Abstract
Hfq, a chaperone for small noncoding RNAs, regulates many processes in Escherichia coli, including the σS-mediated general stress response. Here we used microarray analysis to identify the changes in gene expression resulting from lack of Hfq. We identify several potential new targets for Hfq regulation, including genes encoding outer membrane proteins, enzymes, factors, and transporters. Many of these genes are involved in amino acid uptake and biosynthesis, sugar uptake and metabolism, and cell energetics. In addition, we find altered regulation of the σE- and σ32-mediated stress responses, which we analyze further. We show that cells lacking Hfq induce the σE-mediated envelope stress response and are defective in σE-mediated repression of outer membrane proteins. We also show that the σ32-mediated cytoplasmic stress response is repressed in cells lacking Hfq due to increased expression of DnaK. Furthermore, we show that cells lacking Hfq are defective in the “long-term adaptation” of σ32 to chronic chaperone overexpression. Together, our results indicate that Hfq may play a general role in stress response regulation in E. coli.
Regulation of gene expression by noncoding RNAs is widespread in both prokaryotes and eukaryotes. In prokaryotes, small RNAs (sRNAs) mediate most noncoding RNA regulation by basepairing with their target mRNAs. sRNA-mRNA basepairing inhibits mRNA function, either by increasing mRNA degradation or decreasing mRNA translation or both (reviewed in references 13 and 42). More rarely, basepairing increases translation by removing an inhibitory interaction encoded in the mRNA (22, 24-26, 41) or stabilizes mRNA from degradation (32). Hundreds of sRNAs have been identified, but the targets of many of these sRNAs are currently unknown.
Hfq, an RNA-binding protein initially identified as the host factor required for replication of the RNA phage Qβ, is usually required for sRNA-mRNA transactions. Hfq is a member of the Sm protein family that is widely involved in RNA processing events in eukaryotes. In prokaryotes, Hfq facilitates sRNA-mRNA interactions (reviewed in reference 52). Binding of the sRNA to the hexameric Hfq protein is likely to melt its secondary structure, thereby facilitating sRNA-mRNA interaction. Alternatively, sRNA and mRNA may simultaneously bind Hfq, thereby enhancing interaction between the two RNAs.
As might be expected from the fact that Hfq is required for most sRNA regulation, deletion of Hfq has pleiotropic phenotypes, including slow growth, altered cell division, osmosensitivity, increased oxidation of carbon sources, and altered patterns of protein synthesis (49; reviewed in reference 52). hfq mutant strains are also defective in the σS-mediated general stress response because the sRNAs that promote σS translation are not functional. The role of Hfq in the σE-mediated envelope stress response and the σ32-mediated cytoplasmic heat shock response has not been examined in Escherichia coli. However, there are compelling reasons to do so: Vibrio cholera hfq mutant cells overexpress the σE regulon (8a), and Hfq is encoded in the σ32 regulon (48).
The σE stress response is induced by misfolded envelope proteins, primarily trimeric outer membrane porins (reviewed in references 1, 2, and 34). Porins have a complex assembly pathway, and unassembled monomeric porins accumulate in the envelope when cells are under stress. This activates a proteolytic cascade that degrades RseA, the antisigma factor that negatively regulates σE. Activation of σE-mediated transcription induces the expression of many genes and also downregulates the expression of a subset of outer membrane proteins (OMPs) (37).
The σ32 stress response is induced by misfolded proteins in the cytoplasm and results in the induction of both chaperones and proteases, as well as a number of other proteins that protect various macromolecular processes against heat stress (31; reviewed in reference 58). σ32 is tightly regulated; its translation, degradation, and activity are all modulated. In particular, the major cytoplasmic chaperone machines, DnaKJ and GroELS, regulate both σ32 activity and degradation, thereby allowing σ32 to indirectly sense unfolded proteins by sensing chaperone occupancy (12, 15, 44).
Previous attempts to identify processes regulated by Hfq have relied on identification of sRNAs by bioinformatics and genomic approaches (see, for example, references 54, 55, 59, and 60). Here, we first use microarray analysis of cells lacking Hfq to identify processes that are directly or indirectly regulated by this protein, thus providing potential new target genes for sRNA regulation. We then specifically explore further our finding that Hfq regulates the σE and σ32 stress responses.
MATERIALS AND METHODS
Media, strains, and plasmids.
LB rich medium and M9 minimal medium were prepared as described previously (39). M9 medium was supplemented with 0.2% glucose, 1 mM MgSO4, and 2 μg of thiamine/ml. Complete M9 minimal medium was also supplemented with all amino acids (40 μg/ml), except for methionine and cysteine where indicated. When required, media was supplemented with 100 μg of ampicillin/ml, 30 μg of kanamycin/ml, and/or 20 μg of chloramphenicol/ml.
Bacterial strain and plasmids used in the present study are listed in Table 1. P1 vir-mediated transductions were carried out as described by (29). Hfq parent strains TX2817 and TX2821 (48, 49) contain a kanamycin resistance (Kmr) omega cassette inserted either into the middle of the hfq gene disrupting function (TX2821; Hfq− phenotype), or at the 3′ end of hfq to retain function but still has polarity on the downstream gene, hflX (TX2817; Hfq+ phenotype).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotypea | Source, reference, or construction |
|---|---|---|
| Strains | ||
| MC1061 | E. coli K-12 araD Δ(ara-leu)7697 Δ(codB-lacI) galK16 galE15 mcrA0 relA1 rpsL150 spoT1 mcrB9999 hsdR2 | E. coli Genetic Stock Center (7) |
| MC4100 | E. coli K-12 araD139 Δ(argF-lac)169 rpsL150 thiA1 relA1 flb5301 deoC1 ptsF25 rbsR | E. coli Genetic Stock Center (6) |
| MG1655 | E. coli K-12 (MG1655) rph-1 | E. coli Genetic Stock Center (16, 17) |
| BB7063 | W3110 zad::Tn10 PA1/lacO-1-dnaKJ | 46 |
| TX2817 | MG1655 hfq2::Ω (Km; KpnI) | 48, 49 |
| TX2821 | MG1655 hfq1::Ω (Km; BclI) | 48, 49 |
| CAG2322 | MC4100 [Φ λrpoH′(MluI)-′lacZ] | Lab stocks |
| CAG2323 | MC4100 [Φ λrpoH′(PvuII)-′lacZ] | Lab stocks |
| CAG22357 | MC1061 [Φ λrpoH P3::lacZ] ΔrseA nadB::Tn10; Tetr | 3 |
| CAG25195 | MG1655 ΔlacX74 [Φ λrpoH P3::lacZ] | 37 |
| CAG25196 | MG1655 ΔlacX74 [Φ λrpoH P3::lacZ] pTrc99A | 37 |
| CAG25197 | MG1655 ΔlacX74 [Φ λrpoH P3::lacZ] pLC245 | 37 |
| CAG25198 | MG1655 nadB::Tn10 ΔrseA ΔlacX74 [Φ λrpoH P3::lacZ] | CAG25195 + P1/CAG22357 |
| CAG45146 | MG1655 ΔlacX74 [Φ λhtpG P1::lacZ] | 31 |
| CAG48383 | MG1655 ΩlacX74 [Φ λhtpG P1::lacZ] hfq2::Ω (Km; KpnI) | CAG45146 + P1/TX2817 |
| CAG48400 | MG1655 ΩlacX74 [Φ λhtpG P1::lacZ] hfq2::Ω (Km; KpnI) PA1/lacO-1-dnaKJ | CAG48383 + P1/BB7063 |
| CAG48401 | MG1655 ΩlacX74 [Φ λhtpG P1::lacZ] hfq1::Ω (Km; BclI) PA1/lacO-1-dnaKJ | CAG48422 + P1/BB7063 |
| CAG48402 | MG1655 ΩlacX74 [Φ λhtpG P1::lacZ] hfq2::Ω (Km; KpnI) pGro7 | This study |
| CAG48403 | MG1655 ΩlacX74 [Φ λhtpG P1::lacZ] hfq1::Ω (Km; BclI) pGro7 | This study |
| CAG48408 | MC4100 hfq2::Ω (Km; KpnI) [Φ λrpoH′(MluI)-′lacZ] | CAG2322 + P1/TX2817 |
| CAG48409 | MC4100 hfq1::Ω (Km; BclI) [Φ λrpoH′(MluI)-′lacZ] | CAG2322 + P1/TX2821 |
| CAG48410 | MC4100 hfq2::Ω (Km; KpnI) [Φ λrpoH′(PvuII)-′lacZ] | CAG2323 + P1/TX2817 |
| CAG48411 | MC4100 hfq1::Ω (Km; KpnI) [Φ λrpoH′(PvuII)-′lacZ] | CAG2323 + P1/TX2821 |
| CAG48422 | MG1655 ΔlacX74 [Φ λhtpG P1::lacZ] hfq1::Ω (Km; BclI) | CAG45146 + P1/TX2821 |
| CAG50008 | MG1655 ΔlacX74 [Φ λrpoH P3::lacZ] hfq2::Ω (Km; KpnI) | CAG25195 + P1/TX2817 |
| CAG50009 | MG1655 ΔlacX74 [Φ λrpoH P3::lacZ] hfq1::Ω (Km; BclI) | CAG25195 + P1/TX2821 |
| CAG50010 | MG1655 ΔlacX74 [Φ λrpoH P3::lacZ] hfq2::Ω (Km; KpnI), pTrc99A | This study |
| CAG50011 | MG1655 ΔlacX74 [Φ λrpoH P3::lacZ] hfq2::Ω (Km; KpnI), pLC245 | This study |
| CAG50013 | MG1655 ΔlacX74 [Φ λrpoH P3::lacZ] hfq1::Ω (Km; BclI), pLC245 | This study |
| Plasmids | ||
| pGro7 | Vector, pACYC184 ori; Cmr; ParaB-groELS. Carries groELS induced from PBAD promoter | 30 |
| pTrc99A | Vector, pBR322 ori; Apr; expression vector containing an IPTG inducible trc promoter | Amersham Pharmacia Biotech |
| pLC245 | rpoE cloned in pTrc99A downstream of the IPTG inducible trc promoter; Apr | 37 |
Cmr, chloramphenicol resistance; Tetr, tetracycline resistance; Apr, ampicillin resistance.
DNA microarray procedures.
Two-color DNA microarrays were performed to determine the relative mRNA levels in four experiments (strain details are given in Table 1) as follows: (i) MG1655 hfq+ versus MG1655 hfq mutant (CAG50008 versus CAG50009), eight independent replicates; (ii) MG1655 versus MG1655 ΔrseA (CAG25195 versus CAG25198), seven independent replicates; (iii) MG1655 hfq+ ptrc99A versus MG1655 hfq+ pLC245 in which σE is overexpressed from plasmid pLC245 by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) (CAG50010 versus CAG50011), three independent replicates; and (iv) MG1655 hfq+ ptrc99A versus MG1655 hfq mutant pLC245 in which σE is overexpressed from plasmid pLC245 by the addition of IPTG (CAG50010 versus CAG50013), three independent replicates. With the exception of the ΔrseA strain (CAG25198), cultures for all microarray experiments were prepared by inoculating 500-ml conical flasks containing 100 ml of M9 complete with fresh overnight cultures to a final optical density at 450 nm (OD450) of 0.03. Since CAG25198 lyses in stationary phase, the overnight cultures were inoculated from a range of diluted cell suspensions direct from glycerol stocks so that some cultures were still in exponential growth the following morning. Only these cultures were used for final inoculations of the 100-ml cultures. For all experiments, the 100-ml cultures were grown aerobically at 30°C in a gyratory water bath (model G76 from New Brunswick Scientific, New Jersey) with shaking at 240 rpm. At an OD450 of 0.3, 10-ml samples were harvested from cultures for experiments i and ii, and for experiments iii and iv the cultures were induced with IPTG (1 mM final concentration), and 10-ml samples were harvested 20 min after induction. Since σS is not significantly expressed at an OD450 of 0.3, we performed our comparison at an OD450 of 0.3 to avoid indirect effects resulting from promotion of σS translation by Hfq.
For all experiments, the 10-ml culture samples were harvested by immediately adding to 1.25 ml of ice-cold 5% water-saturated phenol in ethanol and centrifuged at 6,600 × g. Cell pellets were flash frozen in liquid N2 and stored at −80°C. Preparation of labeled probes and microarray procedures were performed exactly as described previously (37). For each experiment, samples from the first listed strain in each pair were labeled with Cy3 (green), and samples from the second listed strain samples were labeled with Cy5 (red). Briefly, relative mRNA levels were determined by scanning parallel two-color hybridization to glass slide cDNA microarrays (40) containing PCR products of 4,110 E. coli open reading frames representing 95.8% of all K-12 open reading frames (http://derisilab.ucsf.edu/core/resources/index.html); the resulting TIFF images were analyzed by using GENEPIX 3.0 software (Axon Instruments, Inc., California), and the data were stored on a NOMAD database (software available from http://derisilab.ucsf.edu/core/resources/index.html).
DNA microarray data analysis.
Expression data were normalized as described earlier (37), assuming mRNA is equivalent in both initial samples (36). Intensity (dye)-dependent biases were corrected by using an MA-plot and Lowess smoothing (47, 57). Raw and normalized microarray expression data are available on the National Center for Biotechnology Information GEO Web site (http://www.ncbi.nih.gov/geo/) under the accession codes GSE3437 for the σE overexpression time course (37) and GSE6444 for all other microarray experiments.
mRNA transcripts present at significantly different levels in the MG1655 hfq+ versus MG1655 hfq mutant microarray experiment were identified from eight independent replicate experiments using by “statistical analysis of microarrays” software (SAM 2.23) (50; http://www-stat.stanford.edu/∼tibs/SAM/index.html). Significantly differentially regulated transcripts were extracted using a stringent cutoff (lowest false discovery rate at the median percentile = 0%) to give 269 transcripts in total; 94 increased and 175 decreased in hfq mutants. This SAM report and transcript list is given in Table S1 in the supplemental material. An additional filter of 1.5-fold change was also applied to give 120 transcripts; 48 increased and 72 decreased in hfq mutants. Note that the change in transcript levels from lacZYA is due to expression of the σE-dependent reporter, rpoHP3::lacZ.
Hierarchical clustering was performed using the software Cluster and visualized by using TreeView (http://rana.lbl.gov/EisenSoftware.htm) (9).
mRNA degradation half-life.
Approximate mRNA degradation half-lives upon overexpression of σE for ompF, ompC, ompX, ompA, fiu, yhcN, lpp, and tsx mRNAs were derived from four time course microarrays of σE overexpression (37). The mRNA half-lives were calculated assuming that σE only affected the rate of mRNA degradation and not synthesis. For each mRNA, an exponential curve of the form y = ae−bx+c was fitted to a plot of expression ratios of wild-type versus σE-overexpressed cells against the time of σE overexpression. The mRNA half-life (t1/2) was calculated from t1/2 = ln(2)/b. Given the variability of microarray expression data, these half-lives should only be regarded as crude estimates.
β-Galactosidase assays.
Overnight cultures of various strains grown in LB medium were diluted 1:100 and grown to an appropriate OD until they reached exponential phase. Standard β-galactosidase assays were performed at least in duplicate from two independent cultures (28). For σ32-dependent activities, assays were performed from exponential-phase cultures and, where analyzed, comparable results were obtained by directly sampling overnight cultures (data not shown). Several different lacZ reporters were utilized: an rpoHP3::lacZ fusion to measure σE-dependent activity; an rpoH::lacZ short fusion to measure σ32 transcription and basal translation (20); an rpoH::lacZ long fusion to measure σ32 transcription, regulated translation, and stability (43); and a PhtpG::lacZ reporter to measure σ32 activity (31).
Protein synthesis measurements.
Overnight cultures grown in complete M9 minimal medium with all amino acids except methionine and cysteine at 30°C were diluted 1:100 and then grown until they reached exponential phase. For each sample, an 800-μl aliquot of cells was pulse-labeled for 1 min with EasyTag Expre35S35S protein labeling mix (NEN), followed by a chase with unlabeled methionine and cysteine. Extracts were then precipitated with a final concentration of 5% trichloroacetic acid (TCA) on ice for at least 30 min, followed by centrifugation. After the TCA was removed, samples were resuspended in 50 μl of 2% sodium dodecyl sulfate and 50 mM Tris (pH 7.5). The extracts were diluted in 750 μl of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.5], 500 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate), and an aliquot was counted in a scintillation counter. To normalize the samples, we used equal the numbers of counts per minute. Immunoprecipitation was done in a total volume of 750 μl containing extract, polyclonal antibodies, 25 μl of a 1:1 suspension of protein A-conjugated Sepharose beads, and RIPA buffer. The samples were rocked at 4°C for at least 1 h, and the beads were washed three times with 900 μl of RIPA buffer. Immunoprecipitated proteins were eluted from the beads with Laemmli sample buffer and boiling. The entire sample was then loaded onto an acrylamide gel, and the proteins were visualized by using a Molecular Dynamics Storm 560 PhosphorImager scanning system.
Western analysis.
Samples for Westerns (900 μl) were collected, and ice-cold TCA was added to a final concentration of 5%. Samples were precipitated on ice for at least 30 min, followed by centrifugation. After the TCA was removed, the samples were resuspended directly in Laemmli sample buffer. An equal number of cells was loaded in each lane of the polyacrylamide gels, and the proteins were transferred to a nitrocellulose membrane. The blots were probed with 1:10,000 dilutions of polyclonal rabbit antibodies and then probed with a 1:10,000 dilution of anti-rabbit horseradish peroxidase-conjugated secondary antibody. Western blots were developed with chemiluminescence (Pierce). Equal loading of Western blots was confirmed by comparing the desired band with either a nonspecific band or by reprobing the blot with a different antibody.
RESULTS AND DISCUSSION
Hfq affects the levels of many transcripts.
We used microarray analysis to compare mRNA levels in cells lacking Hfq (MG1655 hfq mutant) with those in wild-type cells (MG1655 hfq+) to identify genes whose expression is directly or indirectly modulated by Hfq. The hfq mutant strain has a kan::hfq insertion that disrupts Hfq function, whereas the hfq+ control has kan inserted at the 3′ end of hfq and retains Hfq function, thereby controlling for polar effects of the insertion (48, 49). We performed eight independent microarray experiments at an OD450 of 0.3, where hfq mutant strains are in balanced growth and σS expression is low (see Materials and Methods). SAM (50) identified small but significant changes in mRNA from 269 genes, with 94 mRNAs increasing and 175 decreasing in the hfq mutant strain (see Table S1 in the supplemental material). Imposing an additional filter of a 1.5-fold cutoff reduced the set to 120 mRNAs, with 48 increasing and 72 decreasing (Table 3). Importantly, six of the mRNAs in the 120 gene data set (ompF, ompT, fepA, rbsD, oppA, and dppA), and two additional mRNAs in the larger 269 gene data set (ptsG [53] and ompA), are known to be regulated by sRNAs, indicating that our approach can reveal Hfq-dependent mRNA regulation. In addition, since only three σS-dependent genes (56) were differentially regulated in the 269 gene set, we were successful in minimizing indirect effects resulting from reduced σS translation in hfq mutant strains.
TABLE 3.
Genes significantly differentially regulated in hfq+ wild-type versus hfq mutant microarray expression analysis
| Gene product type | Gene(s)a |
|---|---|
| Carrier | cyoB, cyoC, cyoD, gltD |
| Partial information | yeaG, yjdA |
| Enzyme | adhE, aldA, amn, aspS, atpA, atpB, atpD, atpE, atpF, atpG, carA, carB, gatY, gatZ, glmU, gyrB, hslV, lon, map, parE, pgk, ppiB, rnpA, thrA, thrB, thrC, truB, xseB, araB, aspC, cysK, degP, gdhA, lacA, lacZ, lpxD, mdh, mdoG, miaA, sthA |
| Factor | dnaK, efp, ffh, groL, groS, hslO, hslU, infB, priB, rdgC, tsf, rpoE, rseA, sufD, yfiA |
| Phage/IS | nmpC, ompT |
| Unknown function | yhcN |
| Regulator | iscR |
| Transporter | gatA, gatB, gatC, lamB, malE, manX, manY, manZ, mgtA, ptsI, cysP, dppA, dppF, fepA, fhuE, fliY, glnH, kgtP, lacY, metQ, mscS, ompF, oppA, oppB, oppC, shiA, sstT |
| Structural component | fliC, rplA, rplD, rplP, rplV, rplY, rpmD, rpmE, rpmG, rpsF, rpsJ, rpsL, rpsS, rpsU |
| Carrier, predicted | ygcQ |
| Regulator, predicted | yqjI, yrbA, htrG |
| Transporter, predicted | rbsD, sdaC, fiu, sbmA, sufB, ybfM, yliB |
Only genes that were significantly differentially regulated and changed expression at least 1.5-fold are listed (120 genes in all). All genes that were significantly differentially regulated are listed in Table S1 in the supplemental material. Genes in boldface have mRNAs that decrease in hfq mutants; genes in regular typeface have mRNAs that increase in the hfq mutant.
Using the classification of Riley et al. (38), we divided the 120 significantly regulated genes into categories based on the location and function of their products and determined whether any category had more Hfq-regulated genes than expected based on chance alone (Table 2). When categorized by location, only outer membrane β-barrel proteins were significantly over-represented (Table 2). This is consistent with experimental results demonstrating that many OMPs are regulated by multiple sRNAs (reviewed in reference 15). When categorized by function, several gene product types were overrepresented, including enzymes, factors, transporters, and structural components (Table 2). Subdivision of each category into up- and downregulated genes revealed additional significant categories. For example, cytoplasmic genes were over-represented in the downregulated group and under-represented in the upregulated group; the converse was true of periplasmic genes. We note that the 269 gene data set also yielded the same trends captured by the smaller data set, suggesting that the larger data set also captures significant patterns of regulation.
TABLE 2.
Location and function of gene products of significantly differentially regulated mRNAs in hfq+ wild-type versus hfq mutant microarray expression analysis
| Category | Total significant genes (n = 120)c |
Upregulated genes (n = 48)d |
Downregulated genes (n = 72)e |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Observedf | Expectedg | P < 0.05h | Observedf | Expectedg | P < 0.05h | Observedf | Expectedg | P < 0.05h | |
| Locationa | |||||||||
| Cytoplasmic | 79 | 76 | 21 | 30 | 2.5 × 10−3 | 58 | 46 | 7.9 × 10−4 | |
| Integral membrane protein | 20 | 21 | 10 | 8 | 10 | 12 | |||
| Membrane anchored | 2 | 4 | 1 | 2 | 1 | 3 | |||
| Membrane lipoprotein | 1 | 0 | 1 | 0 | 0 | 0 | |||
| Outer membrane β-barrel protein | 8 | 1 | 9.2 × 10−5 | 6 | 1 | 2.2 × 10−5 | 2 | 1 | |
| Periplasmic | 10 | 10 | 9 | 4 | 1.0 × 10−2 | 1 | 6 | 1.3 × 10−2 | |
| Gene product typeb | |||||||||
| Carrier | 4 | 2 | 1 | 1 | 3 | 1 | |||
| Partial information | 2 | 4 | 2 | 2 | 0 | 2 | |||
| Enzyme | 40 | 29 | 6.1 × 10−3 | 12 | 12 | 28 | 17 | 2.2 × 10−3 | |
| Factor | 15 | 4 | 8.5 × 10−6 | 4 | 2 | 11 | 2 | 2.0 × 10−5 | |
| Phage/IS | 2 | 9 | 5.8 × 10−5 | 1 | 3 | 1 | 5 | 2.7 × 10−2 | |
| Unknown function | 1 | 13 | 2.4 × 10−5 | 1 | 5 | 2.8 × 10−2 | 0 | 8 | 3.5 × 10−4 |
| Regulator | 1 | 6 | 9.1 × 10−3 | 1 | 3 | 0 | 4 | 1.9 × 10−2 | |
| Transporter | 27 | 9 | 2.1 × 10−7 | 17 | 4 | 3.5 × 10−8 | 10 | 5 | 2.6 × 10−2 |
| Structural component | 14 | 2 | 9.6 × 10−8 | 0 | 1 | 14 | 1 | 1.1 × 10−10 | |
| Carrier, predicted | 1 | 1 | 0 | 0 | 1 | 1 | |||
| Regulator, predicted | 3 | 4 | 1 | 2 | 2 | 3 | |||
| Transporter, predicted | 7 | 7 | 5 | 3 | 2 | 4 | |||
Location of gene products as described in reference 38. Only categories containing the observed genes are displayed.
Type of gene product as described in reference 38. Only categories containing observed genes are displayed.
Genes with significantly differentially regulated mRNAs from hfq+ versus hfq mutant microarrays as identified by SAM and that also changed levels at least 1.5-fold (120 genes).
Upregulated genes have mRNAs that increased in hfq mutants (48 genes).
Downregulated genes have mRNAs that decreased in hfq mutants (72 genes).
Number of gene products in each category.
Expected number of gene products in each category based on the total number of gene products for the entire genome (38).
Probability of number of observed genes in category based on the expected number using binomial statistics. Only categories with P < 0.05 are listed and deemed significant.
The 120 significantly differentially regulated genes are listed by functional category in Table 3. Pathways from the EcoCyc database (21) with at least two members subject to Hfq regulation are listed in Table 4. Together, these data suggest specific themes in Hfq-dependent regulation.
TABLE 4.
Main metabolic pathways affected by Hfq
| Metabolic pathwaya | Enzyme(s) (genes)b |
|---|---|
| Glycolysis/gluconeogenesis + TCA | Phosphoglycerate kinase (pgk); pyruvate kinase II (pykA); malate dehydrogenase (mdh) |
| Mixed acid fermentation | Pyruvate kinase II (pykA); alcohol dehydrogenase/acetaldehyde dehydrogenase (adhE); malate deydrogenase (mdh) |
| Peptidoglycan biosynthesis via UDP-N-acetyl-d-glucosamine pathway | l-Glutamine:d-fructose-6-phophate aminotransferase (glmS); glucosamine-1-phosphate acetyltransferase/N-acetylglucosamine-1-phosphate uridyltransferase (glmU); UDP-N-acetylglucosmanine enolpyruvoyl transferase (murA) |
| KDO2-lipid A biosynthesis | UDP-3-O-[3-hydroxymyristoyl]glucosamine N-acetyltransferase (lpxD); palmitoleoyl acyltransferase (lpxP) |
| Histidine biosynthesis I | ATP phosphoribosyltransferase (hisG); imidazole glycerol phosphate synthase (hisH subunit); imidazole glycerol phosphate dehydratase/histidinol-phosphatase (hisB); histidinol-phosphate aminotransferase (hisC); histidinol/histidinal dehydrogenase (hisD) |
| Threonine biosynthesis | Aspartate kinase I/homoserine dehydrogenase I (thrA); homoserine kinase (thrB); threonine synthase (thrC) |
| Glutamate biosynthesis | Glutamine synthetase (glnA); glutamate synthase (gltD subunit); glutamate dehydrogenase (gdhA) |
| Aspartate biosynthesis | Aspartate transaminase (aspC) |
| Cysteine biosynthesis | Cysteine synthase A (cysK) |
| Alanine biosynthesis | Cysteine desulfurase (iscS) |
| Leucine biosynthesis | Isopropylmalate isomerase (leuC); 3-isopropylmalate dehydrogenase (leuB) |
| Aminoacyl-tRNA synthetases | (valS, leuS, aspS, thrS) |
Only metabolic pathways are listed that contain at least two genes that are significantly differentially regulated by Hfq or are connected to other affected pathways.
Enzymes in which their encoding mRNAs are significantly regulated by Hfq. Genes in boldface have mRNAs that decrease in hfq mutants; enzymes in regular typeface have mRNAs that increase in hfq mutants.
(i) Amino acid uptake and biosynthesis.
In addition to upregulated amino acid and peptide transporters, enzymes in several amino acid biosynthetic pathways exhibited altered regulation (Table 3). This suggests a significant shift in the utilization and synthesis of amino acids in the hfq mutant strain. For example, increases in a subunit of the glutamine ABC transporter (glnH) and in α-ketoglutarate transporter (kgtP), suggests an increase in glutamine and α-ketoglutarate uptake; glutamate synthase (gltD), which utilizes these compounds to synthesize glutamate, is also upregulated, as is glutamate dehydrogenase (gdhA), which synthesizes glutamate via an alternative route. Glutamine is also a precursor for the methionine, threonine, and lysine biosynthetic pathways. Several enzymes of the threonine biosynthetic pathway (thrABC) are downregulated, providing more flux for methionine and lysine biosynthesis; an increase in a serine/threonine transporter (sstT) could maintain threonine. Increased serine uptake may also provide more substrate for cysteine biosynthesis via cysteine synthase (cysK) and for alanine synthesis from cysteine via cysteine desulfurase (iscS), both of which are upregulated. Many additional enzymes involved in amino acid biosynthesis are also upregulated (leuBC and metQ) or downregulated (shiA and aroB). Although regulation could be indirect, the fact that many of these mRNAs are upregulated in hfq mutant cells raises the possibility that amino acid biosynthesis may be extensively regulated by sRNAs.
(ii) Sugars: uptake, metabolism and energetics.
Most downregulated transporters in the hfq mutant strain are involved in the uptake of phosphotransferase system (PTS) sugars (galactitol and exogenous hexoses) and non-PTS sugars (maltose and other maltodextrins). PTS enzyme I (ptsI) is also downregulated, suggesting a general reduction in PTS-dependent sugar uptake. In addition, enzymes involved in glycolysis and/or mixed acid fermentation (pgk and pykA), UDP-N-acetyl-d-glucosamine biosynthesis (glmS and glum), and ethanol (adhE) are downregulated. We do not know whether this is an indirect consequence of reduced growth rate or whether it results from direct regulation by Hfq.
(iii) Cell energetics.
In addition to downregulation of energy production pathways described above, components of electron transfer chains are also downregulated, including formate dehydrogenase O (fdoG), ubiquinone synthesis (ubiD) and the cytochrome bo terminal oxidation complex (cyoBCD). Since the electron transport chain generates proton motif force across the cytoplasmic membrane, this function may be reduced in the hfq mutant cells. Interestingly, several subunits of the ATP synthase (atpA, atpB, atpD, atpE, atpF, and atpG) that use this energy to generate ATP are also downregulated. Again, it is unclear whether these changes are an indirect effect of growth rate changes or indicate a role for Hfq in adjusting the energy status of the cell to its growth state. In this regard, it would be interesting to know whether the synthesis rate of Hfq increases with growth rate; there is currently conflicting data on this point in the literature (4, 19).
Hfq regulates σE activity through OMP mRNA levels.
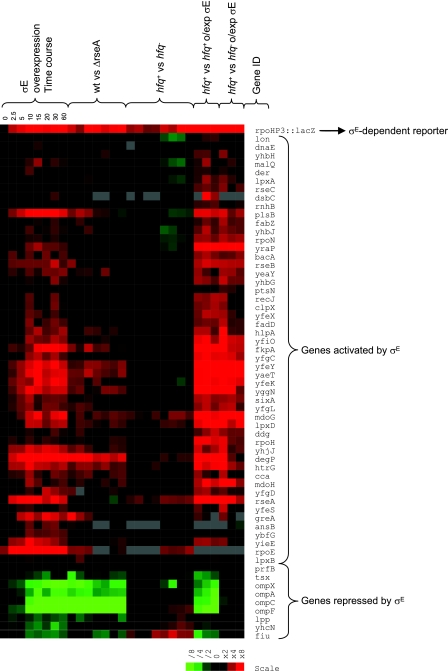
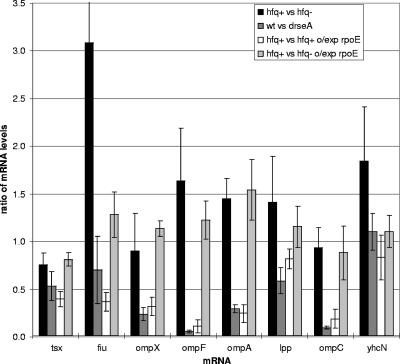
rpoE mRNA encoding σE is upregulated in the E. coli hfq mutant strain (Table 3 and Table S1 in the supplemental material), as has previously been observed for Vibrio cholera (Ding et al., unpublished). σE activity is also elevated in the hfq mutant strain, as evidenced by microarray analysis showing significant induction of several members of the σE regulon and a lacZ reporter fused to a σE-dependent promoter (Fig. 1 and Table S1 in the supplemental material). Induction of the lacZ reporter was confirmed by measuring the β-galactosidase activity to be 10-fold higher in the hfq mutant strain (data not shown), depending upon growth phase, as expected from the σE growth-regulated response (8). However, the downregulation of eight operons encoding one periplasmic and seven OMPs that occurs after σE overexpression is not recapitulated in the hfq mutant strain (Fig. 1 and 2). Lack of downregulation was confirmed for ompF mRNA by Northern analysis (data not shown). Similarly, Figueroa-Bossi et al. recently determined that the downregulation of LamB, OmpA, OmpC, and OmpF require both σE and Hfq in Salmonella enterica (10). This defect in mRNA downregulation does not result from constitutive expression of σE in hfq mutant cells, since cells that constitutively overexpress σE because the RseA antisigma is removed still downregulate these eight mRNAs (Fig. 1 and 2). This suggests that Hfq is required for the rapid downregulation of these mRNAs. Consistent with this idea, when σE was overexpressed in the hfq mutant strain, the induced genes were further upregulated, but the eight mRNAs were not downregulated (Fig. 1 and 2).
FIG. 1.
Expression profiles of the σE regulon. The illustration displays a hierarchical cluster plot of the gene expression patterns of σE regulon members (identified in reference 37) based on an analysis of cDNA microarray data. The first set of columns reproduces a time course microarray in which a wild-type strain (CAG25196) is compared to a wild-type strain after overexpression of σE from an inducible promoter by the addition of IPTG (CAG25197) up to 60 min after overexpression (reproduced from Rhodius et al. [37]). The next set of columns shows seven independent replicates of a steady-state comparison of a wild-type strain (CAG25195) versus a ΔrseA strain (CAG25198); RseA is an antisigma factor for σE. The next set of columns shows eight independent replicates of a steady-state comparison of hfq+ (CAG50008) versus hfq mutant (CAG50009) strains. The next set of columns shows three independent replicates comparing an hfq+ strain (CAG50010) to an hfq+ strain overexpressing σE from an IPTG-inducible promoter (CAG50011) measured 20 min after induction. The final set of columns shows three independent replicates comparing an hfq+ strain (CAG50010) to an hfq mutant strain overexpressing σE from an IPTG-inducible promoter (CAG50011) measured 20 min after induction. For each microarray comparison, red denotes increased mRNA and green indicates decreased mRNA in the experimental strain (control versus experimental); the fold change is indicated by the scale at the bottom of the figure. The time in minutes after the induction of rpoE for the time course is indicated at the top of the figure. Genes are identified by their unique ID and name (Gene ID).
FIG. 2.
Hfq is required for the decrease in OMP RNAs levels after σE overexpression. The averaged mRNA expression ratios for rpoE (σE) and the downregulated RNAs (tsx, fiu, ompX, ompF, ompA, lpp, ompC, and yhcN) derived from the microarray data presented in Fig. 1 are compared in various strains: hfq+ versus hfq mutant, rseA+ versus rseA mutant (wt versus drseA) and 20 min after overexpression of σE in both hfq+ (hfq+ versus hfq+ o/exp rpoE) and hfq mutant strains (hfq+ versus hfq− o/exp rpoE). Error bars represent one standard deviation.
Why are the eight mRNAs not downregulated in hfq mutant strains even though σE activity increases? To answer this question, we examined why overexpression of σE results in rapid downregulation of these mRNAs in wild-type strains. The initial rate of decrease of these mRNAs after σE overexpression (Fig. 3) is equivalent to or greater than the literature values for the half-lives of these RNAs (reported in reference 5). Therefore, the mRNA downregulation is unlikely to result solely from transcriptional repression since even complete and immediate transcriptional repression would result in a decrease in mRNA levels equivalent only to the half-life. Thus, the rapidity of this process suggests that shutoff involves σE-dependent regulated degradation of these mRNAs.
FIG. 3.
Overexpression of σE in wt cells results in a rapid decrease of a subset of mRNAs. A subset of mRNAs shows decreased expression after overexpression of σE. Using the time course data presented in Fig. 1 (wild-type strain CAG25196 compared to the wild type after overexpression of σE [strain CAG25197]; time course data are from Rhodius et al. [37]), the averaged RNA expression ratios are shown as a function of time after σE overexpression. The observed mRNA decay half-lives were estimated by fitting the data to an exponential curve (see Materials and Methods). Previously determined mRNA half-lives are from the data of Bernstein et al. (5).
The simplest explanation for these findings is that Hfq-dependent sRNAs controlled by σE target these eight mRNAs for downregulation; upon σE overexpression, these sRNA increase and downregulate the mRNAs by increasing their degradation and/or inhibiting their translation. Compelling evidence for this idea has recently been presented in work that appeared while this paper was under review. First, micA, which targets ompA mRNA for degradation in both Salmonella and E. coli (10, 18, 33, 35, 51), was shown to be σE dependent (10, 18, 33). Second, rybB, which targets ompA, ompC, tsx, and other mRNAs for degradation, is σE dependent (18, 33). It is likely that a similar mechanism is used to control the additional downregulated genes observed here, but the σE-dependent sRNAs have yet to be identified. We note that for two mRNAs (fiu and yhcN), an Hfq-independent mode of repression is also evident since overexpression of σE in an hfq mutant strain results in decreased expression of these two RNAs (Fig. 2, compare the findings for “hfq+ versus hfq−” and “hfq+ versus hfq− o/exp rpoE”).
We suggest that overexpression of periplasmic proteins and OMPs may be the cause of increased σE activity in hfq mutant cells. The downregulated genes encode for one conserved periplasmic protein (yhcN), one outer membrane murein lipoprotein (lpp), and six outer membrane β-barrel proteins (ompA, ompC, ompF, ompX, fiu, and tsx). Our data demonstrate that fiu, ompF, ompA, lpp, and yhcN mRNAs are induced in the hfq mutant strain relative to wild-type cells (Fig. 2), probably because these mRNAs are no longer subject to sRNA-mediated degradation. Overproduction of each of these proteins is sufficient to induce σE activity (27, R. Chaba and C. Gross, unpublished data), which supports this hypothesis. Interestingly, this mechanism may not explain σE induction in hfq mutant Vibrio cholerae (8a), since there is no significant induction of these homologs or other outer membrane porins in that strain.
Hfq regulates σ32 activity.
Our microarray analysis revealed that several members of the σ32-transcribed stress regulon are downregulated in hfq mutant cells even though there is no decrease in σ32 mRNA (Table 5 and Table S1 in the supplemental material). Regulation results from decreased σ32-dependent transcription because the σ32-dependent PhtpG::lacZ reporter fusion was downregulated two- to threefold in hfq mutant cells (Fig. 4, lane 1). Given the microarray data and the reporter analysis, we suspect that all σ32-dependent transcription is slightly decreased in hfq mutant strains, although the effects are too subtle to be definitive for less well induced members of the regulon.
TABLE 5.
Effects of Hfq on mRNA levels of σ32 and its regulon members
| Gene | Avg expression ratio in hfq+ wild type vs hfq mutanta |
|---|---|
| rpoH | 1.21 (1.09, 1.35) |
| dnaKJb | 0.67 (0.50, 0.89) |
| lon | 0.64 (0.44, 0.92) |
| htpG | 0.71 (0.58, 0.87) |
| topA | 0.77 (0.69, 0.86) |
| hslVUb | 0.59 (0.43, 0.79) |
| groES groELb | 0.48 (0.31, 0.75) |
Average microarray mRNA ratios of hfq+ wild type (CAG50008) versus the hfq mutant (CAG50009). Averages and standard deviations were calculated from the log2-transformed data of eight independent microarrays (the average ±1 standard deviation is given in parentheses). The values are presented as ratios.
Data for first gene of operon.
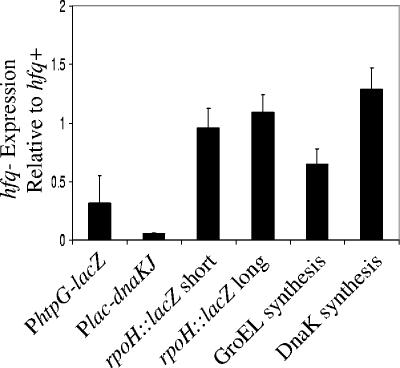
FIG. 4.
Effects of hfq mutant on σ32. Various assays (see the text) were used to compare hfq mutant and hfq+ strains during exponential growth in LB medium. The effects in the hfq mutant strain are shown relative to the hfq+ strain. Lane 1, σ32 activity measured by determining the amount of β-galactosidase (Miller units) from a σ32-dependent PhtpG-lacZ reporter (CAG48422 versus CAG48383); lane 2, σ32 activity (as in lane 1) in strains where the endogenous dnaKJ promoter is replaced by an IPTG-inducible PA1/lac0-1 promoter (CAG48401 versus CAG48400); lane 3, activity of a short rpoH-lacZ translational fusion containing the first 22 amino acids from σ32 (CAG48409 versus CAG48408); lane 4, activity of a long rpoH-lacZ translational fusion containing most of the σ32 coding region (amino acids 1 to 266) (CAG48411 versus CAG48410); lane 5, GroEL protein synthesis rates measured by using [35S]methionine pulse-labeling immunoprecipitation (CAG48422 versus CAG48383); lane 6, DnaK protein synthesis rates measured by using [35S]methionine pulse-labeling immunoprecipitation (CAG48422 versus CAG48383). Error bars represent one standard deviation.
σ32 can be regulated by changing its transcription, translation, stability, or specific activity. We used lacZ fusions susceptible to different regulatory steps to investigate which step(s) were defective in hfq mutant cells. A short rpoH::lacZ translational fusion (amino acids 1 to 22 of σ32 fused to LacZ) driven from the σ32 promoter is sensitive only to altered σ32 transcription (20); a long rpoH::lacZ fusion (amino acids 1 to 266 of σ32 fused to LacZ) is sensitive to altered transcription, translation, and stability of σ32 (43). Neither fusion was affected by the Hfq status of the cell (Fig. 4, lanes 3 and 4), indicating that Hfq does not alter σ32 transcription, translation, or stability. The only remaining regulatory step is at the level of activity; thus, Hfq must affect σ32 activity. Since Hfq affects the activity but not the concentration of σ32, it must affect the specific activity of σ32.
σ32 specific activity is regulated by the GroEL/S and DnaK/J chaperones (15, 45). When these chaperones are in excess, they directly bind to and inhibit σ32 (11, 15, 23). Therefore, we tested whether either chaperone machine was upregulated in hfq mutant cells by measuring chaperone synthesis rates (15). GroEL protein synthesis was downregulated in concert with the downregulation of its mRNA observed in the microarray experiments of hfq mutant cells (Fig. 4, lane 5, and Table 5). In contrast, the synthesis of DnaK protein was upregulated despite downregulation of its mRNA (Fig. 4, lane 6, and Table 5). These results suggest that two competing effects control DnaK level in hfq mutant cells: (i) transcriptional downregulation of dnaK mRNA due to decreased σ32 activity and (ii) translational upregulation of DnaK protein due to lack of Hfq. This model predicts that if DnaK transcriptional downregulation in hfq mutant cells were eliminated, the level of DnaK/J would be even higher and σ32 activity would be further inhibited. We eliminated downregulation by expressing dnaKJ from a PA1/lac0-1 promoter (46), which is not regulated either by Hfq or σ32, using conditions known to give transcription equivalent to that in unstressed wild-type (hfq+) cells. Consistent with this model, such hfq mutant cells exhibit more severe repression of σ32 activity than when dnaKJ is expressed from its endogenous promoter (Fig. 4, lane 2 compared to lane 1).
We next determined the effects of Hfq during heat stress. Because cells lacking Hfq had increased levels of one of the major chaperone machines (DnaKJ) but decreased levels of the other major chaperone machine (GroELS), we were unable to predict the consequences to the stress response. We found that upon a standard heat shock from 30 to 42°C, hfq mutant cells had a bigger heat shock response than hfq+ cells, characterized by a larger maximum increase and longer duration of σ32 induction (Fig. 5).
FIG. 5.
Hfq affects the magnitude and duration of the heat shock response. hfq+ (CAG48383) and hfq mutant (CAG48422) cells were subjected to a temperature upshift from 30 to 42°C during exponential growth in M9 minimal medium supplemented with all amino acids except methionine and cysteine. [35S]methionine pulse-labeling immunoprecipitation was used to measure the synthesis rate of HtpG, a σ32-dependent protein. HtpG synthesis was normalized to the rate at time = 0 for each strain.
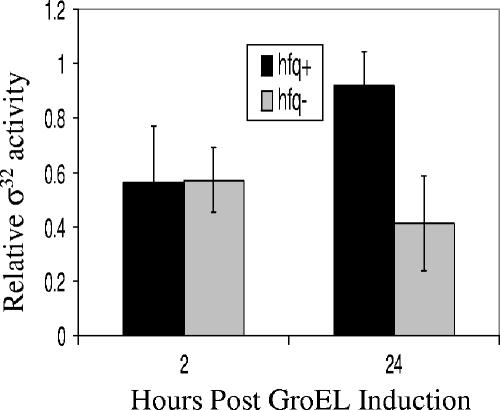
We examined more closely whether the GroEL-mediated negative feedback loop controlling σ32 activity was also compromised by comparing the response of hfq+ cells and hfq mutant cells to GroEL overexpression. As expected, both wild-type hfq+ and hfq mutant cells are inhibited after 2 h of chaperone overexpression (Fig. 6), a finding consistent with previous reports on the effect of GroEL overexpression (15). We note that measuring σ32 activity using Miller units during such a short time course underestimates the amount of σ32 inhibition as this measure includes the β-galactosidase accumulated prior to inhibition.
FIG. 6.
Hfq affects long-term adaptation of σ32 to GroEL. GroEL was overexpressed from an arabinose inducible promoter at time = 0 in both hfq+ (CAG48402) and hfq mutant (CAG48403) cells during exponential growth in LB medium. The σ32 activity was assayed by using a PhtpG-lacZ reporter at 2 and 24 h postinduction. The β- galactosidase activity was measured in Miller units and then normalized to the activity at time = 0. Error bars represent one standard deviation.
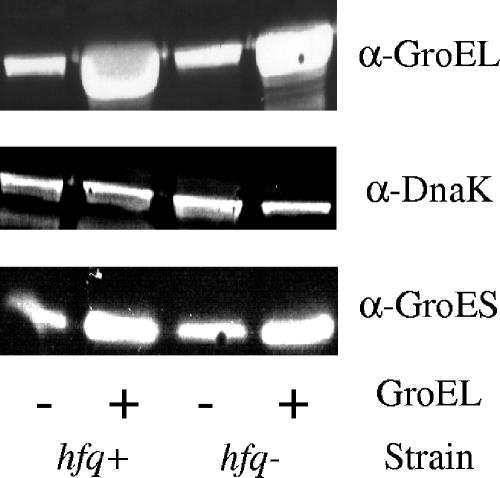
We next examined the role of Hfq in response to chronic GroEL overexpression. wild-type cells recover normal σ32 activity after extended (24 h) exposure to GroEL overexpression, a process we call “long-term adaptation” (15). Interestingly, we found that hfq mutant cells are defective in long-term adaptation, indicating that Hfq plays a role in adaptation in wild-type cells (Fig. 6). We investigated the mechanism of long-term adaptation. First, we showed that cells that have adapted to chronic chaperone overexpression are still sensitive to transient chaperone overexpression, suggesting that adaptation is a reversible process and not due to a genetic change in the cells (data not shown). We also showed that long-term adaptation is independent of the expression system used since it is observed whether GroEL was induced from tetracycline-, IPTG-, or arabinose-based systems (data not shown). In addition, we ruled out the possibility that adaptation is due to a decrease in the levels of GroEL or the GroEL cochaperone GroES by measuring protein levels with Western analysis (Fig. 7). Furthermore, we show that adaptation is not due to altered levels of DnaK, since DnaK levels are slightly higher in hfq mutant cells than in hfq+ cells both before and during adaptation (Fig. 7). These results indicate that modulation of long-term adaptation is a second, independent effect of Hfq on σ32 regulation.
FIG. 7.
The levels of DnaK, GroEL, and GroES are unaffected by Hfq during adaptation. The effects of long-term GroEL overexpression were examined in both hfq+ (CAG48402) and hfq mutant (CAG48403) strains by using Western analysis. GroEL was overexpressed from an arabinose-inducible promoter. Protein levels of DnaK, GroEL, and GroES are shown at 24 h after GroEL induction.
In summary, we showed that Hfq regulates the σ32-mediated cytoplasmic heat shock response in at least two different ways. First, Hfq regulates DnaK translation, thereby indirectly influencing σ32 activity via the DnaK-mediated negative feedback loop. This is the first report indicating that chaperones are subject to posttranscriptional regulation and that this regulation permits differential chaperone expression under certain conditions. In some prokaryotes, groEL and dnaK are controlled by different transcription factors, which permit their differential regulation. In E. coli, Hfq-mediated regulation may accomplish this same goal. Second, Hfq is required for “long-term adaptation” of σ32 to chronic chaperone overexpression, in a process independent of alterations in DnaK level. Further work will be necessary to determine whether either type of regulation is mediated by Hfq-dependent sRNAs.
Summary and future prospects.
Prior to this work, many sRNAs had been identified, but only a few of their targets were known. Our microarray analysis suggests additional targets to test for sRNA regulation, many of which are over-represented for specific cellular functions. In addition, we have expanded the involvement of Hfq in stress responses. Prior to this work, Hfq was implicated only in the regulation of the σS stress response in E. coli. Our work demonstrates that Hfq is involved in at least two additional E. coli stress responses, those mediated by σE and σ32, suggesting that Hfq may play a general role in regulation of stress responses. This may be due to some intrinsic advantage to using noncoding RNAs and/or Hfq in regulation of a stress response, or it may reflect some level of coordination of stress responses by having them utilize a common regulator. Further work will determine whether the involvement of Hfq in stress responses is common in prokaryotic organisms.
Supplementary Material
Acknowledgments
We acknowledge Stacy Chen for strain construction.
This study was supported by National Institutes of Health grants GM57755 and GM32678 (to C.A.G.).
Footnotes
Published ahead of print on 8 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ades, S. E. 2004. Control of the alternative sigma factor sigmaE in Escherichia coli. Curr. Opin. Microbiol. 7:157-162. [DOI] [PubMed] [Google Scholar]
- 2.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52:613-619. [DOI] [PubMed] [Google Scholar]
- 3.Alba, B. M., H. J. Zhong, J. C. Pelayo, and C. A. Gross. 2001. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma activity. Mol. Microbiol. 40:1323-1333. [DOI] [PubMed] [Google Scholar]
- 4.Azam, T., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 8.Costanzo, A., and S. E. Ades. 2006. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J. Bacteriol. 188:4627-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Ding, Y., B. M. Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates σE expression. Mol. Microbiol. 53:345-354. [DOI] [PubMed] [Google Scholar]
- 9.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueroa-Bossi, N., S. Lemire, D. Maloriol, R. Balbontin, J. Casadesus, and L. Bossi. 2006. Loss of Hfq activates the sigma-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 62:838-852. [DOI] [PubMed] [Google Scholar]
- 11.Gamer, J., H. Bujard, and B. Bukau. 1992. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor sigma 32. Cell 69:833-842. [DOI] [PubMed] [Google Scholar]
- 12.Gamer, J., G. Multhaup, T. Tomoyasu, J. S. McCarty, S. Rudiger, H. J. Schonfeld, C. Schirra, H. Bujard, and B. Bukau. 1996. A cycle of binding and release of the DnaK, DnaJ, and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor sigma32. EMBO J. 15:607-617. [PMC free article] [PubMed] [Google Scholar]
- 13.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303-328. [DOI] [PubMed] [Google Scholar]
- 14.Guillier, M., S. Gottesman, and G. Storz. 2006. Modulating the outer membrane with small RNAs. Genes Dev. 20:2338-2348. [DOI] [PubMed] [Google Scholar]
- 15.Guisbert, E., C. Herman, C. Z. Lu, and C. A. Gross. 2004. A chaperone network controls the heat shock response in Escherichia coli. Genes Dev. 18:2812-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guyer, M. S., R. R. Reed, J. A. Steitz, and K. B. Low. 1981. Identification of a sex-factor-affinity site in Escherichia coli as gamma delta. Cold Spring Harbor Symp. Quant. Biol. 45(Pt. 1):135-140. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansen, J., A. A. Rasmussen, M. Overgaard, and P. Valentin-Hansen. 2006. Conserved small non-coding RNAs that belong to the sigma(E) regulon: role in downregulation of outer membrane proteins. J. Mol. Biol. 364:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Kajitani, M., A. Kato, A. Wada, Y. Inokuchi, and A. Ishihama. 1994. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q.β. J. Bacteriol. 176:531-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamath-Loeb, A. S., and C. A. Gross. 1991. Translational regulation of sigma 32 synthesis: requirement for an internal control element. J. Bacteriol. 173:3904-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33:D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lease, R. A., M. E. Cusick, and M. Belfort. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. USA 95:12456-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberek, K., T. P. Galitski, M. Zylicz, and C. Georgopoulos. 1992. The DnaK chaperone modulates the heat shock response of Escherichia coli by binding to the sigma 32 transcription factor. [DOI] [PMC free article] [PubMed]
- 24.Majdalani, N., S. Chen, J. Murrow, K. St John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382-1394. [DOI] [PubMed] [Google Scholar]
- 25.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813-826. [DOI] [PubMed] [Google Scholar]
- 27.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618-2628. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, New York, NY.
- 29.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, New York, NY.
- 30.Nishihara, K., M. Kanemori, M. Kitagawa, H. Yanagi, and T. Yura. 1998. Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 64:1694-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nonaka, G., M. Blankschien, C. Herman, C. A. Gross, and V. A. Rhodius. 2006. Regulon and promoter analysis of the Escherichia coli heat shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 20:1776-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opdyke, J. A., J. G. Kang, and G. Storz. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186:6698-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papenfort, K., V. Pfieffer, F. MIka, S. Lucchini, J. Hinton, and J. Vogel. 2006. Sigma E-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62:1674-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 35.Rasmussen, A. A., M. Eriksen, K. Gilany, C. Udesen, T. Franch, C. Petersen, and P. Valentin-Hansen. 2005. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 58:1421-1429. [DOI] [PubMed] [Google Scholar]
- 36.Rhodius, V., T. K. Van Dyk, C. Gross, and R. A. LaRossa. 2002. Impact of genomic technologies on studies of bacterial gene expression. Annu. Rev. Microbiol. 56:599-624. [DOI] [PubMed] [Google Scholar]
- 37.Rhodius, V. A., W. C. Suh, G. Nonaka, J. West, and C. A. Gross. 2006. Conserved and variable functions of the sigma(E) stress response in related genomes. PLoS Biol. 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riley, M., T. Abe, M. B. Arnaud, M. K. Berlyn, F. R. Blattner, R. R. Chaudhuri, J. D. Glasner, T. Horiuchi, I. M. Keseler, T. Kosuge, H. Mori, N. T. Perna, G. Plunkett III, K. E. Rudd, M. H. Serres, G. H. Thomas, N. R. Thomson, D. Wishart, and B. L. Wanner. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot-2005. Nucleic Acids Res. 34:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 40.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 41.Sledjeski, D. D., C. Whitman, and A. Zhang. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storz, G., J. A. Opdyke, and A. Zhang. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7:140-144. [DOI] [PubMed] [Google Scholar]
- 43.Straus, D. B., W. A. Walter, and C. A. Gross. 1987. The heat shock response of Escherichia coli is regulated by changes in the concentration of sigma 32. Nature 329:348-351. [DOI] [PubMed] [Google Scholar]
- 44.Tatsuta, T., T. Tomoyasu, B. Bukau, M. Kitagawa, H. Mori, K. Karata, and T. Ogura. 1998. Heat shock regulation in the ftsH-null mutant of Escherichia coli: dissection of stability and activity control mechanisms of sigma32 in vivo. Mol. Microbiol. 30:583-593. [DOI] [PubMed] [Google Scholar]
- 45.Tilly, K., N. McKittrick, M. Zylicz, and C. Georgopoulos. 1983. The DnaK protein modulates the heat-shock response of Escherichia coli cell 34:641-646. [DOI] [PubMed]
- 46.Tomoyasu, T., T. Ogura, T. Tatsuta, and B. Bukau. 1998. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol. Microbiol. 30:567-581. [DOI] [PubMed] [Google Scholar]
- 47.Tseng, G. C., M. K. Oh, L. Rohlin, J. C. Liao, and W. H. Wong. 2001. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 29:2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsui, H. C., G. Feng, and M. E. Winkler. 1996. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K-12 from clustered Eσ32-specific promoters during heat shock. J. Bacteriol. 178:5719-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsui, H. C., H. C. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13:35-49. [DOI] [PubMed] [Google Scholar]
- 50.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Udekwu, K. I., F. Darfeuille, J. Vogel, J. Reimegard, E. Holmqvist, and E. G. Wagner. 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 19:2355-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 51:1525-1533. [DOI] [PubMed] [Google Scholar]
- 53.Vanderpool, C. K., and S. Gottesman. 2004. Involvement of a novel transcriptional activator and small RNA in posttranscriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54:1076-1089. [DOI] [PubMed] [Google Scholar]
- 54.Vogel, J., V. Bartels, T. H. Tang, G. Churakov, J. G. Slagter-Jager, A. Huttenhofer, and E. G. Wagner. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31:6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yura, T., M. Kanemori, and M. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 59.Zhang, A., K. M. Wassarman, C. Rosenow, B. C. Tjaden, G. Storz, and S. Gottesman. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50:1111-1124. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, Y., S. Sun, T. Wu, J. Wang, C. Liu, L. Chen, X. Zhu, Y. Zhao, Z. Zhang, B. Shi, H. Lu, and R. Chen. 2006. Identifying Hfq-binding small RNA targets in Escherichia coli. Biochem. Biophys. Res. Commun. 343:950-955. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.