Abstract
Mutations within the −12 and −24 elements provide evidence that the act promoter is recognized by sigma-54 RNA polymerase. Deletion of the −20 base pair, which lies between the two conserved elements of sigma-54 promoters, decreased expression by 90%. In addition, mutation of a potential enhancer sequence, around −120, led to an 80% reduction in act gene expression. actB, the second gene in the act operon, encodes a sigma-54 activator protein that is proposed to be an enhancer-binding protein for the act operon. All act genes, actA to actE, are expressed together and constitute an operon, because an in-frame deletion of actB decreased expression of actA and actE to the same extent. After an initially slow phase of act operon expression, which depends on FruA, there is a rapid phase. The rapid phase is shown to be due to the activation of the operon expression by ActB, which completes a positive feedback loop. That loop appears to be nested within a larger positive loop in which ActB is activated by the C signal via ActA, and the act operon activates transcription of the csgA gene. We propose that, as cells engage in more C signaling, positive feedback raises the number of C-signal molecules per cell and drives the process of fruiting body development forward.
Sensing starvation, myxobacteria stop growing and start building fruiting bodies that fill with many thousands of spores (7). The cell-surface-associated, nondiffusible C signal is essential for both fruiting body formation and sporulation (39, 56). C signal is a morphogen, encoded by the csgA gene (38), translated as a 25-kDa protein, transported to the cell surface, and finally cleaved to the active 17-kDa signal molecule (40). The signal is transmitted from one cell to another by direct end-to-end contact between them (32-34, 52), which links signaling to the arrangement of cells. Mathematical simulation of C signaling in a population of moving cells has shown that C signal controls the shape of the Myxococcus xanthus fruiting body (57, 58). The particular shape of a myxobacterial fruiting body is a species characteristic and is inherited (49, 60).
Cell behavior changes as fruiting bodies form due to changes in the level of C signal, and successive stages of fruiting body development arise from increasing levels of C signal (31, 39). The low initial level of C signal induces traveling wave behavior (20, 37, 55, 70), a higher level produces streaming behavior, and the highest level induces spores to form. Cells in a stream are arranged end to end, all following the same trajectory, which leads to frequent end-to-end contacts (22-24). Streaming cells enlarge tiny initial aggregates by forming an onion-like succession of spherical shells around the aggregate. The cells within each shell stream in roughly circular orbits. This leads to a spherical outer domain of densely packed cells surrounding an inner domain of threefold-lower density, which is observed microscopically (28, 53). The level of C signal also regulates the expression of many genes involved in fruiting body development (35), in a timely way. Sporulation genes have a higher threshold of C-signal intensity than genes for aggregation (31, 39), and the highest level of C signal induces the cells to differentiate into spores within the fruiting body (37). This property confines sporulation to the spherical mass of a Myxococcus fruiting body, while cells around the outside with lower C-signal levels do not sporulate (25, 46).
Thus, cell movement and gene expression are coordinated for M. xanthus fruiting body development by the rising number of C-signal molecules per cell (27). Transcription of csgA is initiated by starvation and the stringent response (5). Later csgA expression is enhanced by the act operon (17). Mutations in act were found to affect the expression of developmental gene reporters that are expressed after 6 h of development and are related to aggregation and sporulation (16). Moreover, deletion of the actC gene led to the premature formation of many small fruiting bodies (17)—an effect similar to a 10-fold overexpression of CsgA (37). By contrast, deletion of the actD gene led to delayed formation of large fruiting bodies. The kinetics of C-signal increase thus appears to control fruiting body size. After initiation of C-signal expression by starvation, how do the Act proteins regulate the rate and magnitude of the coordinating increase in the signal level?
MATERIALS AND METHODS
Cultures.
Bacterial strains and plasmids are listed in Table 1. The conditions for growth; development, including sporulation; and electroporation have previously been described (17). Transduction was carried out as described previously (16).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| M. xanthus strains | ||
| DK1622 | Wild type | 26 |
| DK5208 | Tn5-132::csgA | 35 |
| DK7853 | asgA476 | 15 |
| DK10602 | DK1622::pTG021 | This work |
| DK10603 | ΔactB | 17 |
| DK10604 | ΔactC | 17 |
| DK10605 | ΔactA | 17 |
| DK10627 | Mx8[MS1512] → DK10605a | This work |
| DK10641 | DK1622::pREG1727 | This work |
| DK10642 | DK1622 attMx8::pTG027 | This work |
| DK10643 | Mx8[DK10642] → DK5208 | This work |
| DK10644 | Mx8[DK10642] → DK7853 | This work |
| DK10645 | DK10642::pEE106; SalI-NdeI | This work |
| DK10646 | Mx8[DK10642] → DK10605 | This work |
| DK10647 | Mx8[DK10642] → DK10603 | This work |
| DK10648 | Mx8[DK10642] → DK10604 | This work |
| DK10649 | Mx8[DK10642] → MS1512 | This work |
| DK10650 | Mx8[DK10642] → fruA::TcΩV | 47 and this work |
| DK10651 | Mx8[DK10602] → DK10605 | This work |
| DK10652 | Mx8[DK10602] → DK10603 | This work |
| DK10653 | Mx8[DK10602] → DK10604 | This work |
| DK10654 | Mx8[DK10602] → DK5208 | This work |
| DK10655 | Mx8[DK10602] → DK7853 | This work |
| DK10658 | DK1622 attMx8::pTG033 | Table S2b |
| DK10659 | DK1622 attMx8::pTG037 | enh1-1 (Table S2) |
| DK10660 | DK1622 attMx8::pTG038 | enh2-1 (Table S2) |
| DK10661 | DK1622 attMx8::pTG039 | σ54 5 C→T (Table S2) |
| DK10662 | DK1622 attMx8::pTG040 | σ54 6 C→Δ (Table S2) |
| DK10665 | DK1622::pTG042 | σ54 1 T→C (Table S2) |
| DK10666 | DK1622::pTG043 | σ54 7 T→C (Table S2) |
| DK10667 | DK1622::pTG044 | σ54 2 G→T (Table S2) |
| DK10668 | DK1622::pTG045 | σ54 3 G→T (Table S2) |
| DK10669 | DK1622::pTG046 | σ54 4 C→T (Table S2) |
| DK10670 | DK1622::pTG047 | enh1-2 (Table S2) |
| DK10671 | DK1622::pTG048 | enh2-2 (Table S2) |
| DK10672 | DK1622::pTG049 | enh3 (Table S2) |
| DK10673 | DK1622::pTG050 | C box 1 (Table S2) |
| DK10674 | DK1622::pTG051 | C box 2 (Table S2) |
| DK10675 | DK1622::pTG052 | σ54 8 G→T (Table S2) |
| MS1512 | sdeK (Ω4408) | 11 |
| Plasmids | ||
| pBluescriptII SK+ | 3.0 kb; Ampr (bla) lacZ with multiple cloning site for blue-white screening | Stratagene |
| pEE106 | Promoter and part of fruA gene in pBGS18 (59) | 8 |
| pJBZ281 | 7.2 kb; Kanr; polylinker sites upstream of lacZ for translational fusions | M. R. K. Alley |
| pREG1727 | 20.7 kb; Ampr Kanr Mx8 attachment site attP; transcriptional terminators followed by multiple cloning sites upstream of promoterless lacZYA genes for transcriptional fusions | R. Gill (10) |
| pSWU12 | pBGS18 with Tn903 Kanr deleted and replaced with pKS-Tetr cassette; MluI/EcoRI | 71 |
| pTG021 | pJBZ281::pTG108 (sequence bp 496 to 2192); SmaI-SmaI | This work |
| pTG025 | pTG021::pBluescriptII SK+; EcoRI-BamHI | This work |
| pTG027 | pREG1727::pTG025; EcoRI-XhoI | This work |
| pTG033 | pBluescript/pREG1727::PCR fragment with act promoter region from upstream XhoI to downstream BamHI; BamHI-XhoI | This work |
| pTG108 | Deletion of PstI fragment from pLAG66 (13); PstI-PstI | This work |
Can be read as “a stock of Mx8 grown on MS1512 was used to transduce DK10605.”
Table S2 can be found in the supplemental material.
Preparation of RNA from developing cells.
A culture of 1 × 1010 to 2 × 1010 cells was induced to develop at 32°C on plates consisting of TPM buffer (10 mM Tris-HCl, pH 7.6, 1 mM KPO4, pH 7.6, 8 mM MgSO4) solidified with 1.5% Bacto Agar. Bacteria, or spores, were harvested at the specified time from the surface by scraping, suspension, and washing in TPM buffer. The solid material from harvest was resuspended in lysozyme-containing buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 2 mg/ml lysozyme) and then disrupted with a Branson 450 Tip sonifier, to ensure that both cells and prespores would be lysed and homogenized. RNA was isolated from the homogenate using RNeasy Midi Spin columns (QIAGEN).
Quantifying mRNA.
To prepare slot blots, 25 μl of each developmental RNA sample dissolved in water was diluted by adding 25 μl 2 mM EDTA, 30 μl 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (54), and 20 μl 37% formaldehyde. The entire 100-μl volume of the mixture was then heated at 60°C for 15 min and transferred to a Hybond-N nylon membrane (Amersham) with 10× SSC, according to the protocol for Bio-Dot SF (Bio-Rad). Finally the membranes were irradiated with UV light to cross-link the RNA to them. DNA probes for actA, actB, and actE were labeled with 32P (17). Probes were annealed onto the RNA-loaded membranes, as described for Southern blotting (54).
Site-directed mutagenesis.
A two-step method of overlap extension PCR was employed, whose principles have been described previously (19). Two universal primers designated upstream XhoI and downstream BamHI were designed. Each universal primer had the indicated synthetic restriction site, following a terminal AAAG spacer. The upstream XhoI primer sequence is 5′ AAAGCTCGAGGCGGCTCGTCGGACACCTTC 3′. The downstream BamHI primer is 5′ AAAGGGATCCCACCGCCTCTTCGTCATCCAC 3′, with the act operon sequences underlined in both sequences. From these two primers, the act promoter region starting at nucleotide (nt) 331 in the upstream region of the GenBank sequence AF350253 and running to nucleotide 2501 of that sequence near the beginning of actB was amplified by PCR. The XhoI and BamHI restriction sites oriented the PCR products for cloning into pBluescript or pREG1727.
To introduce particular deletions or point mutations at predetermined sites between nucleotides 331 and 2501, a set of mutagenic primers, shown in Table S2 in the supplemental material, was designed for both template strands. Each primer, located at the center of the cloned promoter region, contained 10 nucleotides of wild-type act sequence, followed by the desired mutant sequence, followed by another 10 nucleotides of the wild-type sequence. The first step of mutation induction involved two sets of 20 to 25 cycles of PCRs with one universal primer (either the upstream XhoI primer or the downstream BamHI primer) and one of each of the mutagenic primers. The mutagenic primer design ensures that pairs of first-step PCR products overlapped by at least 20 bp with the other member of the pair. The PCR products were purified by agarose gel electrophoresis. The second step involved the upstream XhoI primer, the downstream BamHI primer, and the two PCR products from the first step overlapping at the same mutation from the first step, and the complementary strands that formed a duplex in the overlapping mutant region were completed. This second-step PCR product, which contained a duplex version of the mutant sequence at the appropriate internal site, was digested with XhoI and BamHI and then cloned into the XhoI-BamHI site of pBluescript, for structure verification by sequencing. Verified clones were transferred from pBluescript into the XhoI-BamHI site of pREG1727. Plasmid pREG1727, constructed by Ron Gill of the University of Colorado and described in reference 10, forms transcriptional fusions between the cloned fragment and a promoterless lacZ gene. Cloning into the XhoI-BamHI site ensured that the promoter would be oriented properly for transcription of lacZ. The cloned mutant plasmids are described in Table 1. pREG1727 contains the prophage attachment site attP of myxophage Mx8 which promotes site-specific integration of this plasmid into the attB locus of the M. xanthus chromosome (12). pREG1727 also carries the NPTII gene, encoding kanamycin resistance, which can be used for selection of a plasmid integrant into DK1622, since pREG1727 cannot replicate as a plasmid in M. xanthus (12). That each new plasmid had integrated into the attB locus of DK1622 was confirmed by Southern blot hybridization of the products of restriction endonuclease digestion. The mutated sequence was again verified. Finally, each integrant strain was induced to develop, and its β-galactosidase activity was measured relative to that of wild type in the same experiment, as described previously (16). Each expression test was performed three to six times, to obtain a reliable average. Specific activities were measured as nanomoles of o-nitrophenol per minute per milligram of protein.
Nucleotide sequence accession number.
The sequence of the act operon DNA is published in GenBank under accession number AF350253.
RESULTS
Dynamics of act operon expression.
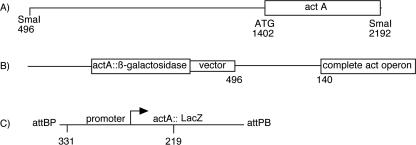
A translational fusion between ActA, the first protein of the operon, and LacZ was constructed for initial measurements of act operon expression. A 1,696-bp SmaI restriction fragment that starts 906 bp upstream of the actA (MXAN3213) translation start, which is located at position 1402 on the fragment shown in Fig. 1A, was inserted into the SmaI site of plasmid pJBZ281. The SmaI site at position 496 lies within MXAN3211; consequently the cloned fragment includes MXAN3211, the 333-bp intergenic region upstream of actA, and the N-terminal 263-amino-acid polypeptide of ActA translationally fused to LacZ. This fusion plasmid integrated by homologous recombination into the act region of the chromosome of DK1622, the strain which provided a standard genetic background for this work. Insertion generated a tandem duplication strain, DK10602; its chromosomal structure, which was confirmed by Southern analysis, is shown in Fig. 1B. The fruiting body sporulation efficiency of DK10602 was equal to that of DK1622: at 3 days, there were 6.4 × 105 viable spores for DK1622 and 6.0 × 105 spores for DK10602. Therefore, the 906-bp upstream segment which includes MXAN3211 and the 333-bp intergenic region in DK10602 contains all the necessary positive elements for act expression (Fig. 1B).
FIG. 1.
Map of the act operon. (A) DNA fragment containing the region upstream of the operon and the N-terminal part of the ActA protein. (B) The fragment shown in panel A was cloned into pJBZ281, and the plasmid was inserted by homologous recombination into the act region of DK10602, a translational fusion. (C) The transcriptional fusion plasmid pTG027 was inserted in strain DK1622, creating DK10642. Position 331 is on the universal PCR primer, described in Materials and Methods.
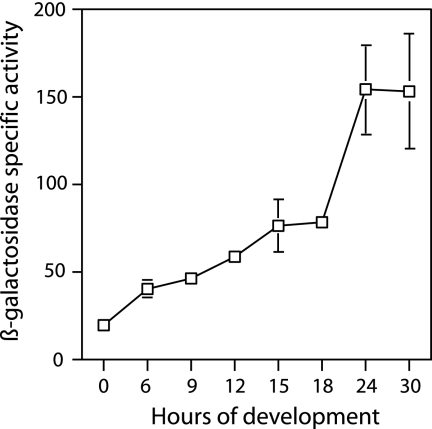
Developmental expression of the act operon, as measured in DK10602, appears to rise in two distinct phases (Fig. 2). It increases gradually during the first 18 h of development and then rises rather steeply to a maximum at 21 to 24 h, when heat-resistant spores begin to appear. A single, continuous rise in expression beginning at 6 h seems incompatible with the experimental data, given the size of measurement errors shown by the bars in Fig. 2. To probe the existence of two distinct phases of act expression, the same 906-bp upstream sequence of strain DK10602 was cloned into pREG1727, to generate a transcriptional fusion of actA to lacZ, and to build a reporter plasmid, pTG027. This fusion plasmid has translational stops in all reading frames ahead of lacZ, ensuring that the fusion of actA to lacZ is transcriptional, not translational. A transcriptional fusion eliminates any effects that translational fusion might have that limit protein synthesis. Plasmid pREG1727 is designed to integrate by site-specific recombination into the Mx8 attB site of M. xanthus (12), which is distant from the chromosomal act region. Figure 1C shows pTG027 inserted into the chromosome of DK1622 at the Mx8 prophage attB site, creating strain DK10642. The chromosomal act region was found to be undisturbed in this strain; its expression should be normal, and pTG027 should be an accurate reporter. Southern blot assays performed on DK10642 supported the idea that insertion had occurred within the attB site and not within the act operon. The spore titer of DK10642 (6.1 × 105) was the same as that of its DK1622 parent (6.4 × 105), confirming that the chromosomal act region was normal. The solid line with filled points in Fig. 3A shows measurement of DK10642 lacZ expression. Again, two phases of lacZ expression are evident. A gradual increase during the first 18 h is followed by a rapid increase to 24 h. Because the expression profiles of Fig. 2 and Fig. 3 are similar, it is likely that the two transcriptional phases seen with DK10642 are responsible for the two phases of translation seen with the fusion DK10602. Thus, Act protein levels are primarily controlled at the level of transcription in these strains, and they can be used as sensitive reporters of transcription.
FIG. 2.
Expression of β-galactosidase in the translational fusion strain DK10602. Specific β-galactosidase activity is shown during the first 30 h of development with error bars showing the standard deviations. Where error bars are not evident, they lie inside the square symbols centered on the average.
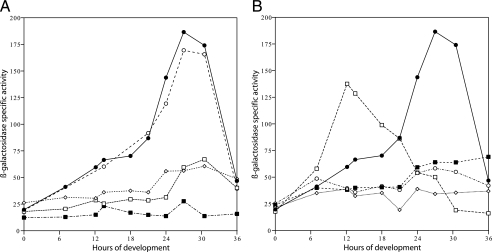
FIG. 3.
A. Transcription of β-galactosidase in DK10642 and mutants defective in asgA, fruA, and sdeK. Closed circles, DK10642; open circles, sdeK mutant; open squares, fruA plasmid insertion mutant; closed squares, fruA Tn5 insertion mutant; open diamonds, asgA mutant. B. Transcription of β-galactosidase in DK10627 (closed circles) and mutants defective in csgA (open diamonds), actA (closed squares), actB (open circles), and actC (open squares). Specific activity is shown as nanomoles of o-nitrophenol per minute per milligram of protein.
To find transcriptional regulators that are needed for act operon expression, pTG027 with the act insert was transduced or transfected into a series of candidate regulatory mutant strains derived from DK1622. Figure 3A shows evidence for two similar phases of act expression in an sdeK mutant background. The very similar time course of the ΔsdeK strain compared with DK10627, which has the wild-type allele of sdeK, indicates that act transcription does not depend on sdeK. The sdeK gene encodes a histidine protein kinase that has been shown to be essential for expression of many developmental genes (11, 36, 48). The substrate(s) for the putative sdeK kinase has not yet been identified, and the data in Fig. 3 imply that neither FruA, for which there is evidence of phosphorylation correlated with activity (8), nor any of the act products—among which ActB is likely to be phosphorylated—is a substrate for the sdeK kinase. However, it is evident from Fig. 3A that transcription of act depends on asgA, since expression is greatly depressed in that mutant. Expression of act also depends on the response regulator protein FruA, which has been verified with two different fruA insertion mutations (8, 47). The Ogawa fruA plasmid insertion mutant happens to be in a DZF1 background that contains a pilQ1 motility mutation (47), which may account for the slightly lower level of act expression than in the Ellehauge Tn5 insertion mutant, which is pilQ+. Expression of fruA depends on the A signal (8). Together, the asgA and fruA defects in act operon expression imply a FruA requirement for the transcription of act.
The second phase of act expression that is evident in Fig. 2 and Fig. 3A rises steeply at 20 h when CsgA protein is reaching its highest specific activity under the same conditions (17). The data of Fig. 3B indicate that transcriptional expression of act directly depends on C signal for several reasons. First, there is very little, if any, increase in act expression in a csgA mutant. Second, the ΔactC mutant, which gives precocious csgA expression (16), shows precocious expression of the actA transcriptional reporter. This argues that the level of C signal normally limits actA expression. The steepness of the early rise in the ΔactC expression profile in Fig. 3B recalls the second phase of act expression in DK10642 cells. For that reason, it appears that in the ΔactC mutant phase two is precocious while phase one is truncated. Together, the effect of the null csgA and of the ΔactC mutants implies that the steep phase of act expression depends on C signaling. The early phase is depressed in both the asgA mutant and in two fruA mutants (Fig. 3A).
A promoter for act.
Figure 3B shows that act reporter expression is substantially and equally depressed in the ΔactA and ΔactB mutants. Inasmuch as actB encodes a sigma-54 activator protein (15), and actA encodes a response regulator that works with actB (17), the dependence of act expression on actB strongly suggests that the act operon is expressed from a sigma-54 promoter, and that inference has been tested by site-directed mutagenesis.
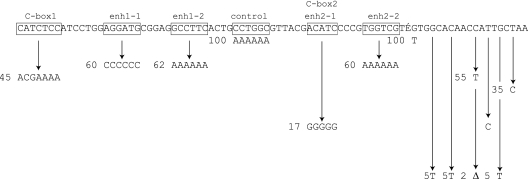
When the actA, actB, actC, and actD genes are expressed as an operon, they are transcribed from a single adenosine residue 57 nt ahead of the presumed actA translational start (17). The sequence ahead of the transcription start site, taken from the GenBank sequence with accession number AF350253, is shown in Fig. 4. The entire sequence was verified in the complete sequence of the M. xanthus genome, GenBank accession number CP000113. A proposed ribosome binding site (TGAGGAAGT) and the ATG translation start codon of actA (nucleotide +1) that lie to the right of the sequence shown in Fig. 4 are evident in the published sequence. Relative to the transcription start, the upstream region from −167 to −12 is spelled out in Fig. 4. It includes −15 to −29 with a TGGCAC-N5-TTGC sequence resembling the sigma-54 consensus from a compilation of sigma-54 promoters (2). That compilation shows that transcription initiates from 8 to 21 nt downstream from the highly conserved GC element in the proximal (−12) element of a sigma-54 promoter (2). In the act operon, transcription starts 15 nt from that GC element. This resembles the extensively studied M. xanthus sigma-54 pilA promoter (71). It also resembles the M. xanthus Ω4521 promoter, for which sequence mutations have indicated sigma-54 specificity (29). The sigma-54 gene (rpoN), which is in single copy, is essential for M. xanthus development. Uniquely among bacteria checked, rpoN of M. xanthus is also essential for growth (30).
FIG. 4.
DNA sequence of the act promoter region from residues −167 to −12. The transcription start, AGC, is residue +1, and it is located to the right of the sequence shown. The −12 and −24 regions of a sigma-54 promoter, two potential enhancer sequences, and two potential C boxes are included. Boxed elements in the line were replaced by the sequences shown below them at the arrow tips, and the numbers next to the new sequence give the resulting level of actA::lacZ fusion reporter expression, expressed as a percentage of the unmutated sequence. Specific β-galactosidase activities relative to the wild-type promoter were measured as described in Materials and Methods. Site-specific mutations were introduced with the PCR primers presented in Table S2 in the supplemental material.
Individual nucleotides in the promoter-like sequence were altered and then tested for level of act expression. Mutant promoters were generated by sequence overlap extension PCR (Materials and Methods), using the oligonucleotides shown in Table S2 in the supplemental material. The mutated segments were then individually cloned into pREG1727, replacing the normal sequence with its mutant versions in pTG027. This cloning incorporated each of them into the same transcriptional fusion segment so that the activity of each mutant promoter could be quantified by measuring the β-galactosidase activity relative to that of the sequence without mutation. β-Galactosidase specific activity was measured 24 h after initiation of development, the time at which maximum act expression was observed from the normal promoter, as indicated in Fig. 2 and Fig. 3. To compensate for the biological variation in the level of gene expression, each mutant-to-wild-type comparison was made within the same experiment. Figure 4 presents the relative β-galactosidase activities observed in each mutant as relative promoter activity in percent. Changes in the −14 to −20 region, which is the downstream or proximal oligonucleotide element of the promoter (the −12 element), show that those bases are important for expression. By the change of residue −14 from T to C, activity fell to 34% of the original level; changing residue −16 from G to a nonconsensus T reduced activity to 7% of that of the wild-type promoter. Changing residue −18 from T to C reduced activity to 20% of the original level, and at −20 changing C to T decreased activity to 50% of the original level. Changes in the upstream promoter element (the −24 element) at residue −24 or −27 decreased activity to less than 10% of that of the wild-type promoter. By contrast, changing the −30 G residue to T had no effect, suggesting that −30 does not have a significant interaction with sigma-54 holoenzyme (Fig. 4). If the act promoter were recognized by a member of the sigma-70 family of factors, assuming that −30 is part of the −35 region, an effect would have been expected (18). M. xanthus is known to employ several members of the sigma-70 family of factors (67).
One of the largest mutant defects (93%) was obtained by deleting the residue at position −21, which lies between the upstream and downstream elements of a sigma-54 promoter. This defect exemplifies a requirement for proper spacing between the −12 and −24 elements, as has been observed elsewhere (3, 29, 43-45). Strict spacing between promoter elements helps to distinguish sigma-54 promoters from sigma-70 promoters (2, 18). Considering the conserved residues, the interelement spacing, and the distance from the act transcription start, the data strongly support recognition by sigma-54.
Upstream regulatory sequences.
Sigma-54 promoters often have upstream regulatory elements. To identify regulatory elements, five to seven base pairs were replaced in each of the potential regulatory sites, indicated at the top of Fig. 4. Kroos and coworkers have identified a set of septanucleotide sequences, named C boxes, near the transcription start of a number of established C-signal-dependent promoters (9, 41, 61, 66, 73-75). Expression of the act operon is C signal dependent (17), but C boxes have yet to be reported with sigma-54 promoters. Two sequences that resemble the C-box consensus CAYYCCY (where Y = C or T) (41) are evident −167 and −120 nt upstream of actA (Fig. 4). Both sequences were mutated, replacing all seven or four of seven bases with nonconsensus residues. Replacing all five residues of potential box 2 lowered activity to 17% of the wild-type level, a substantial decrease (Fig. 4). Changing all seven residues of potential box 1 significantly decreased the activity to 45% of the wild-type level (Fig. 4). These changes may be contrasted with mutation of a hexanucleotide sequence, “control,” in the same general area, centered at −130. Replacement of all six bases of this sequence, which lies between the two C boxes and between the two enhancer-like sequences described below, was found to have no detrimental effect (Fig. 4).
Holoenzyme Eσ54 forms physically detectable closed-promoter complexes at known σ54 promoters, but the enzyme is unable to initiate transcription spontaneously. To separate the polynucleotide strands of an Eσ54-DNA complex requires an enhancer sequence in the DNA (42). A promoter-specific activator protein binds the enhancer to interact, by DNA looping, with holoenzyme bound at the promoter (51, 62, 68). Two candidate sequences, which are boxed in Fig. 4, were identified by comparing the sequence upstream of the act promoter with several sigma-54 enhancers that have been studied—the enhancers for glnAp2 and pspA in Escherichia coli (50, 69) and for nodD3 in Sinorhizobium meliloti (6). Two sequences upstream of the act promoter are indicated, one located around −150 (candidate enh1) and another sequence located around −110 (candidate enh2). Candidate enhancer 2 includes C box 2 (Fig. 4). Because the glnAp2 enhancers have dyad symmetry, the two half-sequences were separately mutated. Localized multibase mutations were obtained by substituting either A6, C6, or G5 for the five or six resident bases (see Table S2 in the supplemental material). Candidate half-sequence 2-1 proved important for expression of the act operon, since expression in the substitution mutant fell to 17% of the wild-type level (Fig. 4). Mutation of candidate elements 1-1, 1-2, and 2-2 showed expression reduced to 60%, 62%, and 60% of wild-type levels, respectively (Fig. 4). The possibility that A6 bends DNA should be noted.
actE.
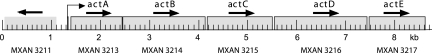
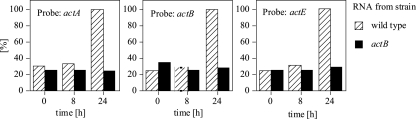
Examination of the act DNA sequence revealed an open reading frame that starts just downstream of actD and that could encode a polypeptide of 753 amino acids. Figure 5 presents a fine-scale map of the whole region. The newly recognized gene, MXAN3217, named actE, is cooriented with the other act genes and is likely to be cotranscribed with actA and actB, according to the time pattern of hybridization of developmental RNA to open reading frame-specific probes (Fig. 6). Deletion of actB decreases the amount of actE transcript to the same extent as it decreases actA and actB transcript levels, as if all were transcribed together. The actE stop codon is at nucleotide residue 8591 in the GenBank sequence with the accession number AF350253. A 7.3-kb act mRNA has been reported elsewhere (17)—just long enough to extend from the beginning of actA through the end of actE (Fig. 5). Mutations in actE have not yet been sought, and no sequence homologs have been found to suggest function.
FIG. 5.
The act operon, showing the location of all act genes, including actE. Horizontal arrows indicate the promoter and direction of transcription of each gene.
FIG. 6.
Relative levels of act operon mRNA at 0, 8, and 24 h of development are shown expressed as percentages. The 24-h level was set as 100%. DNA probes specific for actA and actB were described in reference 17, and an ApaI fragment was employed for actE. The mRNA levels were quantified by measuring 32P on slot blots as described in Materials and Methods.
DISCUSSION
The effects of sequence changes in the act operon promoter of M. xanthus are strong evidence that the promoter is recognized by sigma-54. Considering the large number of sigma-54 activator proteins encoded by the genome (13, 21), and with this addition to the list of established sigma-54 promoters, it is evident that sigma-54 regulates a substantial part of fruiting body development. The mutations reported here lie in the right half of the 333-bp segment that separates the actA (MXAN3213) transcription start from the start of MXAN3211, an open reading frame which, if expressed, is expected to be transcribed in the opposite direction from act (Fig. 5). Two candidate sigma-54 enhancer elements were found, one around −150 and one around −110. Site-specific mutagenesis of those two elements led to an 80% activity reduction from the wild type when the −110 element was mutated and a smaller, but significant, reduction when the one at −150 was mutated. Direct evidence for enhancer function is lacking.
The product of actB is shown here to be an activator of act operon expression, since deletion of actB decreases expression of actA (Fig. 3B) and of actE (Fig. 6). ActB might interact with one or both of the potential enhancers, but no binding studies have been carried out. ActA and ActB appear to be part of a signal transduction pathway that receives input from C signal on the transmitting cell. The output of this signal transduction pathway would be a transcriptionally active ActB (perhaps phosphorylated ActB [ActB∼P]), whose absence would account for the reduced act operon expression in a C-signal-deficient or an ActA-deficient mutant (Fig. 3B). Thus, the rapidly rising late phase of ActA and ActB expression evident in Fig. 2 and Fig. 3 could be explained by ActB binding to the enhancer of its own promoter. Because a positive feedback loop would thereby be created, an accelerated rise in expression would be expected.
Expression of the act operon starts by 6 h of development, after the traffic jams that nucleate aggregates have formed (28). Ellehauge et al. have shown that A signaling, but not C signaling, is required for fruA expression (8). FruA protein is a DNA-binding transcriptional activator that is necessary for expression of a number of developmentally regulated genes, devRS, Ω4400, Ω4469, Ω4273, Ω4500, and fdgA (8, 47, 64, 75), as well as for act. FruA belongs to the FixJ family, and it begins to be expressed between 3 and 6 h (8, 47), in preparation for aggregation gene expression, including the act genes. ActB belongs to the NtrC family of transcriptional activators, which have an ATPase domain located between their receiver and DNA-binding domains that FruA lacks. It is attractive to think that FruA is a positive transcriptional regulator of the act operon for the early phase of act operon expression that occurs at a lower rate than the late phase because ActB has an ATPase activity to drive promoter opening while FruA does not.
Early-phase expression of the act operon would produce some ActB protein. ActB could then bind one or more specific enhancers. Using its ATPase to open the sigma-54 promoter, ActB would initiate the second, rapidly rising phase of act transcription. In addition to actB, csgA and actA are also needed for the second, more rapid phase of act expression (Fig. 3B). p17 CsgA on the transmitting cell, via a (currently unknown) receptor on the receiving cell, would signal ActA, which, in turn, would activate ActB, presumably to ActB∼P. With active ActB protein, expression of the entire act operon would accelerate, rapidly producing all the Act proteins, including ActC and ActD for timing.
Recently it has been suggested that the actD gene, MXAN3216, is a cysteine protease homolog that belongs to clan CD, which includes family C14 of caspase-like enzymes EC 3.4.22.36 and which is directed toward particular proteins (1, 4, 63, 65). Members of this family have a His/Cys catalytic dyad around which sequence is conserved. In actD the His/Cys residues are found in the consecutive sequences LVYYSGHS and LDSCASG (with putative catalytic residues underlined). Because deletion of actD delays C-signal-dependent gene expression, the normal timing of csgA expression is likely to involve proteolysis. Since deletion of actC leads to precocious C-signal-dependent gene expression, actC protein appears to inhibit the rise of csgA expression. One scheme for the temporal control of csgA expression would start with production of comparable amounts of ActC and ActD proteins from their coexpression, and ActC (MXAN3215) would inhibit csgA expression. ActD caspase could degrade ActC, releasing the inhibition and allowing the expression of csgA to rise at the appropriate time. The final level of csgA expression is set by actA and actB (17). Since C signaling stimulates csgA expression, ActB∼P might be a transcription factor or an activator of a transcription factor for the csgA promoter.
Supplementary Material
Acknowledgments
Lisa Gorski was the first to identify actB, and we thank her for help.
This investigation was supported by U.S. Public Health Service grant GM 23441 to D.K. from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 22 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2002. Classification of the caspase-hemoglobinase fold: detection of new families and implications for the origin of the eukaryotic separins. Proteins Struct. Funct. Genet. 45:355-367. [DOI] [PubMed] [Google Scholar]
- 2.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck, M. 1986. Deletion analysis of the Klebsiella pneumoniae nitrogenase promoter: importance of spacing between conserved sequences around positions −12 and −24 for activation by the nifA and ntrC (glnG) products. J. Bacteriol. 166:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, G. M. 1997. Caspases: the executioners of apoptosis. Biochem. J. 326:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford, E. W., Jr., and L. J. Shimkets. 2000. The Myxococcus xanthus socE and csgA genes are regulated by the stringent response. Mol. Microbiol. 37:788-799. [DOI] [PubMed] [Google Scholar]
- 6.Dusha, I., S. Austin, and R. Dixon. 1999. The upstream region of the nodD3 gene of Sinorhizobium meliloti carries enhancer sequences for the transcriptional activator NtrC. FEMS Microb. Lett. 179:491-499. [DOI] [PubMed] [Google Scholar]
- 7.Dworkin, M. 1996. Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 60:70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellehauge, E., M. Norregaard-Madsen, and L. Søgaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal coordination of intercellular signals in M. xanthus development. Mol. Microbiol. 30:807-813. [DOI] [PubMed] [Google Scholar]
- 9.Fisseha, M., D. Biran, and L. Kroos. 1999. Identification of the Ω4499 regulatory region controlling developmental expression of a Myxococcus xanthus cytochrome P-450 system. J. Bacteriol. 181:5467-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisseha, M., M. Gloudemans, R. E. Gill, and L. Kroos. 1996. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J. Bacteriol. 178:2539-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garza, A. G., J. S. Pollack, B. Z. Harris, A. Lee, I. M. Keseler, E. F. Licking, and M. Singer. 1998. SdeK is required for early fruiting body development in Myxococcus xanthus. J. Bacteriol. 180:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill, R. E., and L. J. Shimkets. 1993. Genetic approaches for analysis of bacterial behavior, p. 129-155. In M. Dworkin and D. Kaiser (ed.), Myxobacteria II. American Society for Microbiology, Washington, DC.
- 13.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. MacKensie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorski, L., T. Gronewold, and D. Kaiser. 2000. A σ54 activator protein necessary for spore differentiation within the fruiting body of Myxococcus xanthus. J. Bacteriol. 182:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorski, L., and D. Kaiser. 1998. Targeted mutagenesis of σ54 activator proteins in Myxococcus xanthus. J. Bacteriol. 180:5896-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gronewold, T. M. A., and D. Kaiser. 2002. act operon control of developmental gene expression in Myxococcus xanthus. J. Bacteriol. 184:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gronewold, T. M. A., and D. Kaiser. 2001. The act operon controls the level and time of C-signal production for M. xanthus development. Mol. Microbiol. 40:744-756. [DOI] [PubMed] [Google Scholar]
- 18.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 20.Igoshin, O., A. Mogilner, R. Welch, D. Kaiser, and G. Oster. 2001. Pattern formation and traveling waves in myxobacteria: theory and modeling. Proc. Natl. Acad. Sci. USA 98:14913-14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jelsbak, L., M. Girskov, and D. Kaiser. 2005. Enhancer-binding proteins with a forkhead-associated domain and the sigma54 regulon in Myxococcus xanthus fruiting body development. Proc. Natl. Acad. Sci. USA 102:3010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jelsbak, L., and L. Søgaard-Andersen. 1999. The cell-surface associated C-signal induces behavioral changes in individual M. xanthus cells during fruiting body morphogenesis. Proc. Natl. Acad. Sci. USA 96:5031-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jelsbak, L., and L. Søgaard-Andersen. 2002. Pattern formation by a cell-surface associated morphogen in M. xanthus. Proc. Natl. Acad. Sci. USA 99:2032-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jelsbak, L., and L. Søgaard-Andersen. 2000. Pattern formation: fruiting body morphogenesis in Myxococcus xanthus. Curr. Opin. Microbiol. 3:637-642. [DOI] [PubMed] [Google Scholar]
- 25.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser, A. D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 1:45-54. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser, D., and R. Welch. 2004. Dynamics of fruiting body morphogenesis. J. Bacteriol. 186:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keseler, I. M., and D. Kaiser. 1995. An early A-signal-dependent gene in Myxococcus xanthus has a σ54-like promoter. J. Bacteriol. 177:4638-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keseler, I. M., and D. Kaiser. 1997. Sigma-54, a vital protein for Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 94:1979-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S. K., and D. Kaiser. 1991. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J. Bacteriol. 173:1722-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, S. K., and D. Kaiser. 1990. Cell alignment required in differentiation of Myxococcus xanthus. Science 249:926-928. [DOI] [PubMed] [Google Scholar]
- 33.Kim, S. K., and D. Kaiser. 1990. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 4:896-905. [DOI] [PubMed] [Google Scholar]
- 34.Kroos, L., P. Hartzell, K. Stephens, and D. Kaiser. 1988. A link between cell movement and gene expression argues that motility is required for cell-cell signalling during fruiting body development. Genes Dev. 2:1677-1685. [DOI] [PubMed] [Google Scholar]
- 35.Kroos, L., and D. Kaiser. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1:840-854. [DOI] [PubMed] [Google Scholar]
- 36.Kroos, L., A. Kuspa, and D. Kaiser. 1990. Defects in fruiting body development caused by Tn5 lac insertions in Myxococcus xanthus. J. Bacteriol. 172:484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kruse, T., S. Lobendanz, N. M. S. Bertheleson, and L. Søgaard-Andersen. 2001. C-signal: a cell surface-associated morphogen that induces and coordinates multicellular fruiting body morphogenesis and sporulation in M. xanthus. Mol. Microbiol. 40:156-168. [DOI] [PubMed] [Google Scholar]
- 38.Lee, B.-U., K. Lee, J. Mendez, and L. J. Shimkets. 1995. A tactile sensory system of Myxococcus xanthus involves an extracellular NAD(P)+-containing protein. Genes Dev. 9:2964-2973. [DOI] [PubMed] [Google Scholar]
- 39.Li, S., B. U. Lee, and L. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 40.Lobedanz, S., and L. Søgaard-Andersen. 2003. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 17:2151-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loconto, J., P. Viswanathan, S. J. Nowak, M. Gloudemans, and L. Kroos. 2005. Identification of the Ω4406 regulatory region, a developmental promoter of Myxococcus xanthus, and a DNA segment responsible for chromosomal position-dependent inhibition of gene expression. J. Bacteriol. 187:4149-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morett, E., and L. Segovia. 1993. The σ54 bacterial enhancer-binding protein family: mechanics of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullin, D. A., and A. Newton. 1989. Ntr-like promoters and upstream regulatory sequence ftr are required for transcription of a developmentally regulated Caulobacter crescentus flagellar gene. J. Bacteriol. 171:3218-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullin, D. A., and A. Newton. 1993. A σ54 promoter and downstream sequence elements ftr2 and ftr3 are required for regulated expression of divergent transcription units flaN and flbG in Caulobacter crescentus. J. Bacteriol. 175:2067-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ninfa, A. J., D. A. Mullin, G. Ramakrishnan, and A. Newton. 1989. Escherichia coli σ54 RNA polymerase recognizes Caulobacter crescentus flbG and flaN flagellar gene promoters in vitro. J. Bacteriol. 171:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Connor, K. A., and D. R. Zusman. 1991. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J. Bacteriol. 173:3318-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22:757-767. [DOI] [PubMed] [Google Scholar]
- 48.Pollack, J. S., and M. Singer. 2001. SdeK, a histidine kinase required for Myxococcus xanthus development. J. Bacteriol. 183:3589-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reichenbach, H. 1993. Biology of the myxobacteria: ecology and taxonomy, p. 13-62. In M. Dworkin and D. Kaiser (ed.), Myxobacteria II. American Society for Microbiology, Washington, DC.
- 50.Reitzer, L. J., and B. Magasanik. 1986. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell 45:785-792. [DOI] [PubMed] [Google Scholar]
- 51.Reitzer, L. J., B. Movsas, and B. Magasanik. 1989. Activation of glnA transcription by nitrogen regulator I (NRI)-phosphate in Escherichia coli: evidence for a long-range physical interaction between NRI-phosphate and RNA polymerase. J. Bacteriol. 171:5512-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sager, B., and D. Kaiser. 1994. Intercellular C-signaling and the traveling waves of Myxococcus. Genes Dev. 8:2793-2804. [DOI] [PubMed] [Google Scholar]
- 53.Sager, B., and D. Kaiser. 1993. Two cell-density domains within the Myxococcus xanthus fruiting body. Proc. Natl. Acad. Sci. USA 90:3690-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 55.Shimkets, L., and D. Kaiser. 1982. Induction of coordinated movement of Myxococcus xanthus cells. J. Bacteriol. 152:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Søgaard-Andersen, L., F. Slack, H. Kimsey, and D. Kaiser. 1996. Intercellular C-signaling in Myxococcus xanthus involves a branched signal transduction pathway. Genes Dev. 10:740-754. [DOI] [PubMed] [Google Scholar]
- 57.Sozinova, O., Y. Jang, D. Kaiser, and M. Alber. 2006. A three-dimensional model of myxobacterial fruiting body formation. Proc. Natl. Acad. Sci. USA 103:17255-17259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sozinova, O., Y. Jang, D. Kaiser, and M. S. Alber. 2005. Three-dimensional model of myxobacterial aggregation by contact-mediated interaction. Proc. Natl. Acad. Sci. USA 102:11308-11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spratt, B. G., P. J. Hedge, S. teHessen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 60.Sproer, C., H. Reichenbach, and E. Stackebrandt. 1999. Correlation between morphological and phylogenetic classification of myxobacteria. Int. J. Syst. Bacteriol. 49:1255-1262. [DOI] [PubMed] [Google Scholar]
- 61.Srinivasan, D., and L. Kroos. 2004. Mutational analysis of the fruA promoter region demonstrates that C-box and 5-base-pair elements are important for expression of an essential developmental gene of Myxococcus xanthus. J. Bacteriol. 186:5961-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su, W., S. Porter, S. Kustu, and H. Echols. 1990. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc. Natl. Acad. Sci. USA 87:5504-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szallies, A., B. Kubata, and M. Duszenko. 2002. A metacaspase of Trypanosoma brucei causes loss of respiration competence and clonal death in the yeast Saccharomyces cerevisiae. FEBS Lett. 517:144-150. [DOI] [PubMed] [Google Scholar]
- 64.Ueki, T., and S. Inouye. 2005. Identification of a gene involved in polysaccharide export as a transcription target of FruA, an essential factor for Myxococcus xanthus development. J. Biol. Chem. 280:32279-32284. [DOI] [PubMed] [Google Scholar]
- 65.Uren, A. G., K. O'Rourke, L. Aravind, M. T. Pisabarro, S. Seshagiri, E. V. Koonin, and V. M. Dixit. 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6:961-967. [DOI] [PubMed] [Google Scholar]
- 66.Viswanathan, P., and L. Kroos. 2003. cis elements necessary for developmental expression of a Myxococcus xanthus gene that depends on C signaling. J. Bacteriol. 185:1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viswanathan, P., and L. Kroos. 2006. Role of σD in regulating genes and signals during Myxococcus xanthus development. J. Bacteriol. 188:3246-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wedel, A., D. S. Weiss, D. Popham, P. Droge, and S. Kustu. 1990. A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science 248:486-490. [DOI] [PubMed] [Google Scholar]
- 69.Weiner, L., J. L. Brissette, N. Ramani, and P. Model. 1995. Analysis of the proteins and cis-acting elements regulating the stress-induced phage shock protein operon. Nucleic Acids Res. 23:2030-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Welch, R., and D. Kaiser. 2001. Cell behavior in traveling wave patterns of myxobacteria. Proc. Natl. Acad. Sci. USA 98:14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, S. S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109-121. [DOI] [PubMed] [Google Scholar]
- 73.Yoder, D. R., and L. Kroos. 2004. Mutational analysis of the Myxococcus xanthus Ω4400 promoter region provides insight into developmental gene regulation by C signaling. J. Bacteriol. 186:661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoder, D. R., and L. Kroos. 2004. Mutational analysis of the Myxococcus xanthus Ω4499 promoter region reveals shared and unique properties in comparison with other C-signal-dependent promoters. J. Bacteriol. 186:3766-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoder-Himes, D. R., and L. Kroos. 2006. Regulation of the Myxococcus xanthus C-signal-dependent Ω4400 promoter by the essential developmental protein FruA. J. Bacteriol. 188:5167-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.