATCACCTTGACCAGCAG |
| 5 | Used for RT-PCR | CGATCAAGGAATGGATCCTG |
| 6 | Used for RT-PCR | GGCACACCGATCTGAATCTT |
| 7 | Used for RT-PCR | CGAGTTGTGGAGCTTTTACG |
| 8 | Used for RT-PCR | CAGGGCGTTGGAGAAGAAC |
| 54 | Forward primer used to verify transcription of foxZ from pfoxZ in SB1003 | TTTCATCAACTCGCAACTGG |
| 55 | Reverse primer used to verify transcription of foxZ from pfoxZ in SB1003 | ATAAAGCTTGCCGTCAAAGG |
| 29 | Forward primer used to verify transcription of foxY from pfoxY in SB1003 | GACCCTGTGCTATGCCATC |
| 51 | Reverse primer used to verify transcription of foxY from pfoxY in SB1003 | GGCACCTGGTAGTTCGACAG |
Nucleotides in lowercase denote engineered restriction site sequences. Oligonucleotides 9, 13, 17, and 21 have MfeI and PstI restriction sites at the 5′ end. Oligonucleotides 10, 14, and 18 have an SpeI restriction site at the 5′ end.
For RT-PCR experiments, total RNA was extracted from H2-grown SW2 cells and H2-grown SW2 cells incubated in the light in assay buffer containing 0.5 mM Fe(II)Cl2·H2O and 20 mM NaHCO3 for ∼30 min (Fe-induced cells) with the RNeasy Protect Bacteria Mini Kit (QIAGEN). cDNA was synthesized with the Bio-Rad iScript cDNA Synthesis Kit. Control reaction mixtures contained no reverse transcriptase. These cDNAs, as well as genomic DNA from SW2, served as templates for PCR amplification with the primers described in Table 2. The same cDNA template was used for the data presented in Fig. 5B and C, and the same cDNA template was used for the data presented in Fig. 5D and E.
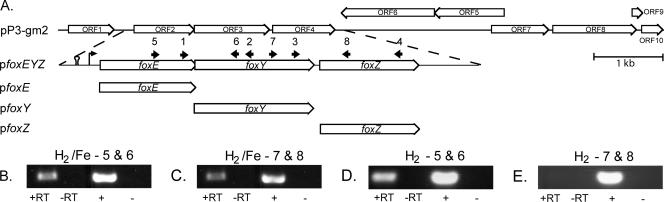
FIG. 5.
In SW2, foxE, foxY, and foxZ are cotranscribed in the presence of Fe(II). (A) Cloning strategy to identify foxE, foxY, and foxZ, genes from SW2 that enhance light-dependent Fe(II) oxidation activity in SB1003. Numbered arrows identify the positions of the oligonucleotides used for RT-PCR experiments (described in Table 2). The 1-kb scale bar applies to the P3-gm2 segment only. With total RNA extracted from H2-grown, Fe(II)-induced SW2 as a template for cDNA synthesis and oligonucleotides 5 and 6 (B) and 7 and 8 (C), PCR products were obtained for both of the regions between the fox genes (+RT lanes). With total RNA extracted from H2-grown SW2 as a template for cDNA synthesis and oligonucleotides 5 and 6 (D) and 7 and 8 (E), a PCR product was obtained for the region between foxE and foxY but not for that between foxY and foxZ. Additional experiments with oligonucleotides 1, 2, 3, and 4 confirmed the results in panels B, C, D, and E (data not shown). −RT lanes, controls with no reverse transcriptase added to the cDNA reaction mixtures; + lanes, controls with SW2 genomic DNA; − lanes, no-template controls.
To confirm expression of the pfoxY and pfoxZ inserts, RT-PCR was performed on total RNA extracted from YP medium-grown cultures of SB1003 carrying these plasmids with primers integral to these two genes. A product was obtained in both cases (data not shown).
Biochemical methods.
For total protein extraction, cells from 1-liter cultures of acetate- or H2-grown SW2 and H2-grown SB1003/pfoxE and SB1003/ pBBRMCS2 were harvested in exponential phase by centrifugation and washed once with assay buffer. Because the ferric precipitates that form during phototrophic growth of SW2 on Fe(II) alone preclude the harvesting of cells for protein extraction, to approximate Fe(II)-growth conditions and obtain sufficient cells for total protein extraction, cells from a 1-liter culture of H2-grown SW2 were harvested, washed with assay buffer, and resuspended in 20 ml of assay buffer containing 2 mM Fe(II)Cl2·H2O and 20 mM NaHCO3. This suspension was incubated in the light until all of the Fe(II) was oxidized, and cells were then harvested by centrifugation. Cell pellets from all conditions were resuspended in 3 ml of assay buffer to which a protease inhibitor cocktail (EDTA-free Complete; Roche) was added. Cells were lysed via passage three times though a French pressure cell at 16,000 lb/in2. DNase (Sigma) was added to the extracts, followed by a 30-min incubation on ice. Cell debris and unbroken cells were removed by low-speed centrifugation (10,000 × g for 20 min). Crude membrane and soluble fractions were separated by ultracentrifugation (200,000 × g for 90 min), and the membrane pellet was resuspended in assay buffer. Protein concentration was measured with the Bio-Rad protein assay. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed by standard procedures according to the Laemmli method (2). Cytochrome c bands were detected according to the in-gel peroxidase activity assay of Francis and Becker (22). Bands of interest were cut from the heme-stained gels and submitted to the Protein/Peptide MicroAnalytical Laboratory/Facility at Caltech for liquid chromatography-tandem mass spectrometry analysis.
Nucleotide sequence accession number.
The sequence of the fox operon has been deposited in GenBank under accession number DQ381537.
RESULTS
Identification of four cosmids that enhance light-dependent Fe(II) oxidation activity in R. capsulatus SB1003.
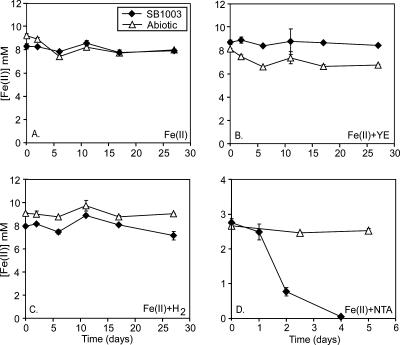
The Fe(II)-oxidizing phototroph Rhodobacter sp. strain SW2 is not amenable to traditional genetic analysis (14) but is closely related to R. capsulatus SB1003, which is genetically tractable (56). SB1003 is not able to grow photoautotrophically on Fe(II) alone (Fig. 1A) or in the presence of additional substrates such as 0.1% yeast extract (Fig. 1B) or H2 (Fig. 1C). However, in the presence of Fe(II)-NTA, both Fe(II) oxidation (Fig. 1D) and growth occur (data not shown); NTA does not support phototrophic growth in the absence of Fe(II) (data not shown). Therefore, to identify genes from SW2 that conferred Fe(II) oxidation activity, we expressed a genomic cosmid library of SW2 in SB1003 under conditions where Fe(II) oxidation by SB1003 was kinetically slow. Under the conditions used, SW2 oxidized, on average, sixfold more Fe(II) than SB1003. This difference in activity could be clearly visualized via our colorimetric endpoint ferrozine assay allowing for the identification of putative SB1003 clones with enhanced Fe(II) oxidation activity (Fig. 2).
FIG. 1.
Fe(II) oxidation activity by R. capsulatus SB1003 under different conditions. Cultures of SB1003 containing Fe(II)Cl2 as the sole electron donor show no Fe(II) oxidation over a 27-day period (A). Addition of 0.1% yeast extract (YE) or H2 as a cosubstrate does not stimulate Fe(II) oxidation by SB1003 (B and C, respectively), as has been found for other R. capsulatus strains (20, 25). When SB1003 is inoculated into medium containing Fe(II)Cl2 plus NTA, however, rapid and complete oxidation of Fe(II) is observed (D). The data in panels A, B, and C are representative of multiple independent cultures, and the error bars are the standard deviation for triplicate ferrozine assays. Data shown in panel D are the average of three independent SB1003 cultures, and error bars represent the standard deviation of this average. The symbol definitions in panel A apply to all of the graphs.
FIG. 2.
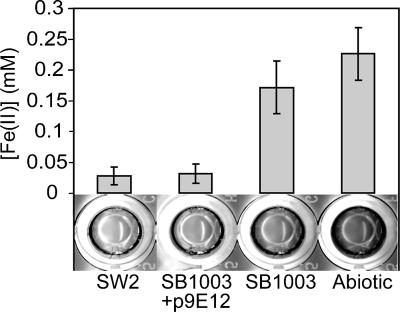
Screen for enhanced Fe(II) oxidation activity in SB1003. After a ∼20-h incubation in assay buffer containing 0.2 mM Fe(II), SW2 oxidizes an average of about sixfold more Fe(II) than SB1003. This difference in Fe(II) oxidation is clearly visualized upon the addition of the colorimetric reagent ferrozine, where wells containing Fe(II) turn purple and wells with no Fe(II) remain clear. Likewise, during the screen, Fe(II)-oxidizing clones like SB1003/9E12 are identified as clear wells upon addition of ferrozine.
In our screening of 1,536 cosmid-carrying SB1003 clones, 4 clones with enhanced Fe(II) oxidation activity were identified. To further characterize this activity, Fe(II) and total Fe concentrations were followed for 3 h in cell suspensions of these strains incubated in the light and in the dark. This shortened incubation time, compared to that of the screening (∼20 h), magnified the differences in Fe(II) oxidation activity between the putative clones and the control strains. As shown by the representative clone, SB1003/9E12, there is an ∼1,000-fold increase in the amount of Fe(II) oxidized after 3 h in light-incubated SB1003/9E12 cell suspensions compared to control strain SB1003/pLAFR5 (Fig. 3A). In terms of rate, in the light-incubated cell suspensions of SB1003/9E12, the amount of Fe(II) decreased at a rate ∼45 times faster than that of SB1003/pLAFR5 (Fig. 3A) while the total amount of Fe stayed constant (data not shown). In comparison, Fe(II) and the total amount of Fe remained constant when cell suspensions of SB1003/9E12 were incubated in the dark (Fig. 3B and total Fe data not shown), demonstrating the light dependence of this reaction. The light dependence of this Fe(II) oxidation activity may be due to the involvement of the photosynthetic reaction center complex in generating a proton gradient across the cytoplasmic membrane, which in turn drives electron flow from Fe(II) to NAD(P)+; it is not due to a need for light in accepting electrons from Fe(II) per se.
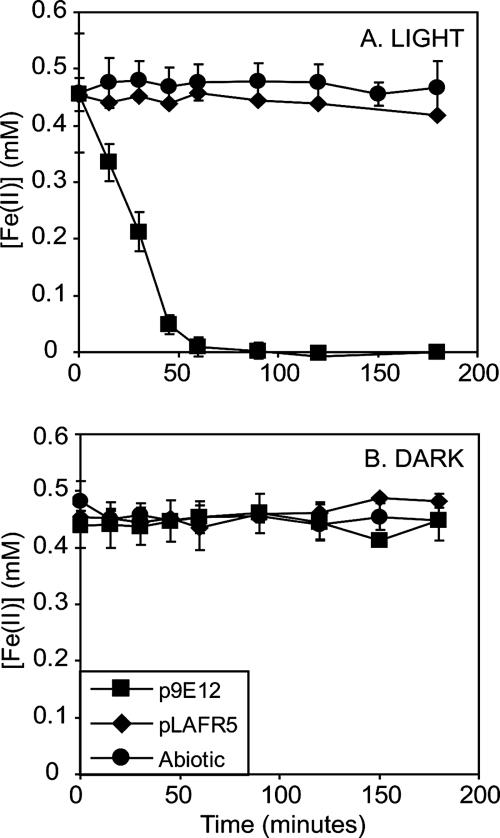
FIG. 3.
Cosmid 9E12 confers enhanced light-dependent Fe(II)-oxidizing activity to SB1003. When cell suspensions of SB1003/9E12 are incubated in the light, the concentration of Fe(II) decreases at a rate ∼45 times faster than that of SB1003 carrying control cosmid pLAFR5 (A). In contrast, no decrease in Fe(II) is observed when SB1003/9E12 is incubated in the dark (B). This demonstrates that cosmid 9E12 confers enhanced light-dependent Fe(II) oxidation to SB1003. The symbol definitions in panel B also apply to panel A. Data shown are the averages of biological triplicates for each strain, and error bars represent the standard deviation.
Although the 9E12 cosmid significantly enhanced light-dependent Fe(II) oxidation activity in SB1003 in the concentrated cell suspension assay, this cosmid did not enable this strain to grow photoautotrophically on Fe(II) alone over a period of 40 days, nor did the other cosmids identified (data not shown).
Identification and characterization of the fox operon.
Restriction site mapping of the four cosmids revealed an ∼9-kb PstI restriction fragment (designated P3) common to three of the cosmids, a portion of which was found in the fourth cosmid (data not shown). P3 was cloned from 9E12 into the broad-host-range vector pBBR1MCS5 in two transcriptional orientations to create pP3-gm1 and pP3-gm2. Both of these constructs conferred equivalent enhanced light-dependent Fe(II) oxidation activity to SB1003 (data shown for P3-gm2 in Fig. 4A and B), suggesting that the gene(s) responsible for the observed activity was expressed from its endogenous promoter. The rates of Fe(II) oxidation by these clones were equivalent to that of SB1003/9E12, indicating that P3 contained all the genes necessary to confer maximal Fe(II) oxidation activity (Fig. 3A and 4A).
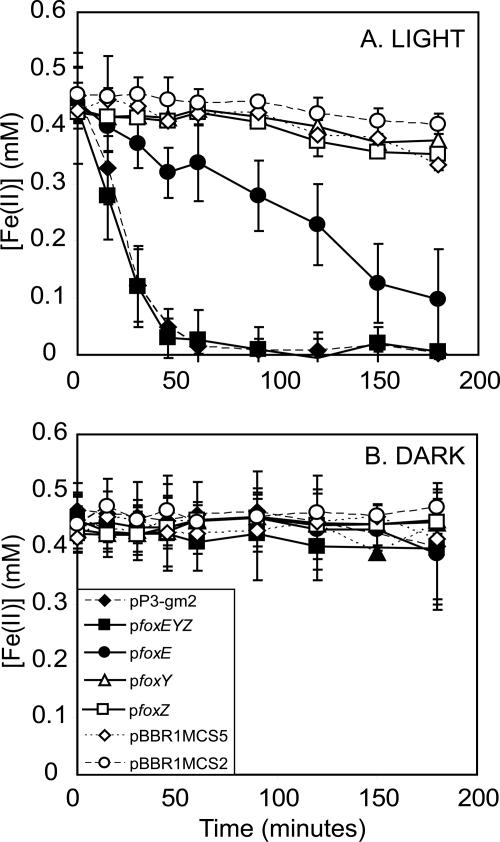
FIG. 4.
Stimulation of light-dependent Fe(II) oxidation activity in SB1003 by the fox genes. When incubated in the light, cell suspensions of strain SB1003 carrying 9E12 subclones pP3-gm2, pfoxEYZ, and pfoxE show a significant increase in light-dependent Fe(II) oxidation activity whereas versions of strain SB1003 carrying pfoxY, pfoxZ, and vector controls pBBR1MCS2 and pBBR1MCS5 do not (A). When cell suspensions of all of the 9E12 subclone-carrying versions of strain SB1003 are incubated in the dark, the concentration of Fe(II) remains constant, showing the light dependence of this Fe(II) oxidation (B). The symbol definitions in panel B also apply to panel A. Data shown are the averages of biological triplicates for each strain, and error bars represent the standard deviation.
In purple nonsulfur bacteria, of which Rhodobacter is one type, the capacity to use inorganic electron donors such as sulfide or H2 as reductants for NAD(P)+ is enabled by redox-active enzymes that are able to accept electrons from these substrates and subsequently donate them to the cyclic electron transport chain (23, 49). Thus, our primary criterion to identify ORFs in the P3 sequence encoding putative Fe(II)-oxidizing proteins was the predicted presence of redox cofactor binding sites. Among the 10 ORFs identified in P3, ORF2 and -3 were the most promising candidates (Fig. 5A). ORF2 is 876 bp and is predicted to encode a soluble, 291-amino-acid (aa) protein with two C-X-X-C-H peptide motifs—the classic sequence suggestive of covalent heme attachment (47). A signal sequence characteristic of secreted proteins is found at the N terminus of this protein, predicting transport across the cytoplasmic membrane. The predicted product of ORF2 shows no significant similarity to other proteins in the BLAST database or the SB1003 genome. ORF3 is 1,089 bp and appears to encode a soluble 362-aa protein with conserved repetitive domains similar to those found in eukaryotic and bacterial WD repeat regulatory proteins, as well as bacterial dehydrogenases and serine/threonine kinases containing the redox cofactor pyrroloquinoline quinone (PQQ) (3, 42, 44). A putative signal sequence is also found in this protein.
ORF2 and -3 are predicted to be part of the same transcriptional unit, along with downstream ORF4. ORF4 is 906 bp and is predicted to encode a 301-aa cytoplasmic membrane protein with 10 transmembrane domains, a signal peptide, and two domains of unknown function conserved among known drug/metabolite transporters like PecM from Erwinia chrysanthemi (DUF6, Pfam accession number PF00892). A putative σ70 promoter consensus sequence resides 113 bp upstream of ORF2 (TTACCG[12 bp]CGGTATATT), and a predicted Rho-independent bacterial terminator is found upstream of this promoter (Fig. 5A). ORF2, -3, and -4 could be PCR amplified from all four of the identified cosmids (data not shown). By RT-PCR with RNA from Fe(II)-induced, H2-grown cells of SW2, we verified that these three genes are cotranscribed and thus form an operon (Fig. 5B and C). We designate ORF2, -3, and -4 foxE, foxY, and foxZ [Fe(II) oxidation], respectively. ORF5 and 6 are predicted to be transcribed in the opposite direction relative to ORF4 and are thus segregated from the fox operon.
The vectors pfoxEYZ, pfoxE, pfoxY, and pfoxZ were constructed (Fig. 5A) to test the gene products for their effects on light-dependent Fe(II) oxidation activity in SB1003. SB1003/pfoxEYZ showed light-dependent Fe(II) oxidation activity at a rate equivalent to that of SB1003/pP3-gm2, suggesting that foxEYZ are sufficient to confer this activity (Fig. 4A and B). Of the individually cloned genes, only SB1003/pfoxE showed significant light-dependent Fe(II) oxidation activity, at a rate ∼20% that of SB1003/pfoxEYZ, while SB1003/pfoxY and SB1003/pfoxZ behaved similarly to the control, SB1003/pBBR1MCS2, showing little Fe(II) oxidation activity (Fig. 4A and B). These experiments show that foxE is sufficient to promote Fe(II) oxidation activity in SB1003, but they do not demonstrate that the fox genes are required for phototrophic growth of SW2 on Fe(II). Because we cannot delete genes in SW2, the closest we can come to addressing whether the fox genes are involved in phototrophic Fe(II) oxidation in SW2 is to determine whether they are differentially expressed under different growth conditions. RT-PCR revealed that the fox genes are cotranscribed in SW2 in the presence of Fe(II) (Fig. 5B and C). In contrast, under H2-grown conditions, foxZ is not transcribed whereas foxE and foxY are (Fig. 5D and E). Although sequences with weak similarity to promoters are predicted 305 and 805 bp upstream of foxZ, no corresponding Rho-independent termination sequences are identified, making it unlikely that this difference in foxZ transcription results from its independent transcription in the presence of Fe(II). The cause of this layered transcriptional regulation is not clear.
FoxE is a c-type cytochrome.
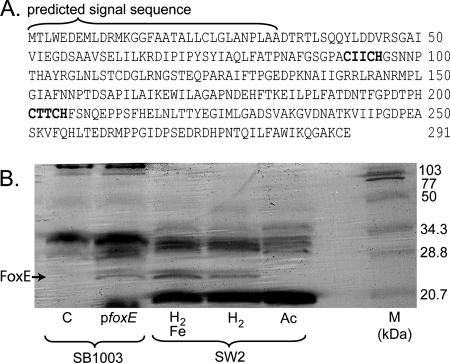
c-type cytochromes are characterized by the covalent attachment of heme(s). In these proteins, heme is bound via thioether bonds to the two cysteines of a conserved sequence motif. Although less common sequence motifs have been identified, the classic sequence motif for heme-binding sites in a c-type cytochrome is C-X-X-C-H (6, 47). The predicted protein sequence of foxE has two of these classic heme-binding motifs (Fig. 6A). To test if foxE encodes a c-type cytochrome, we performed in-gel heme-peroxidase activity staining of crude protein extracts from H2-grown SB1003/pfoxE. These stainings revealed the presence of an ∼25-kDa c-type cytochrome that was not present in extracts of the control strain, SB1003/pBBR1MCS2, when an equivalent amount of total protein was loaded (Fig. 6B). A c-type cytochrome of the same approximate size was identified in crude protein extracts of SW2 grown photoautotrophically on H2 with Fe(II) induction (Fig. 6B). Mass spectrometry analysis confirmed that these bands from SW2 and SB1003/pfoxE contained peptide fragments that matched fragments predicted from the FoxE sequence and that these fragments showed no similarity to any protein in the BLAST database or the SB1003 genome. Heme stain analyses of the soluble and membrane fractions of SW2 crude extracts showed FoxE to be present in the soluble fraction (data not shown). FoxE was also present in crude cell extracts of H2-grown SW2 cells without an Fe(II) induction, as identified by heme stains and confirmed by mass spectrometry, but was not detected in extracts of cells grown photoheterotrophically on acetate (Fig. 6B). Detection of FoxE in H2-grown SW2 cells with and without an Fe(II) induction is consistent with our RT-PCR data showing that foxE is transcribed under both conditions.
FIG. 6.
FoxE is a c-type cytochrome. (A) Amino acid sequence of FoxE. In bold are two consensus motifs suggestive of covalent heme binding sites, and the predicted signal sequence of this protein is 32 aa long. (B) Heme-peroxidase staining of crude cell extracts separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. SB1003 lanes pfoxE and C, SB1003/pfoxE and SB1003/pBBR1MCS2, respectively, grown phototrophically on H2 and induced with Fe(II). SW2 lanes: H2/Fe, SW2 grown phototrophically on H2 with an Fe(II) induction; H2, SW2 grown phototrophically on H2; Ac, SW2 grown phototrophically on acetate. The c-type cytochrome FoxE is indicated by the black arrow. Approximately 120 μg of total protein was loaded per lane, and the molecular weight marker (lane M) is the broad-range standard from Bio-Rad.
DISCUSSION
With a heterologous expression system, we identified an operon from genetically intractable Rhodobacter sp. strain SW2 that confers a significant increase in light-dependent Fe(II) oxidation activity when heterologously expressed in R. capsulatus SB1003. This operon, which we designate the fox operon, contains the following three genes: foxE, which encodes a novel c-type cytochrome; foxY, predicted to encode a protein with repetitive domains similar to WD repeat family proteins and proteins that bind the redox cofactor PQQ; and foxZ, predicted to encode a protein with transport function.
Given the precedent for a c-type cytochrome Fe(II) oxidoreductase in A. ferrooxidans and our observation that the foxE gene from SW2 encodes a novel c-type cytochrome that significantly enhances light-dependent Fe(II) oxidation activity in SB1003 when expressed alone, it is possible that FoxE is the native Fe(II) oxidoreductase in SW2. Although we are unable to prove this via deletion of the foxE gene in SW2, it is perhaps not surprising that mutational analysis of R. palustris strain TIE-1 demonstrates that a soluble c-type cytochrome (PioA) is essential for phototrophic growth on Fe(II) in that organism (27). Given that ours is a heterologous expression system, it is possible that FoxE stimulates Fe(II) oxidation indirectly in SB1003. By drawing an analogy to PioA, however, the simplest interpretation is to predict that FoxE stimulates Fe(II) oxidation directly by serving as the Fe(II) oxidoreductase. In both strains, c-type cytochromes are found in an operon with another putative electron carrier (FoxY in SW2, PioC in TIE-1). Although potentially similar in function, the fox and pio gene products are not homologs. Interestingly, the expression of foxZ in SW2 correlates specifically with the presence of Fe(II). More work needs to be done to understand the regulation of the fox genes, and biochemical experiments are required to elucidate their functions.
Electrons derived from Fe(II) must ultimately make their way to NAD(P)+ via electron carriers in the intracytoplasmic membrane of Fe(II) phototrophs. Because these bacteria oxidize Fe(II) at neutral pH, where the mineral product of this metabolism is insoluble, the question of where the Fe(II) oxidoreductase is localized is important to consider because intracellular precipitation of the Fe(III) mineral product of this metabolism presents a hazard to these organisms. We have previously reported that Fe(III) does not precipitate inside SW2 during growth on Fe(II) (29), and in line with this, others have proposed that Fe(II) oxidation might occur on the cell surface (20). This prediction is consistent with the model for Fe(II) oxidation by the acidophilic Fe(II)-oxidizing bacterium A. ferrooxidans ATCC 33020, where the purported Fe(II)-oxidizing enzyme, Cyc2, is localized to the outer membrane (55). FoxE, however, appears to lack β-sheets or lipoprotein attachments characteristic of known integral or outer membrane-associated c-type cytochromes like Cyc2 or OmcA and OmcB from Shewanella oneidensis MR-1 (39, 55). In addition, our detection of FoxE only in the soluble fraction of cell extracts suggest that it resides either in the cytosol or in the periplasmic space. Although the localization of FoxE remains to be experimentally determined, because c-type cytochrome maturation occurs in the periplasm in gram-negative bacteria, and we know of no examples of bacterial c-type cytochromes that function in the cytosol, we hypothesize that FoxE functions in the periplasm of SW2 (1).
If FoxE functions as the Fe(II) oxidoreductase in the periplasm of SW2, this implies that SW2 has a mechanism for preventing the precipitation of Fe(III) in the periplasmic space. While Fe(III) may be released as a soluble inorganic species (45) and quickly transported out of the cell, it is also possible that the cell produces organic ligands that aid in this process. This hypothesis has been previously put forth to explain, in part, the Fe isotope fractionations produced by Fe(II)-oxidizing phototrophs (15, 29). If such cell-associated ligands exist and are stable over geological time scales, they may provide a means to identify traces of this metabolism in the rock record, enabling studies directed at understanding the environmental impact of these organisms over time (15, 28). For this to be possible, these ligands would need to be extractable from organic remains in ancient rocks, and their structures would need to be recognizable. While, of course, this cannot be guaranteed, the fact remains that organic geochemists have unearthed a plethora of “orphan” biomarkers whose functions are unknown (10). As the structures of molecules required for Fe(II) oxidation in contemporary organisms are determined, with luck, perhaps a match will be found. Additionally, if SW2 has a ligand-based mechanism to avert Fe(III) precipitation in the periplasm that SB1003 does not, this may help explain why SB1003 can only use Fe(II) as an electron donor for growth when an Fe chelator, such as NTA, is added to the medium (Fig. 1A, B, C, and D). This is not the only scenario, however. Addition of NTA to the medium will decrease the redox potential of iron (38), which could facilitate Fe(II) oxidation by an Fe(II) oxidoreductase optimized for substrates with low redox potentials. Elucidation of the native Fe(II) oxidoreductase from SB1003 should be possible by traditional genetic analysis and is one of our goals.
The roles of foxY and foxZ in Fe(II) oxidation by SW2 are unclear. Sequence analyses predict that FoxY contains motifs suggestive of PQQ binding. If FoxY does bind PQQ, its role in phototrophic Fe(II) oxidation by SW2 may be to assist FoxE in electron transfer to a component of the cyclic electron transfer chain. Recent findings support a role for a quinoprotein in Mn(II) oxidation by Erythrobacter sp. strain SD21, providing precedent for the involvement of PQQ-containing enzymes in metabolisms involving metal oxidation (H. Johnson and B. Tebo, personal communication). Sequence analysis of FoxZ predicts that it is a cytoplasmic membrane protein with transport function. The involvement of a putative cytoplasmic membrane transporter is consistent with the finding that a homolog of the permease subunit of an ABC-type transporter is necessary for full Fe(II) oxidation activity by R. palustris TIE-1 (26).
The discovery of a putative Fe(II) oxidoreductase from the neutrophilic, anoxygenic phototroph SW2 that is different in its predicted cellular localization from that involved in Fe(II) oxidation by the acidophilic aerobe A. ferrooxidans is curious, as one might have expected the opposite on the basis of the respective growth environments of these organisms [e.g., Fe(III) is more soluble at acidic pH compared to neutral pH]. Moreover, evidence from studies with R. palustris strain TIE-1 suggests that the putative Fe(II) oxidoreductase from this strain also functions in the periplasm (27). While an emerging theme among the Fe(II) oxidoreductases from both phototrophs and acidophilic bacteria is that they involve c-type cytochromes, it seems that the topology of the components involved is quite different. Whether these differences are reflective of differing evolutionary origins for biologically catalyzed Fe(II) oxidation remains to be explored. For such studies, more knowledge of the mechanisms of Fe(II) oxidation by diverse organisms is needed so that comparative studies can be conducted. Our work demonstrates that a heterologous complementation approach can be used to identify genes involved in Fe phototrophy even from genetically “intractable” species.
Acknowledgments
We thank Doug Lies, Nicky Caiazza, Lars Dietrich, and anonymous reviewers for comments on the manuscript and the members of the Newman lab for helpful discussion.
This work was supported by grants from the Packard Foundation and Howard Hughes Medical Institute to D.K.N. and an NSF graduate fellowship to L.R.C.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Allen, J. W. A., O. Daltrop, J. M. Stevens, and S. J. Ferguson. 2003. C-type cytochromes: diverse structures and biogenesis systems pose evolutionary problems. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 358:255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2003. Denaturing (SDS) discontinuous PAGE: Laemmli method, p. 10-4-10-7. In J. E. Coligan, B. M. Dunn, D. W. Speicher, and P. T. Wingfield (ed.), Short protocols in protein science. John Wiley & Sons, Inc., New York, N.Y.
- 3.Anthony, C. 2004. The quinoprotein dehydrogenases for methanol and glucose. Arch. Biochem. Biophys. 428:2-9. [DOI] [PubMed] [Google Scholar]
- 4.Appia-Ayme, C., N. Guiliani, J. Ratouchniak, and V. Bonnefoy. 1999. Characterization of an operon encoding two c-type cytochromes, an aa(3)-type cytochrome oxidase, and rusticyanin in Thiobacillus ferrooxidans ATCC 33020. Appl. Environ. Microbiol. 65:4781-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bader, K. P. 1994. Physiological and evolutionary aspects of the O2/H2O2-cycle in cyanobacteria. Biochim. Biophys. Acta 1188:213-219. [Google Scholar]
- 6.Barker, P. D., and S. J. Ferguson. 1999. Still a puzzle: why is haem covalently attached in c-type cytochromes? Structure 7:R281-R290. [DOI] [PubMed] [Google Scholar]
- 7.Blankenship, R. E. 2001. Molecular evidence for the evolution of photosynthesis. Trends Plant Sci. 6:4-6. [DOI] [PubMed] [Google Scholar]
- 8.Blankenship, R. E., and H. Hartman. 1998. The origin and evolution of oxygenic photosynthesis. Trends Biochem. Sci. 23:94-97. [DOI] [PubMed] [Google Scholar]
- 9.Brocks, J. J., G. A. Logan, R. Buick, and R. E. Summons. 1999. Archaean molecular fossils and the early rise of eukaryotes. Science 285:1033-1036. [DOI] [PubMed] [Google Scholar]
- 10.Brocks, J. J., and A. Pearson. 2005. Building the biomarker tree of life. Rev. Miner. Geochem. 59:233-258. [Google Scholar]
- 11.Buick, R. 1992. The antiquity of oxygenic photosynthesis: evidence from stromatolites in sulphate-deficient Archaean lakes. Science 255:74-77. [DOI] [PubMed] [Google Scholar]
- 12.Cavalier-Smith, T., M. Brasier, and T. M. Embley. 2006. Introduction: how and when did microbes change the world? Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 361:845-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobley, J. G., and B. A. Haddock. 1975. The respiratory chain of Thiobacillus ferrooxidans: the reduction of cytochromes by Fe2+ and the preliminary characterization of rusticyanin, a novel ‘blue’ copper protein. FEBS Lett. 60:29-33. [DOI] [PubMed] [Google Scholar]
- 14.Croal, L. R., J. A. Gralnick, D. Malasarn, and D. K. Newman. 2004. The genetics of geochemistry. Annu. Rev. Genet. 38:175-202. [DOI] [PubMed] [Google Scholar]
- 15.Croal, L. R., C. J. Johnson, B. L. Bread, and D. K. Newman. 2004. Iron isotope fractionation by Fe(II)-oxidizing photoautotrophic bacteria. Geochim. Cosmochim. Acta 68:1227-1242. [Google Scholar]
- 16.Darbre, P. D. 1999. Basic molecular biology: essential techniques. John Wiley & Sons, Inc., Chichester, N.Y.
- 17.Davis, J., T. J. Donohue, and S. Kaplan. 1988. Construction, characterization and complementation of a Puf− mutant of Rhodobacter sphaeroides. J. Bacteriol. 170:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dietrich, L. E. P., M. M. Tice, and D. K. Newman. 2006. The co-evolution of life and earth. Curr. Biol. 16:R395-R400. [DOI] [PubMed] [Google Scholar]
- 19.Dismukes, G. C., V. V. Klimov, S. V. Baranov, Y. N. Kozlov, J. DasGupta, and A. Tyryshkin. 2001. The origin of atmospheric oxygen on Earth: the innovation of oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 98:2170-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehrenreich, A., and F. Widdel. 1994. Anaerobic oxidation of ferrous iron by purple bacteria, a new type of phototrophic metabolism. Appl. Environ. Microbiol. 60:4517-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falkowski, P. G. 2006. Tracing oxygen's imprint on Earth's metabolic evolution. Science 311:1724-1725. [DOI] [PubMed] [Google Scholar]
- 22.Francis, R. T., and R. R. Becker. 1984. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal. Biochem. 136:509-514. [DOI] [PubMed] [Google Scholar]
- 23.Griesbeck, C., G. Hauska, and M. Schutz. 2000. Biological sulfide oxidation: sulfide quinone reductase (Sqr), the primary reaction, p. 179-203. In S. G. Pandalai (ed.), Recent research developments in microbiology, vol. 4. Research Signpost, Trivadrum, India. [Google Scholar]
- 24.Haselkorn, R., A. Lapidus, Y. Kogan, C. Vlcek, J. Paces, V. Paces, P. Ulbrich, T. Pecenkova, D. Rebrekov, A. Milgram, M. Mazur, R. Cox, N. Kyrpides, N. Ivanova, V. Kapatral, T. Los, A. Lykidis, N. Mikhailova, G. Reznik, O. Vasieva, and M. Fonstein. 2001. The Rhodobacter capsulatus genome. Photosynth. Res. 70:43-52. [DOI] [PubMed] [Google Scholar]
- 25.Heising, S., and B. Schink. 1998. Phototrophic oxidation of ferrous iron by a Rhodomicrobium vannielii strain. Microbiology 144:2263-2269. [DOI] [PubMed] [Google Scholar]
- 26.Jiao, Y., A. Kappler, L. R. Croal, and D. K. Newman. 2005. Isolation and characterization of a genetically tractable photoautotrophic Fe(II)-oxidizing bacterium, Rhodopseudomonas palustris strain TIE-1. Appl. Environ. Microbiol. 71:4487-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao, Y., and D. K. Newman. 2007. The pio operon is essential for photrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 189:1765-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, C. M., B. L. Beard, N. J. Beukes, C. Klein, and J. M. O'Leary. 2003. Ancient geochemical cycling in the Earth as inferred from Fe isotope studies of banded iron formations from the Transvaal Craton. Contrib. Mineral. Petrol. 144:523-547. [Google Scholar]
- 29.Kappler, A., and D. K. Newman. 2004. Formation of Fe(III)-minerals by Fe(II)-oxidizing photoautotrophic bacteria. Geochim. Cosmochim. Acta 68:1217-1226. [Google Scholar]
- 30.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 31.Knoll, A. H. 2003. The geological consequences of evolution. Geobiology 1:3-14. [Google Scholar]
- 32.Komeili, K., H. Vali, T. J. Beveridge, and D. K. Newman. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc. Natl. Acad. Sci. USA 101:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 34.Kusano, T., T. Takeshima, K. Sugawara, C. Inoue, T. Shiratori, T. Yano, Y. Fukumori, and T. Yamanaka. 1992. Molecular cloning of the gene encoding Thiobacillus ferrooxidans Fe(II) oxidase. High homology of the gene product with HiPIP. J. Biol. Chem. 267:11242-11247. [PubMed] [Google Scholar]
- 35.Mackenzie, C., M. Choudhary, F. W. Larimer, P. F. Predki, S. Stilwagen, J. P. Armitage, R. D. Barber, T. J. Donohue, J. P. Hosler, J. E. Newman, J. P. Shapleigh, R. E. Sockett, J. Zeilstra-Ryalls, and S. Kaplan. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth. Res. 70:19-41. [DOI] [PubMed] [Google Scholar]
- 36.Mansch, R., and W. Sand. 1992. Acid-stable cytochromes in ferrous iron oxidizing cell-free preparations from Thiobacillus ferrooxidans. FEMS Microbiol. Lett. 92:83-88. [Google Scholar]
- 37.Mjoli, N., and C. F. Kulpa. 1988. Identification of a unique outer membrane protein required for Fe(II)-oxidation in Thiobacillus ferrooxidans, p. 89-102. In P. R. Norris and D. P. Kelly (ed.), Biohydrometallurgy. Science and Technology Letters, Kew, United Kingdom.
- 38.Morel, F. M. M., and J. G. Hering. 1993. Principles and applications of aquatic chemistry. John Wiley & Sons, Inc., New York, N.Y.
- 39.Myers, C. R., and J. M. Myers. 2004. The outer membrane cytochromes of Shewanella oneidensis MR-1 are lipoproteins. Lett. Appl. Microbiol. 39:466-470. [DOI] [PubMed] [Google Scholar]
- 40.Olson, J. M. 2001. ‘Evolution of photosynthesis’ (1970), re-examined thirty years later. Photosynth. Res. 68:95-112. [DOI] [PubMed] [Google Scholar]
- 41.Olson, J. M., and R. E. Blankenship. 2004. Thinking about the evolution of photosynthesis. Photosynth. Res. 80:373-386. [DOI] [PubMed] [Google Scholar]
- 42.Petricková, K., and M. Petricek. 2003. Eukaryotic-type protein kinases in Streptomyces coelicolor: variations on a common theme. Microbiology 149:1609-1621. [DOI] [PubMed] [Google Scholar]
- 43.Rutherford, A. W. 1989. Photosystem-II, the water-splitting enzyme. Trends Biochem. Sci. 14:227-232. [DOI] [PubMed] [Google Scholar]
- 44.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 45.Sobolev, D., and E. E. Roden. 2001. Suboxic deposition of ferric iron by bacteria in opposing gradients of Fe(II) and oxygen at circumneutral pH. Appl. Environ. Microbiol. 67:1328-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stookey, L. L. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42:779-781. [Google Scholar]
- 47.Thöny-Meyer, L. 2000. Haem-polypeptide interactions during cytochrome c maturation. Biochim. Biophys. Acta 1459:316-324. [DOI] [PubMed] [Google Scholar]
- 48.Valkova-Valchanova, M. B., and S. H. Chan. 1994. Purification and characterization of two new c-type cytochromes involved in Fe2+ oxidation from Thiobacillus ferrooxidans. FEMS Microbiol. Lett. 121:61-69. [DOI] [PubMed] [Google Scholar]
- 49.Vignais, P. M., and A. Colbeau. 2004. Molecular biology of microbial hydrogenases. Curr. Issues Mol. Biol. 6:159-188. [PubMed] [Google Scholar]
- 50.Walker, J. C. G. 1984. Suboxic diagenesis in banded iron formations. Nature 309:340-342. [DOI] [PubMed] [Google Scholar]
- 51.Weaver, P. F., J. D. Wall, and H. Gest. 1975. Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 105:207-216. [DOI] [PubMed] [Google Scholar]
- 52.Widdel, F., S. Schnell, S. Heising, A. Ehrenreich, B. Assmus, and B. Schink. 1993. Ferrous iron oxidation by anoxygenic phototrophic bacteria. Nature 362:834-836. [Google Scholar]
- 53.Xiong, J., W. M. Fischer, K. Inoue, M. Nakahara, and C. E. Bauer. 2000. Molecular evidence for the early evolution of photosynthesis. Science 289:1724-1730. [DOI] [PubMed] [Google Scholar]
- 54.Yarzábal, A., G. Brasseur, and V. Bonnefoy. 2002. Cytochromes c of Acidithiobacillus ferrooxidans. FEMS Microbiol. Lett. 209:189-195. [DOI] [PubMed] [Google Scholar]
- 55.Yarzábal, A., G. Brasseur, J. Ratouchniak, K. Lund, D. Lemesle-Meunier, J. A. DeMoss, and V. Bonnefoy. 2002. The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J. Bacteriol. 184:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yen, H. C., and B. Marrs. 1976. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J. Bacteriol. 126:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]