Abstract
α-Synuclein is a major component of Lewy bodies in Parkinson's disease. Although no signal sequence is apparent, α-synuclein expressed in Escherichia coli is mostly located in the periplasm. The possibilities that α-synuclein translocated into the periplasm across the inner membrane by the SecA or the Tat targeting route identified in bacteria and that α-synuclein was released through MscL were excluded. The signal recognition particle-dependent pathway is involved in the translocation of α-synuclein. The C-terminal 99-to-140 portion of the α-synuclein molecule plays a signal-like role for its translocation into the periplasm, cooperating with the central 61-to-95 section. The N-terminal 1-to-60 region is not required for this translocation.
Parkinson's disease is the second most common neurodegenerative disease resulting from the loss of dopaminergic neurons and affects approximately 2% of the population over the age of 65 (17). α-Synuclein (AS) in fibrillar form has been found to be a major component of proteinacous Lewy bodies in diseased neurons (33, 43, 52). AS is an acidic protein composed of 140 amino acid residues and exists as a natively unfolded protein under physiological conditions (65). The N-terminal 1-to-60 region, containing five imperfect repeats of KTK(E/Q)GV, is highly conserved, while the C-terminal 96-to-140 portion, with Ca2+-binding activity, is not well conserved (37). The central 61-to-95 section is termed nonamyloid β component protein (NAC) (60), as it can also be isolated from senile plaques in the brains of Alzheimer's disease patients. The hydrophobic 71-to-82 stretch has been found to be essential for filament assembly (19). A few AS-transgenic models, such as fly (3), mouse (50), and yeast (39) models, have been constructed for in vivo Parkinson's disease study. However, the physiological functions of AS and the link between Parkinson's disease and AS remain obscure (41, 64).
Recombinant AS has been produced in Escherichia coli since 1994 by a number of different methods (24) which, without exception, use a whole-cell extract as a protein source. It surprised us to find that the expressed AS in E. coli bearing a cloned AS cDNA without any signal sequence was located not in the cytoplasm, as expected, but instead in the periplasm. Proteins synthesized in the cytoplasm must be translocated to the correct compartment to exert their specific functions. Three main targeting routes for protein export into the periplasm in E. coli have been identified. (i) The SecA-dependent pathway, the most common, exports unfolded proteins with an N-terminal signal peptide posttranslationally. The key component of the system is the ATPase SecA, which supplies energy for preprotein translocation by mediating repeated cycles of ATP binding and hydrolysis (46). (ii) The signal recognition particle (SRP)-dependent pathway, a cotranslational protein export pathway, functions mainly in the targeting and integration of membrane proteins from the cytoplasm (61, 47, 31), although it is also involved in the translocation of some secreted proteins (30, 49). The above two pathways converge at the translocon SecYEG (62). (iii) The twin-arginine-targeting (Tat)-dependent pathway is responsible for the translocation of polypeptides with a twin-arginine motif as a signature located near the N terminus of the leader peptide (56) and is also used by a subset of preproteins that are folded prior to translocation (7). Five proteins, TatA, TatB, TatC, TatD, and TatE, are required for the operation of the Tat transport system, which is generally incompatible with the Sec machinery (40). In addition, E. coli has mechanosensitive channels (Msc) which can open large pores in the plasma membrane in response to membrane tension or osmotic shifts (53). According to conductance, Msc proteins are classified into three families, MscM (mini), MscS (small), and MscL (large). Nonspecific protein leaking into the periplasm was reported to occur through MscL as a response to osmotic shock (2, 8).
In this study, we have examined the pathway by which expressed AS translocates to the periplasm and the component parts of the AS molecule responsible for this translocation. The results show that AS translocation is neither through the SecA and Tat routes nor through MscL but appears to be mediated by the SRP-dependent pathway. We have found that the C-terminal 99-to-140 portion is a major part responsible for the translocation of AS into the periplasm and the N-terminal 1-to-60 region is not required.
MATERIALS AND METHODS
Materials.
The E. coli strains and plasmids used in this study are listed in Table 1. PB104 and PB106, kindly donated by Paul Blount, are PB102 and PB107, respectively, with mscL completely deleted. Both the wild-type and deletion strains have been made recA negative (53). The mutant strains have been well characterized and used in a number of studies (2, 8, 10). B1LK0 and DADE, generously given by Tracy Palmer, are MC4100 with gene deletions that completely block the Tat system (11, 66). The Ffh conditional strain WAM121 (16) was generously provided by Eitan Bibi. Anti-AS serum was a kind gift from Qihong Sun. HtrA, a protease in the E. coli periplasm, was generously provided by Zengyi Chang. The plasmid pQE60-RNase I contains the full-length coding sequence of the E. coli RNase I precursor cloned from JM109 genomic DNA. Chemicals were local products of analytical grade unless specified otherwise.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | F−ompT hsdS(rB− mB−) gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | Novagen |
| M15(pREP4) | NaIs Strs Rifsthi lac ara gal mtl F−recA+uvr+lon+ pREP4 | QIAGEN |
| JS7131 | MC1060 ΔyidC attB::R6Kori ParaBAD-yidC+ (Specr) | R. Dalbey |
| DH5α | deoR endA1 gyrA96 hsdR17(rK− mK+) recA1 relA1 supE44 thi-1 Δ(lacZYA-argF)169 φ80dlacZΔM15 F− λ− | Invitrogen |
| MC4100 | araD139 Δ(argF-lac)U169 rpsL150 thiA relA1 flbB5301 deoC1 ptsF25 rbsR | T. Palmer |
| B1LK0 | MC4100 ΔtatC | |
| DADE | MC4100 ΔtatABCDE | |
| WAM121 | MC4100 ffh-1::kan attB::R6Kori ParaBAD-ffh+ (Cmr) | E. Bibi |
| PB102 | AW405 ΔrecA | P. Blount |
| PB104 | AW405 ΔrecA ΔmscL | |
| PB106 | AW737 ΔrecA ΔmscL | |
| PB107 | AW737 ΔrecA | |
| Plasmids | ||
| pET-3a-AS | pET-3a, AS | 21 |
| pQE-AS | pQE60, AS | This work |
| pQE-ASΔ(1-60) | pQE60, ASΔ(1-60) | This work |
| pQE-ASΔ(121-140) | pQE60, ASΔ(121-140) | This work |
| pQE-ASΔ(111-140) | pQE60, ASΔ(111-140) | This work |
| pQE-ASΔ(99-140) | pQE60, ASΔ(99-140) | This work |
| pQE-ASΔ(99-110) | pQE60, ASΔ(99-110) | This work |
| pQE-ASΔ(61-95) | pQE60, ASΔ(61-95) | This work |
| pQE-ASΔ(61-95, 121-140) | pQE60, ASΔ(61-95, 121-140) | This work |
| pQE-ASΔ(61-95, 111-140) | pQE60, ASΔ(61-95, 111-140) | This work |
| pQE-DsbC | pQE30, DsbC precursor | 29 |
| pQE30-DsbCn-DsbA | pQE30, DsbCn-DsbA | 67 |
| pQE30-sDsbCn-AS | pQE30, DsbCn-AS with signal peptide of DsbC | This work |
| pQE30-DsbCn-AS | pQE30, DsbCn-AS without signal peptide of DsbC | This work |
| pQE16 | DHFR | QIAGEN |
| pQE16-AS(1-95) | DHFR-AS(1-95) | This work |
| pQE16-AS(96-140) | DHFR-AS(96-140) | This work |
| pQE16-AS(96-120) | DHFR-AS(96-120) | This work |
| pQE16-AS(96-110) | DHFR-AS(96-110) | This work |
| pQE30-Trx | pQE30, Trx | 54 |
| pOE30-Trx-AS(99-140) | pQE30, Trx-AS(99-140) | 28 |
| pQE-DsbA | pQE30, MRGSH6GS-DsbA precursor | 29 |
| pQE60-RNase I | pQE60, RNase I precursor | This work |
AS mutations.
All mutations of AS (Table 1) were constructed with the plasmid pET-3a-AS containing human AS cDNA (21) as a template and pQE-60 (QIAGEN) as an expression vector, unless otherwise specified. The sequences of the constructs were verified by DNA sequencing.
Cell growth.
M15(pREP4) and BL21(DE3) cells were used with pQE-60 and pET-3a (Novagen), respectively. WAM121 was grown in Luria-Bertani medium containing 100 μg/ml ampicillin, 25 μg/ml kanamycin, and 0.2% arabinose or glucose. JS7131 was grown in Luria-Bertani medium containing 100 μg/ml ampicillin, 25 μg/ml spectinomycin, and 0.2% arabinose or glucose. Unless otherwise specified, all other cell strains were cultured in Luria-Bertani medium with 100 μg/ml ampicillin. At an optical density at 600 nm of 0.3 to 0.4, the cells were induced with 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG; Merck) for 4 h at 37°C. NaN3 was included in the culture at 3 mM before induction with IPTG (38) to inhibit the ATPase activity of SecA and thus to block SecA-dependent translocation.
Cell fractionation.
Osmotic shock was carried out basically as described by Shevchik et al. (48). Briefly, the cell pellet from 1 ml of culture was resuspended in 100 μl ice-cold osmotic shock buffer (30 mM Tris-HCl, 40% sucrose, 2 mM Na2-EDTA, pH 7.2) and incubated for 10 min at room temperature. The pellet collected by centrifugation at 10,000 × g for 10 min was resuspended rapidly with 90 μl ice-cold water, followed by adding 10 μl of 20 mM MgCl2, and kept on ice for 3 min. The supernatant collected after centrifugation at 12,000 × g for 10 min contained periplasmic proteins, and the pellet was resuspended in 100 μl water for either sonication or electrophoresis. After sonication, the supernatant collected after centrifugation at 12,000 × g for 10 min at 4°C contained soluble proteins in the cytoplasm, and the pellet contained inclusion bodies.
Chloroform treatment was performed essentially as described by Thorstenson et al. (57). The cell pellet from 1 ml of culture was resuspended in 10 μl Tris-EDTA buffer, vortexed for 10 s after addition of 10 μl chloroform, and incubated on ice for 30 min with an additional 80 μl Tris-EDTA buffer. The supernatant after centrifugation at 6,000 × g for 20 min was the periplasm fraction.
Lysozyme-EDTA-sucrose treatment was performed principally as described by Birdsell and Cota-Robles (9). The cell pellet from 1 ml of culture was resuspended in 100 μl of 10 mM Tris-HCl buffer containing 0.5 M sucrose and 20 μg/ml lysozyme (pH 8.0), incubated for 10 min at room temperature, diluted 1:1 with 10 mM Tris-HCl containing 2 mM EDTA, and incubated on ice for 5 min to ensure complete formation of spheroplasts. The supernatant collected after centrifugation at 12,000 × g for 10 min contained periplasmic proteins. Aliquots of 100 μl lysozyme-treated cells before centrifugation were incubated with 20 μg/ml proteinase K (Merck) in the presence of 1 mM CaCl2 for 1 h at 4°C. The reaction was stopped by incubation with 5 mM phenylmethylsulfonyl fluoride for 3 min on ice and analyzed by electrophoresis to check the proteinase K accessibility of AS.
Electrophoresis was carried out by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-PAGE), and the bands were visualized with Coomassie blue and anti-AS serum as specified. Coomassie blue-stained bands were scanned with a Flatbed scanner13 (Acer), and Western blot profiles were pictured by a Kodak Z7590 digital camera. The bands were quantitatively analyzed by using Totallab version 1.10. The sum of all bands in a lane was taken as the total proteins. The ratio of the AS band to the total proteins of the whole cell was taken as the expression level. The ratio of the band for AS or mutant proteins in the periplasm fraction to that in the whole cell was taken as the translocation efficiency of the AS proteins.
β-Galactosidase, a marker enzyme for the cytoplasm, hydrolyzes o-nitrophenol-β-d-galactopyranoside to produce o-nitrophenol, which can be determined quantitatively by spectrometry (45). The whole-cell extract or periplasm fraction was diluted 10-fold with phosphate-buffered saline, and 40 μl of sample was added to a cuvette containing 590 μl Z buffer (phosphate-buffered saline with 1 mM MgSO4 and 50 mM β-mercaptoethanol) and 120 μl of 4 mg/ml o-nitrophenol-β-d-galactopyranoside incubated at 37°C. Absorbance at 420 nm was immediately monitored for 1 min with a Shimadzu UV2501 spectrophotometer. The slope of the linear part of the absorbance curve was taken as the activity.
Protein preparation.
The overnight culture of the transformed M15(pREP4) cells grown in Luria-Bertani medium with 100 μg/ml ampicillin and 25 μg/ml kanamycin was incubated for another 4 h after 100-fold dilution and then induced with 1 mM IPTG for 4 h at 37°C. AS and ASΔ(61-95) were purified by chromatography of the supernatant after osmotic shock on a Q-Sepharose fast-flow column. For ASΔ(61-95, 121-140) and ASΔ(61-95, 111-140), the cells were lysed in 50 mM Tris-HCl, pH 8.0, (hereafter referred to as Tris buffer) containing 5 M urea in a boiling water bath for 10 min. The supernatant was dialyzed overnight against Tris buffer and subjected to chromatography on a Q-Sepharose fast-flow column with a gradient of 0 to 0.2 M NaCl in Tris buffer. The peak at around 0.07 M for ASΔ(61-95, 121-140) or at around 0.05 M NaCl for ASΔ(61-95, 111-140) was collected. For ASΔ(99-140), the supernatant of sonicated cells was incubated with 0.2 M NaCl in a boiling water bath for 10 min. The supernatant was chromatographed on a Q-Sepharose fast-flow column after thorough dialysis, and the flowthrough was further purified by CM-Sephadex C-25 with a gradient of 0 to 0.15 M NaCl. The peak collected at 0.08 to 0.1 M NaCl was dialyzed, lyophilized, and stored at −20°C.
The overnight-cultured seed cells bearing pQE30-DsbA and pQE60-RNase I were incubated for another 4 h after 100-fold dilution and then induced with 50 μM IPTG for 8 h at 25°C. DsbA was purified according to Liu et al. (29). RNase I was purified with a nickel-nitrilotriacetic acid column, and N-terminal sequencing was performed to confirm the procession of the signal peptide.
Apocytochrome c was prepared mainly as described by Tong et al. (58). Horse heart cytochrome c (Sigma) at 3 mg/ml in buffer A (Tris buffer containing 8 mg/ml Ag2SO4, 8 M urea, and 1.2 M acetic acid) was incubated at 40°C for 4 h to dissociate heme, subsequently desalted with a Sephadex G-25 column, and lyophilized. The protein was then incubated in buffer B (Tris buffer with 6 M guanidine hydrochloride and 1 M dithiothreitol) at 25°C for 2 h to remove Ag+ and then desalted by using a Sephadex G-75 column. Finally, the protein was fully unfolded to remove all of the remaining heme and Ag+ by incubation in buffer C (Tris buffer with 8 M urea and 1 mM dithiothreitol) at 37°C for 30 min. After thorough dialysis, the lyophilized protein was stored at −20°C. All incubations for apocytochrome c preparation were carried out in the dark.
Protein concentrations were determined by the method of Bradford (12) with bovine serum albumin as the standard.
Preparation of trypsin-entrapped large unilamellar vesicles (LUV) and analysis of protein translocation.
A mixture of 2 ml ether containing 2.4 μM phospholipid preparation (Sigma) and 240 μl trypsin (Sigma) at 2 mg/ml in Tris buffer was sonicated in an ice-water bath for 20 min until the water phase disappeared. Reverse-phase evaporation was then performed to remove the ether at room temperature (55). The prepared liposomes were dialyzed against 500 ml Tris buffer for 10 h to remove the residual ether and then centrifuged at 15,000 × g for 15 min. The supernatant was further centrifuged at 35,000 × g for 25 min, and the precipitated LUV was dissolved in 200 μl Tris buffer. The concentration of LUV was defined as (Atotal − Aoutside)V0/A0, where Atotal is the total trypsin activity in the LUV solution, Aoutside is the trypsin activity outside of LUV, A0 is the trypsin activity at 2 mg/ml, and V0 is an experiential constant of the inclusive volume of LUV, 20 μl/μM (Qi Miao, personal communication). Trypsin activity was assayed with Nα-benzoyl-l-arginine ethyl ester (Sigma) as the substrate (6). In this work, two phospholipid preparations were used: 75% 1,2-dioleoyl-sn-glycerol-3-phosphatidylethanolamine, 20% 1,2-dioleoyl-sn-glycerol-3-[phospho-rac-(1-glycerol)], and 5% cardiolipin (44) for LUVI and 60% 1,2-dioleoyl-sn-glycerol-3-phosphatidylethanolamine and 40% 1,2-dioleoyl-sn-glycerol-3-[phospho-rac-(1-glycerol)] (1) for LUVII.
Protein translocation in LUV was determined basically as described by Tong et al. (58). A protein sample of 3.6 nmol was incubated at 37°C for 4 h in 100 μl Tris buffer containing 108 nmol LUV and 10 μg soybean trypsin inhibitor (Sigma) for full inhibition of the remaining trypsin activity outside of LUV, and aliquots were removed at different times for SDS-PAGE analysis.
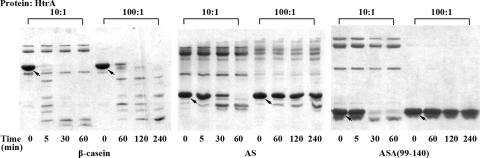
Limited digestion of AS and ASΔ(99-140) by E. coli HtrA.
AS or ASΔ(99-140) at 2.0 mg/ml was incubated with HtrA at a molar ratio of 10:1 for 1 h and 100:1 for 4 h in Tris buffer at 37°C with natively unfolded β-casein as a positive control. Aliquots were removed at different times for SDS-PAGE analysis.
RESULTS
AS expressed in E. coli is located in the periplasm.
As shown in Fig. 1A, AS with an apparent molecular mass of 17 kDa was overexpressed in E. coli BL21(DE3) and approximately 85% of the expressed AS was detected in the supernatant fraction after osmotic shock, indicating that the expressed AS was mostly transported into the periplasm. This result was unexpected, because the constructed expression vector pET-3a containing AS cDNA does not have any recognizable signal peptide sequence according to a search of the PSORT database (http://psort.nibb.ac.jp/form.html). We therefore verified that the translocation of AS into the periplasm was not simply caused by experimental artifacts. Firstly, to exclude the possibility that massive overproduction of AS caused membrane permeabilization during cell growth, we checked the localization of AS at a low expression level by shortening the duration of induction with IPTG to 30 min so that many native periplasmic components but few AS were detected by SDS-PAGE and Coomassie blue staining (Fig. 1B, left). The expression levels of bands 1 and 2 were estimated to be 3% and 1% of the total cellular proteins, respectively; the expression level of AS is therefore very likely less than 1%. The localization of AS was then identified by Western blotting with anti-AS serum, and the result showed that nearly 70% of the expressed AS was transported to the periplasm (Fig. 1B, right). The results indicate that the location of AS is not an artifact of massive overproduction.
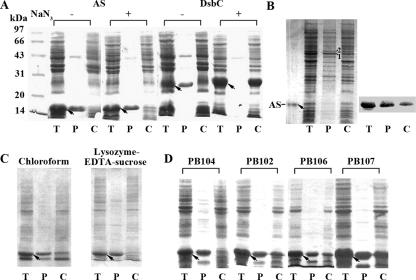
FIG. 1.
Subcelluar location of AS expressed in E. coli determined by SDS-PAGE. (A) AS and DsbC was expressed in BL21(DE3) and M15(pREP4), respectively, in the absence and presence of 3 mM NaN3. (B) AS expressed in BL21(DE3) at a low level induced with 100 μM IPTG for 30 min was determined by Coomassie blue staining (left) and Western blotting with anti-AS serum (right). Bands 1 and 2 in lane P in the left panel are two native periplasmic proteins. (C) AS expressed in BL21(DE3) treated with chloroform or lysozyme-EDTA-sucrose. (D) AS expressed in MscL− (PB104 and PB106) and MscL+ strains (PB102 and PB107). Cell fractionation was carried out by osmotic shock except for panel C. Apart from the right part of panel B, all of the gels were stained with Coomassie blue and the bands were quantitatively analyzed by using Totallab version 1.10 as described in the text. T denotes total cellular proteins; P and C denote periplasmic proteins in the supernatant fraction and cytoplasmic proteins in the pellet after cell fractionation, respectively. Arrows indicate the positions of expressed AS or DsbC.
It was suggested that some cytoplasmic proteins in E. coli, depending on the size of the molecules, can be released into the periplasm upon osmotic shock through a molecular-sieve mechanism (63). To exclude the possible roles of osmotic-shock treatment in AS translocation, we examined the localization of AS by using another two procedures for cell fractionation, i.e., chloroform treatment and lysozyme-EDTA-sucrose treatment. As shown in Fig. 1C, the same percentage of AS, ∼85%, was released into the periplasm fraction when cells were treated with either chloroform or lysozyme-EDTA-sucrose. The leakage of β-galactosidase was determined to be lower than 5% in the above cases. The three methods used for cell fractionation do not harm β-galactosidase activity determination (data not shown). The accessibility of the lysozyme-treated cells to protease showed that AS was mostly degraded by proteinase K under very mild conditions, confirming the periplasmic location of AS (data not shown). In addition, to rule out the possibility that the translocation of AS occurred by nonspecific leakage through MscL, AS was expressed in cells of strains PB102 and PB107 (MscL+) and strains PB104 and PB106 (MscL−) (10). As shown in Fig. 1D, AS was located in the periplasm of both wild-type (MscL+) and mutant (MscL−) strains. The above results indicate that translocation of AS to the periplasm is not caused by osmotic shock treatment and MscL is not involved in the translocation of AS.
Thirdly, AS was expressed in other E. coli strains with different expression vectors to see whether specific properties of the host cell strains affected protein transport. We found that in M15(pREP4), JS7131, and JM109 with pQE60-AS and in DH5α with pET-3a-AS, AS was mainly translocated into the periplasm, similar to that in BL21(DE3) with pET-3a-AS (data not shown). One exception was the MC4100 strain, in which the release of AS was only 60%. Consistent with this, it was noted that a natively periplasmic protein, DsbC, was released into the periplasm at a level of 90% when expressed in most E. coli strains but at only 70% in strain MC4100 (data not shown). Therefore, translocation of AS into the periplasm is not a specific property of strain BL21(DE3) with plasmid pET-3a-AS.
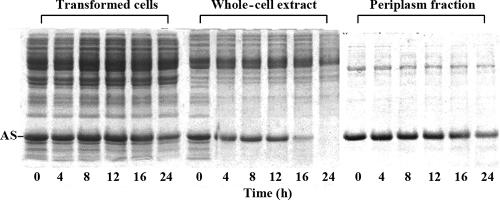
As shown in Fig. 2, half of the expressed AS in the whole-cell extract was degraded after incubation at 37°C for 16 h whereas the amount of AS protein in the periplasm fraction or in living cells did not change with the same treatment, indicating that the periplasm is a safer compartment for AS than is the cytoplasm.
FIG. 2.
The periplasm is a safer compartment for the life of AS than is the cytoplasm. AS was expressed in strain BL21(DE3). Transformed cells, the whole-cell extract obtained by sonication, and the periplasm fraction obtained by osmotic shock were incubated at 37°C for different times as indicated and were then analyzed by SDS-PAGE.
AS does not translocate through LUV.
Some small proteins are able to insert themselves into membrane autonomously. Apocytochrome c has been reported to translocate into mitochondria by following a unique pathway (20) and to be able to translocate through LUV in vitro freely via protein-acidic phospholipid interactions (36). Considering the ability of AS to bind membrane lipids (15, 25), we examined the possibility that AS translocates through interactions with membrane lipids, like apocytochrome c does. Trypsin-entrapped LUVI were made based on the composition of E. coli inner membrane lipid (44). As shown in Fig. 3, at a molar ratio of AS to trypsin-entrapped LUVI of 1:30, the majority of AS was not degraded by trypsin entrapped within LUVI after 240 min, with only 10% partially degraded, resulting in a cleaved fragment of about 8 kDa. Apocytochrome c, as a positive control, was fully degraded under the same conditions. When incubated with 0.1% Triton-treated LUVI, AS was fully degraded within 30 min (data not shown). Considering that translocation of secretory proteins via proteinaceous pores in membrane is also accompanied by direct interaction of signal peptides with membrane lipids, LUVII with a large portion of acidic phospholipids, which are favorable for direct interaction with signal peptides (1), was also examined, and the same results were obtained (data not shown). The above findings indicate that AS does not translocate through LUV.
FIG. 3.
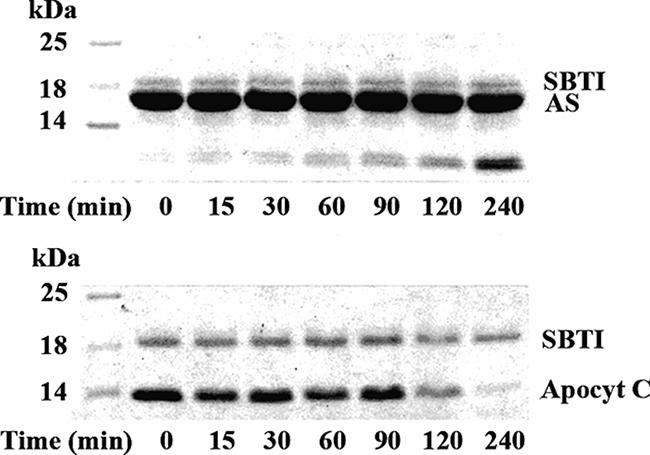
Translocation of AS into trypsin-entrapped LUV. A 3.6-nmol sample of AS and apocytochrome c (Apocyt C) was incubated in 100 μl of Tris buffer containing 108 nmol of trypsin-entrapped LUV and 10 μg soybean trypsin inhibitor (SBTI) at 37°C, respectively. Reaction mixture samples of 12 μl were taken at different times as indicated for SDS-PAGE analysis.
Translocation of AS across the inner membrane is not through the SecA or Tat pathway.
SecA is a key component of the most common targeting route for protein export into the periplasm in E. coli. As shown in Fig. 1A, the presence of 3 mM NaN3 did not affect the translocation of AS but did impede the transport into the periplasm of E. coli DsbC, which translocation is SecA dependent (57). The results indicate that the translocation of AS across the inner membrane is not SecA dependent.
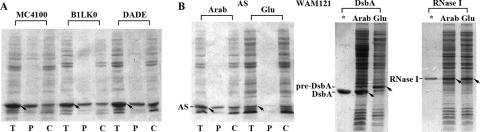
As neither a signal peptide nor an RR motif exists in the AS sequence and the constructed vector, AS does not appear to be transported through the Tat-dependent pathway. Nevertheless, we examined the translocation of AS in two Tat-negative strains, B1LK0 (ΔtatC) (11) and DADE (ΔtatABCDE) (66). In these two strains, AS translocated into the periplasm as in control strain MC4100 (Fig. 4A), confirming that AS is not transported to the periplasm through the Tat-dependent pathway.
FIG. 4.
Subcelluar location of AS expressed in Tat-negative strains and strain WAM121. AS was expressed (A) in MC4100 (wild type), B1LK0 (ΔtatC), and DADE (ΔtatABCDE) and (B) in WAM121 in the presence of 0.2% arabinose (Arab) or glucose (Glu). The cell fractions after osmotic shock were examined by SDS-PAGE. T, P, and C mean the same as in Fig. 1. DsbA and RNase I were expressed in WAM121 (B) to verify effectively controlled expression of Ffh. Arrows indicate the positions of expressed AS, DsbA, Pre-DsbA (DsbA precursor), and RNase I. Stars denote the input of the purified proteins.
The SRP-dependent pathway is involved in the translocation of AS.
E. coli SRP consists of an Ffh protein (48 kDa) and a 4.5S RNA. WAM121, a strain with the expression of Ffh controlled by an arabinose-inducible promoter, was used to examine the translocation of AS. To verify whether the expression of Ffh in strain WAM121 was effectively controlled, E. coli DsbA, an SRP pathway-dependent substrate (23), was used as a positive control. As shown in Fig. 4B, DsbA was detected as a mature protein in the culture containing 0.2% arabinose but as a precursor, which failed to translocate into the periplasm, in the culture containing 0.2% glucose, indicating effectively controlled expression of Ffh. AS translocated into the periplasm of WAM121 in the presence of 0.2% arabinose but was located in the cytoplasm in the presence of 0.2% glucose. In addition, RNase I, an RNase located in the periplasm (35), was detected as a mature protein no matter whether cells were cultured in arabinose or in glucose, indicating that RNase I translocated via an SRP-independent pathway. These results indicate that the SRP-dependent pathway is involved in the translocation of AS.
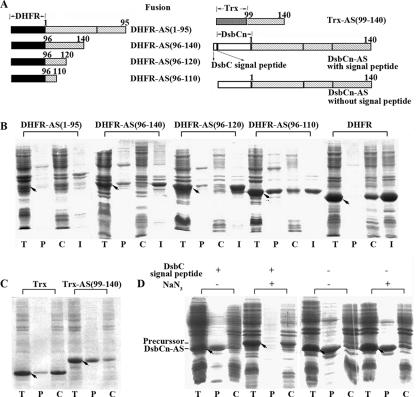
The N-terminal 1-to-60 region is not required for translocation of AS into the periplasm.
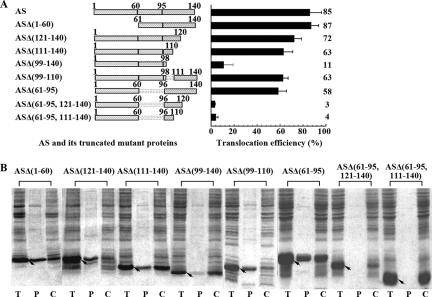
A series of truncated AS mutations were constructed (Fig. 5A) to examine whether any sequence within the AS molecule plays a signal-like role and is responsible for its translocation. As shown in Fig. 5, the mutant protein ASΔ(1-60) with the N-terminal region deleted translocated into the periplasm to an extent similar to that of intact AS, indicating that the N-terminal 60 amino acid residues are not required for the export of AS.
FIG. 5.
Subcellular location of truncated AS mutant proteins expressed in M15(pREP4). (A) Schematic representation of intact AS and truncated mutant proteins (left) and their corresponding translocation efficiencies (right, estimated from panel B). Data are expressed as the mean ± the standard error (n ≥ 3). (B) Subcellular location of the AS mutant proteins was examined by SDS-PAGE after osmotic shock. T, P, and C mean the same as in Fig. 1, and arrows indicate the positions of the corresponding mutant proteins.
The C-terminal 99-to-140 sequence is the main part responsible for the translocation of AS.
The two mutant proteins ASΔ(121-140) and ASΔ(111-140), which have the C-terminal 20 and 30 residues deleted, respectively, were translocated into the periplasm but with lower efficiency compared to intact AS (Fig. 5). When more residues at the C-terminal end were removed, only 10% or so of the ASΔ(99-140) was exported to the periplasm. As the translocation of ASΔ(111-140) increased to 65% from 10% for ASΔ(99-140), the 99-to-110 segment seemed to be important for the translocation of AS. However, ASΔ(99-110) was exported into the periplasm to an extent similar to that of ASΔ(111-140) (Fig. 5), suggesting that the 111-to-140 part can substitute for the 99-to-110 segment in the translocation of AS. The above findings showed that the entire C-terminal 99-to-140 fragment is required for translocation of AS molecules across the inner membrane with the highest efficiency.
The mutant protein ASΔ(61-95), in which the whole NAC section is deleted, was translocated into the periplasm with an efficiency of 60%, leaving the remainder in the osmotic-shock pellet. However, the other two mutant proteins, ASΔ(61-95, 121-140) and ASΔ(61-95, 111-140), with shorter C-terminal fragments were located in the cytoplasm (Fig. 5). Considering that only 10% of the ASΔ(99-140) was translocated into the periplasm, the above suggests that the NAC section plays a role in the translocation of the AS molecule but a weaker role than that of the C-terminal fragment.
Purified AS, ASΔ(99-140), ASΔ(61-95), ASΔ(61-95, 121-140), and ASΔ(61-95, 111-140) are not highly polymerized.
Mutant AS proteins truncated from the C terminus have been reported to assemble into filaments more readily than wild-type AS in vitro (13). We examined whether the mutant proteins ASΔ(99-140), ASΔ(61-95, 121-140), ASΔ(61-95, 111-140), and ASΔ(61-95) translocate across the membrane with low efficiency because they are highly polymerized. Wild-type AS and the four mutant proteins were purified and subjected to size exclusion chromatography on a SuperdexTM 200 column. The molecular mass was determined to be 48.5 kDa for AS, 25.3 kDa for ASΔ(99-140), 30.2 kDa for ASΔ(61-95), 23.5 kDa for ASΔ(61-95, 121-140), and 19.1 kDa for ASΔ(61-95, 111-140) (data not shown). According to Weinreb et al. (65) wild-type AS, with an apparent molecular mass of 58 kDa determined by size exclusion chromatography, is actually a monomer identified by nondissociating gel electrophoresis and sedimentation analysis as well. Recently it has been reported that wild-type AS in the periplasm of E. coli exists as a disordered monomer determined by using in-cell nuclear magnetic resonance spectroscopy (34). The calculated molecular masses of a protomer of wild-type AS and the four mutant proteins are 14.4, 9.8, 11.2, 8.9, and 7.8 kDa, respectively. Thus, the apparent molecular mass of wild-type AS determined by chromatography is about 3.5 to 4 times the calculated value, which was attributed to the unfolded conformation (65), and the apparent molecular masses of the four mutant proteins determined by size exclusion chromatography are about 2.5 times the calculated values. Therefore, wild-type AS and the four mutant proteins are not highly polymerized.
To examine another possibility, that a large proportion of ASΔ(99-140) was transported to the periplasm but was then degraded by proteases in the periplasm, ASΔ(99-140) was incubated with HtrA, a main protease in the periplasm catalyzing the degradation of unfolded or misfolded proteins (26). At a molar ratio to HtrA of 100:1, ASΔ(99-140) and AS were basically not degraded after 240 min whereas β-casein, also a naturally unfolded protein used as a control, was degraded almost completely in 60 min under the same conditions (Fig. 6). These results indicated that ASΔ(99-140) is resistant to HtrA in vitro similar to wild-type AS. Therefore, the low level of ASΔ(99-140) in the periplasm may not account for its lower stability in the periplasm.
FIG. 6.
Degradation of AS and ASΔ(99-140) by HtrA. AS, ASΔ(99-140), and β-casein as a control at 2.0 mg/ml were incubated with HtrA at a molar ratio of 10:1 or 100:1 at 37°C, respectively, and aliquots were removed at different times as indicated for SDS-PAGE analysis.
Fusion with the C-terminal fragments of AS directs the translocation of dihydrofolate reductase (DHFR) and thioredoxin (Trx) into the periplasm.
To further test whether the C-terminal fragments have a signal-like role, we linked DHFR to the N termini of four different fragments of AS to make four fusion proteins, DHFR-AS(1-95), DHFR-AS(96-140), DHFR-AS(96-120), and DHFR-AS(96-110) (Fig. 7A). As shown in Fig. 7B, DHFR alone was entirely located in the cytoplasm, with a major proportion as inclusion bodies and a minor proportion as soluble protein. The fusions DHFR-AS(96-140), DHFR-AS(96-120), and DHFR-AS(96-110) turned out to be partially translocated into the periplasm, whereas DHFR-AS(1-95) was located in the cytoplasm like DHFR itself. It was also found that 60% of Trx, which alone was located mostly in the cytoplasm as a soluble protein, was directed into the periplasm when linked to the N terminus of the AS(99-140) fragment (Fig. 7C). The above results strongly suggest that the C-terminal fragments of AS, but not the N-terminal 1-to-95 fragment, is able to direct the translocation of DHFR or Trx into the periplasm.
FIG. 7.
Subcellular location of fusion proteins expressed in M15(pREP4). (A) Schematic representation of fusion proteins. The subcellular locations of the fusions with (B) DHFR, (C) Trx, and (D) DsbCn were examined by SDS-PAGE after osmotic shock, respectively. T, P, and C mean the same as in Fig. 1, and I in panel B represents inclusion bodies. Arrows indicate the positions of the corresponding fusion proteins.
Comparison of the C-terminal fragment of AS and the Sec-dependent signal sequence of DsbC.
To further evaluate the signal-like role of the C-terminal fragment of AS, fusions of the N-terminal domain of DsbC (DsbCn) with and without a Sec-dependent signal sequence linked to the N terminus of AS were prepared to compare the signal role of the SecA-dependent signal of DsbC with that of the C-terminal fragment of AS (Fig. 7A). The results showed that the fusion of DsbCn-AS with the Sec-dependent signal sequence was detected in the periplasm in the absence of NaN3, but in the cytoplasm with an unprocessed signal peptide in the presence of 3 mM NaN3. The fusion without the Sec-dependent signal translocated to the periplasm no matter whether NaN3 was present or absent (Fig. 7D). The above suggested that the fusion with the signal peptide of DsbC was transported into the periplasm through the SecA-dependent pathway like DsbC itself. The fusion without the DsbC signal peptide also translocated into the periplasm but in a SecA-independent way. The SecA-dependent pathway appears to be preferred, and the C-terminal fragment of AS worked as a signal in the absence of a SecA-dependent signal.
DISCUSSION
We report here for the first time that recombinant AS with no signal sequence produced in E. coli mostly localizes in the periplasm. We demonstrated that the translocation of AS into the periplasm is not an artificial phenomenon caused by massive overproduction or the experimental method of osmotic shock.
AS is very sensitive to protease degradation in vitro. More than 95% of AS was degraded in 5 min at 25°C by 1% trypsin or chymotrypsin or 0.1% proteinase K (data not shown). It is reasonable to ascribe the high accessibility of AS to proteases to its unfolded conformation (65). In SH-SY5Y cells with transiently transfected AS, the half time for degradation of AS was determined to be only 3 h (5). However, AS in the periplasm of E. coli exists stably in disordered monomer form but not in aggregated form at a high concentration of ∼1 mM (34) and does not inhibit cell growth (data not shown), unlike in the yeast model (39). In fact, recombinant AS has been purified at high yields from the periplasm of E. coli (80 mg AS is usually purified in 1 liter of culture) in our laboratory (22). Our data obtained with E. coli indicated that the periplasm is a safe compartment for AS.
Two main targeting routes for protein export into the periplasm of E. coli, SecA dependent and Tat dependent, have been excluded as being responsible for the translocation of AS into the periplasm. Involvement of MscL in the translocation of AS was also excluded. In addition, interaction with membrane phospholipid is unlikely to play a role in the translocation of AS. The fact that AS was retained in the cytoplasm of WAM121 when Ffh was not expressed indicates the involvement of SRP in the translocation of AS. In general, the SRP-dependent targeting signals are either an N-terminal signal sequence or the first transmembrane segment in membrane proteins, which lack discrete signal peptides (27). AS does not contain any recognizable SRP-dependent signal sequence, and the N-terminal 1-to-60 region of AS was actually not required for translocation. Therefore, it appears that SRP mediates the translocation of AS but not via the known mechanisms involving interactions with the N-terminal fragments of proteins.
The C-terminal 99-to-140 portion as a main part, cooperating with the central 61-to-95 section, is responsible for the translocation of AS into the periplasm. Furthermore, this C-terminal fragment of AS is able to play a signal-like role to direct DHFR and Trx, which are cytoplasmic proteins, to translocate into the periplasm. Therefore, it is suggested that the C-terminal portion of AS might contain an unorthodox signal sequence. It would be interesting to inspect the possibility that SRP recognizes targeting signals that reside in the C-terminal portion and/or the middle NAC section of the AS molecule. Another possibility is that SRP mediates the translocation of AS by an indirect pathway through some other SRP-dependent proteins, whose membrane localization requires SRP and may be responsible for the translocation of AS. In this respect, the secretion of β-lactamase was inhibited by depletion of Ffh (42), but no cross-linking between β-lactamase and Ffh was detected (4).
Although presynaptic localization of AS has been frequently observed, the precise subcellular distribution of AS has long been a subject of dispute (24, 32, 59). Expression of murine AS-enhanced green fluorescent protein fusions and deletion variants in hippocampal neurons indicated that the presynaptic localization of AS is determined by the N-terminal and core domains while the nuclear localization is determined by the C-terminal domain (51). In a yeast model, wild-type AS and a natural mutant protein, A53T, were found to be concentrated at the plasma membrane and to completely inhibit the growth of cells carrying two copies whereas another natural mutant protein, A30P, was found to be dispersed within the cytoplasm with no influence on cell viability (39). Recently, AS expressed in yeast was found to be translocated from the endoplasmic reticulum to the Golgi and further to the plasma membrane by a classical secretion pathway (18), but the details of the whole pathway are not yet apparent. A pentapeptide sequence (95VKKDQ99) within the AS molecule was reported to guide selective translocation of the protein into lysosomes for degradation (14). These observations suggest that the AS molecule very likely contains unorthodox translocation signals for specific subcelluar localization. In this respect, our data obtained with E. coli also suggest unorthodox translocation signals in the AS molecule.
Acknowledgments
This work was supported by the Chinese Ministry of Science and Technology (2006CB910903 and 2006CB806508), the China National Science Foundation (30470363), and the Chinese Academy of Sciences (KSCX2-SW214-2).
We are extremely grateful to Paul Blount (University of Texas Southwestern Medical Center) for the generous gifts of PB102, PB104, PB106, and PB107; to Tracy Palmer (John Innes Centre, United Kingdom) for MC4100, B1LK0, and DADE; to Eitan Bibi (Weizmann Institute of Science, Israel) for WAM121; to R. Dalbey (Ohio State University) for JS7131; to Zengyi Chang (Peking University, China) for E. coli HtrA; to Qihong Sun (Beijing Institute of Radiation Medicine, China) for anti-AS serum; and to Shengjian Li (this group) for plasmid pOE30-Trx-AS(99-140). We thank Qi Miao (this laboratory) for very helpful advice about LUV experiments. We most sincerely thank Robert J. Kadner for extremely helpful advice and suggestions about the manuscript. His dedicated spirit and warm kindness in his editorial work impressed us deeply.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Ahn, T., D. B. Oh, H. Kim, and C. Park. 2002. The phase property of membrane phospholipids is affected by the functionality of signal peptides from the Escherichia coli ribose-binding protein. J. Biol. Chem. 277:26157-26162. [DOI] [PubMed] [Google Scholar]
- 2.Ajouz, B., C. Berrier, A. Garrigues, M. Besnard, and A. Ghazi. 1998. Release of thioredoxin via the mechanosensitive channel MscL during osmotic downshock of Escherichia coli cells. J. Biol. Chem. 273:26670-26674. [DOI] [PubMed] [Google Scholar]
- 3.Auluck, P. K., H. Y. E. Chan, J. Q. Trojanowski, V. M.-Y. Lee, and N. M. Bonini. 2002. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson's disease. Science 295:865-868. [DOI] [PubMed] [Google Scholar]
- 4.Beha, D., S. Deitermann, M. Muller, and H. G. Koch. 2003. Export of beta-lactamase is independent of the signal recognition particle. J. Biol. Chem. 278:22161-22167. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, M. C., J. F. Bishop, Y. Leng, P. B. Chock, T. N. Chase, and M. M. Mouradian. 1999. Degradation of α-synuclein by proteasome. J. Biol. Chem. 274:33855-33858. [DOI] [PubMed] [Google Scholar]
- 6.Bergmeyer, H. U., K. Gawehn, and M. Grassl. 1974. Trypsin, p. 515-516. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, vol. I, 2nd ed. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 7.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 8.Berrier, C., M. Besnard, B. Ajouz, A. Coulombe, and A. Ghazi. 1996. Multiple mechanosensitive ion channels from Escherichia coli, activated at different thresholds of applied pressure. J. Membr. Biol. 151:175-187. [DOI] [PubMed] [Google Scholar]
- 9.Birdsell, D. C., and E. H. Cota-Robles. 1967. Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli. J. Bacteriol. 93:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blount, P., S. I. Sukharev, M. J. Schroeder, S. K. Nagle, and C. Kung. 1996. Single residue substitutions that change the gating properties of a mechanosensitive channel in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:11652-11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogsch, E. G., F. Sargent, N. R. Stanley, B. C. Berks, C. Robinson, and T. Palmer. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273:18003-18006. [DOI] [PubMed] [Google Scholar]
- 12.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 13.Crowther, R. A., R. Jakes, M. G. Spillantini, and M. Goedert. 1998. Synthetic filaments assembled from C-terminally truncated alpha-synuclein. FEBS Lett. 436:309-312. [DOI] [PubMed] [Google Scholar]
- 14.Cuervo, A. M., L. Stefanis, R. Fredenburg, P. T. Lansbury, Jr., and D. Sulzer. 2004. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305:1292-1295. [DOI] [PubMed] [Google Scholar]
- 15.Davidson, W. S., A. Jonas, D. F. Clayton, and J. M. George. 1998. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 273:9443-9449. [DOI] [PubMed] [Google Scholar]
- 16.de Gier, J. W., P. Mansournia, Q. A. Valent, G. J. Phillips, J. Luirink, and G. von Heijne. 1996. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 399:307-309. [DOI] [PubMed] [Google Scholar]
- 17.de Rijk, M. C., C. Tzourio, M. M. Breteler, J. F. Dartigues, L. Amaducci, S. Lopez-Pousa, J. M. Manubens-Bertran, A. Alperovitch, and W. A. Rocca. 1997. Prevalence of parkinsonism and Parkinson's disease in Europe: the EUROPARKINSON Collaborative Study. European Community concerted action on the epidemiology of Parkinson's disease. J. Neurol. Neurosurg. Psychiatry 62:10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon, C., N. Mathias, R. M. Zweig, D. A. Davis, and D. S. Gross. 2005. α-Synuclein targets the plasma membrane via the secretory pathway and induces toxicity in yeast. Genetics 170:47-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giasson, B. I., I. V. J. Murray, J. Q. Trojanowski, and M. Y. V. Lee. 2001. A hydrophobic stretch of 12 amino acid residues in the middle of alpha-synuclein is essential for filament assembly. J. Biol. Chem. 276:2380-2386. [DOI] [PubMed] [Google Scholar]
- 20.Hannavy, K., S. Rospert, and G. Schatz. 1993. Protein import into mitochondria: a paradigm for the translocation of polypeptides across membranes. Curr. Opin. Cell Biol. 5:694-700. [DOI] [PubMed] [Google Scholar]
- 21.Hu, H. Y., Q. Li, H. Q. Cheng, and H. N. Du. 2001. β-Sheet structure formation of proteins in solid state as revealed by circular dichroism spectroscopy. Biopolymers 62:15-21. [DOI] [PubMed] [Google Scholar]
- 22.Huang, C., G. Ren, H. Zhou, and C. C. Wang. 2005. A new method for purification of recombinant human alpha-synuclein in Escherichia coli. Protein Expr. Purif. 42:173-177. [DOI] [PubMed] [Google Scholar]
- 23.Huber, D., D. Boyd, Y. Xia, M. H. Olma, M. Gerstein, and J. Beckwith. 2005. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J. Bacteriol. 187:2983-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakes, R., M. G. Spillantini, and M. Goedert. 1994. Identification of two distinct synucleins from human brain. FEBS Lett. 345:27-32. [DOI] [PubMed] [Google Scholar]
- 25.Jensen, P. H., M. S. Nielsen, R. Jakes, C. G. Dotti, and M. Goedert. 1998. Binding of alpha-synuclein to brain vesicles is abolished by familial Parkinson's disease mutation. J. Biol. Chem. 273:26292-26294. [DOI] [PubMed] [Google Scholar]
- 26.Kim, K. I., S. C. Park, S. H. Kang, G. W. Cheong, and C. H. Chung. 1999. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli. J. Mol. Biol. 294:1363-1374. [DOI] [PubMed] [Google Scholar]
- 27.Lee, H. C., and H. D. Bernstein. 2001. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. USA 98:3471-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, S. J., X. G. Hong, Y. Y. Shi, H. Li, and C. C. Wang. 2006. Annular arrangement and collaborative actions of four domains of protein-disulfide isomerase: a small angle X-ray scattering study in solution. J. Biol. Chem. 281:6581-6588. [DOI] [PubMed] [Google Scholar]
- 29.Liu, X. Q., S. Zhang, X. M. Pan, and C. C. Wang. 1999. A novel method for increasing production of mature proteins in the periplasm of Escherichia coli. Protein Sci. 8:2085-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luirink, J., C. M. ten Hagen-Jongman, C. C. van der Weijden, B. Oudega, S. High, B. Dobberstein, and R. Kusters. 1994. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 13:2289-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacFarlane, J., and M. Muller. 1995. Functional integration of a polytopic membrane protein of E. coli requires the bacterial signal recognition particle. Biochem. Soc. Trans. 23:560S. [DOI] [PubMed] [Google Scholar]
- 32.Masliah, E., E. Rockenstein, I. Veinbergs, M. Mallory, M. Hashimoto, A. Takeda, Y. Sagara, A. Sisk, and L. Mucke. 2000. Dopaminergic loss and inclusion body formation in α-synuclein mice: implications for neurodegenerative disorders. Science 287:1265-1269. [DOI] [PubMed] [Google Scholar]
- 33.McKeith, I. G., E. K. Perry, and R. H. Perry. 1999. Report of the second dementia with Lewy body international workshop: diagnosis and treatment. Consortium on dementia with Lewy bodies. Neurology 53:902-905. [DOI] [PubMed] [Google Scholar]
- 34.McNulty, B. C., G. B. Young, and G. J. Pielak. 2006. Macromolecular crowding in the Escherichia coli periplasm maintains alpha-synuclein disorder. J. Mol. Biol. 355:893-897. [DOI] [PubMed] [Google Scholar]
- 35.Meador, J., III, B. Cannon, V. J. Cannistraro, and D. Kennell. 1990. Purification and characterization of Escherichia coli RNase I. Comparisons with RNase M. Eur. J. Biochem. 187:549-553. [DOI] [PubMed] [Google Scholar]
- 36.Miao, Q., X. Han, and F. Yang. 2001. Phosphatidic acid-phosphatidylethanolamine interaction and apocytochrome c translocation across model membranes. Biochem. J. 354:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen, M. S., H. Vorum, E. Lindersson, and P. H. Jensen. 2001. Ca2+ binding to alpha-synuclein regulates ligand binding and oligomerization. J. Biol. Chem. 276:22680-22684. [DOI] [PubMed] [Google Scholar]
- 38.Oliver, D. B., R. J. Cabelli, K. M. Dolan, and G. P. Jarosik. 1990. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc. Natl. Acad. Sci. USA 87:8227-8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Outeiro, T. F., and S. Lindquist. 2003. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science 302:1772-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer, T., F. Sargent, and B. C. Berks. 2005. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 13:175-180. [DOI] [PubMed] [Google Scholar]
- 41.Perez, R. G., and T. G. Hastings. 2004. Could a loss of alpha-synuclein function put dopaminergic neurons at risk. J. Neurochem. 89:1318-1324. [DOI] [PubMed] [Google Scholar]
- 42.Phillips, G. J., and T. J. Silhavy. 1992. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359:744-746. [DOI] [PubMed] [Google Scholar]
- 43.Polymeropoulos, M. H., C. Lavedan, E. Leroy, S. E. Ide, A. Dehejia, A. Dutra, B. Pike, H. Root, J. Rubenstein, R. Boyer, E. S. Stenroos, S. Chandrasekharappa, A. Athanassiadou, T. Papapetropoulos, W. G. Johnson, A. M. Lazzarini, R. C. Duvoisin, G. Di Iorio, L. I. Golbe, and R. L. Nussbaum. 1997. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science 276:2045-2047. [DOI] [PubMed] [Google Scholar]
- 44.Raetz, C. R. 1978. Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol. Rev. 42:614-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Schiebel, E., A. J. Driessen, M. F.-U. Hartl, and W. Wickner. 1991. Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell 64:927-939. [DOI] [PubMed] [Google Scholar]
- 47.Seluanov, S., and E. Bibi. 1997. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem. 272:2053-2055. [DOI] [PubMed] [Google Scholar]
- 48.Shevchik, V. E., G. Condemine, and J. R. Baudouy. 1994. Characterization of DsbC, a periplasmic protein of Erwinia chrysanthemi and Escherichia coli with disulfide isomerase activity. EMBO J. 13:2007-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sijbrandi, R., M. L. Urbanus, C. M. Hagen-Jongman, H. D. Bernstein, B. Oudega, B. R. Otto, and J. Luirink. 2003. Signal recognition particle (SRP)-mediated targeting and Sec-dependent translocation of an extracellular E. coli protein. J. Biol. Chem. 278:4654-4659. [DOI] [PubMed] [Google Scholar]
- 50.Song, D. D., C. W. Shults, A. Sisk, E. Rockenstein, and E. Masliah. 2004. Enhanced substantia nigra mitochondrial pathology in human alpha-synuclein transgenic mice after treatment with MPTP. Exp. Neurol. 186:158-172. [DOI] [PubMed] [Google Scholar]
- 51.Specht, C. G., C. M. Tigaret, G. F. Rast, A. Thalhammer, Y. Rudhard, and R. Schoepfer. 2005. Subcellular localisation of recombinant α- and γ-synuclein. Mol. Cell. Neurosci. 28:326-334. [DOI] [PubMed] [Google Scholar]
- 52.Spillantini, M. G., R. A. Crowther, R. Jakes, M. Hasegawa, and M. Goedert. 1998. Alpha-synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with Lewy bodies. Proc. Natl. Acad. Sci. USA 95:6469-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sukharev, S. I., P. Blount, B. Martinac, F. R. Blattner, and C. Kung. 1994. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature 368:265-268. [DOI] [PubMed] [Google Scholar]
- 54.Sun, X. X., Y. Dai, H. P. Liu, S. M. Chen, and C. C. Wang. 2000. Contributions of protein disulfide isomerase domains to its chaperone activity. Biochim. Biophys. Acta 1481:45-54. [DOI] [PubMed] [Google Scholar]
- 55.Szoka, F., and D. Papahadjopoulos. 1978. Procedure for preparation of liposomes with large internal aqueous space and high capture by reverse-phase evaporation. Proc. Natl. Acad. Sci. USA 75:4194-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teter, S. A., and D. J. Klionsky. 1999. How to get a folded protein across a membrane. Trends Cell Biol. 9:428-443. [DOI] [PubMed] [Google Scholar]
- 57.Thorstenson, Y. R., Y. Zhang, P. S. Olson, and D. Mascarenhas. 1997. Leaderless polypeptides efficiently extracted from whole cells by osmotic shock. J. Bacteriol. 179:5333-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tong, J. C., L. Q. Zhu, and F. Y. Yang. 1996. V92A mutation altered the folding propensity of chicken apocytochrome c and its interaction with phospholipids. Biochemistry 35:9460-9468. [DOI] [PubMed] [Google Scholar]
- 59.Totterdell, S., D. Hanger, and G. E. Meredith. 2004. The ultrastructural distribution of alpha-synuclein-like protein in normal mouse brain. Brain Res. 1004:61-72. [DOI] [PubMed] [Google Scholar]
- 60.Uéda, K., H. Fukushima, E. Masliah, Y. Xia, A. Iwai, M. Yoshimoto, D. A. Otero, J. Kondo, Y. Ihara, and T. Saitoh. 1993. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc. Natl. Acad. Sci. USA 90:11282-11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ulbrandt, N. D., J. A. Newitt, and H. D. Bernstein. 1997. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187-196. [DOI] [PubMed] [Google Scholar]
- 62.Valent, Q. A., P. A. Scotti, S. High, J. W. de Gier, G. von Heijne, G. Lentzen, W. Wintermeyer, B. Oudega, and J. Luirink. 1998. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17:2504-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vázquez-Laslop, N., H. Lee, R. Hu, and A. A. Neyfakh. 2001. Molecular sieve mechanism of selective release of cytoplasmic proteins by osmotically shocked Escherichia coli. J. Bacteriol. 183:2399-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Volles, M. J., and P. T. Lansbury, Jr. 2003. Zeroing in on the pathogenic form of alpha-synuclein and its mechanism of neurotoxicity in Parkinson's disease. Biochemistry 42:7871-7878. [DOI] [PubMed] [Google Scholar]
- 65.Weinreb, P. H., W. Zhen, A. W. Poon, K. A. Conway, and P. T. Lansbury, Jr. 1996. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry 35:13709-13715. [DOI] [PubMed] [Google Scholar]
- 66.Wexler, M., F. Sargent, R. L. Jack, N. R. Stanley, E. G. Bogsch, C. Robinson, B. C. Berks, and T. Palmer. 2000. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J. Biol. Chem. 275:16717-16722. [DOI] [PubMed] [Google Scholar]
- 67.Zhao, Z., Y. Peng, S. F. Hao, Z. H. Zeng, and C. C. Wang. 2003. Dimerization by domain hybridization bestows chaperone and isomerase activities. J. Biol. Chem. 278:43292-43298. [DOI] [PubMed] [Google Scholar]