Abstract
PDZ domains are modular protein interaction domains that are present in metazoans and bacteria. These domains possess unique structural features that allow them to interact with the C-terminal residues of their ligands. The Escherichia coli essential periplasmic protein DegP contains two PDZ domains attached to the C-terminal end of the protease domain. In this study we examined the role of each PDZ domain in the protease and chaperone activities of this protein. Specifically, DegP mutants with either one or both PDZ domains deleted were generated and tested to determine their protease and chaperone activities, as well as their abilities to sequester unfolded substrates. We found that the PDZ domains in DegP have different roles; the PDZ1 domain is essential for protease activity and is responsible for recognizing and sequestering unfolded substrates through C-terminal tags, whereas the PDZ2 domain is mostly involved in maintaining the hexameric cage of DegP. Interestingly, neither of the PDZ domains was required for the chaperone activity of DegP. In addition, we found that the loops connecting the protease domain to PDZ1 and connecting PDZ1 to PDZ2 are also essential for the protease activity of the hexameric DegP protein. New insights into the roles of the PDZ domains in the structure and function of DegP are provided. These results imply that DegP recognizes substrate molecules targeted for degradation and substrate molecules targeted for refolding in different manners and suggest that the substrate recognition mechanisms may play a role in the protease-chaperone switch, dictating whether the substrate is degraded or refolded.
PDZ domains represent a common protein interaction motif, and their name was derived from the first three proteins in which such domains were identified, namely PSD-95, Drosophila melanogaster Disc large protein, and zonula occludens protein 1. Bacterial PDZ domains are homologous to the metazoan PDZ domains (23, 24, 26); however, their topology is different (20), and thus they are designated “PDZ-like” domains.
PDZ domains are approximately 90 residues long and have a common structure consisting of six β-strands and two α-helices, which fold in an overall six-stranded β-sandwich. The C-terminal ends of a protein substrate (7) usually bind in a groove of the domain formed between one of the α-helices and the adjacent β-strand, which thus serves as an extra β-strand added to the β-sheet (8). In this manner, the C-terminal peptide backbone participates in an extensive hydrogen-bonding pattern with the main chain atoms of the PDZ domain β-strand. The terminal carboxylate group is also stabilized by a series of hydrogen bonds with the highly conserved “carboxylate-binding loop” (3). However, the side chain of the C-terminal residue and the side chain of the residue at position −2 are the structures that are most critical for the specificity of substrate recognition by the PDZ domain, rather than the extensive hydrogen bonds with the main chain of the PDZ domain β-strand (29). In this context, the C-terminal residue of the substrate molecule is referred to as P0, and upstream residues are designated P−1, P−2, etc.
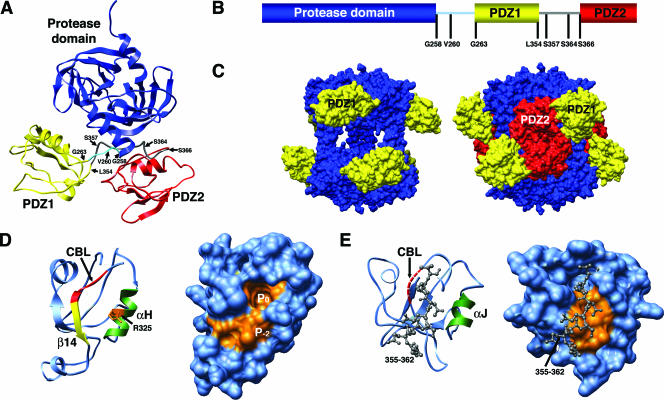
The Escherichia coli DegP protein (also called HtrA or protease Do) is an essential periplasmic protein (21) that functions as both a chaperone and a protease in a temperature-dependent fashion. DegP contains two PDZ domains, PDZ1 and PDZ2, following the N-terminal protease domain (2). One flexible loop links the protease domain to the PDZ1 domain, and another flexible loop links the PDZ1 domain to the PDZ2 domain (Fig. 1A and B) (19). X-ray crystallography studies have shown that DegP oligomerizes into a hexamer with a central chamber containing the proteolytic sites. The PDZ domains extend out from the ends of the cage, but they can also adopt a closed conformation blocking the lateral routes of access to the internal chamber (Fig. 1C) (19).
FIG. 1.
Structure of DegP and peptide-binding clefts of the PDZ1 and PDZ2 domains. (A) Ribbon representation of the DegP monomer (PDB identification no. 1KY9), showing the protease domain (blue), the PDZ1 domain (yellow), and the PDZ2 domain (red). The loop between the protease and the PDZ1 domain is cyan, and the loop between the two PDZ domains is dark gray. Residues involved in generating the DegP deletion mutants in this study are indicated. (B) Linear diagram of the DegP protein. Residues involved in generating the DegP deletion mutants are indicated. (C) Side views of the open conformation (left) and closed conformation (right) of the molecular surfaces of the DegP hexamer. The PDZ2 domain is not defined in the open conformation of the original structure of DegP. (D and E) Ribbon representations (left) and molecular surface representations (right) of the PDZ1 (D) and PDZ2 (E) domains, showing the substrate-binding cleft. In the ribbon representations elements important for substrate binding, including the carboxylate-binding loop (CBL), are indicated by different colors. The carboxylate-binding loop is not defined in the X-ray structure of the PDZ2 domain, as indicated by the red dashed line. The hydrophobic pockets of the binding cleft are orange in the surface representations. Residues 355 to 358 in the loop separating the PDZ domains are indicated by a ball-and-stick representation in panel E, filling the binding cleft of the PDZ2 domain.
The PDZ1 domain of DegP contains a deep binding cleft formed by β-strand 14, the “carboxylate-binding loop,” and helix H (Fig. 1D, left panel) (19). This domain is predicted to be involved in substrate binding through recognition of the C-terminal residues of the substrate molecule. Hydrophobic residues form the pockets for the P0 and P−2 residues of the substrate (Fig. 1D, right panel). In this manner, the PDZ1 domain is proposed to bind substrates with hydrophobic C-terminal residues.
Interestingly, residues 358 to 362 in the loop separating the two PDZ domains fill the corresponding binding cleft of PDZ2 (Fig. 1E) (19) and adopt an extended conformation identical to the conformation exhibited by C-terminal peptide substrates bound to other PDZ domains. Thus, this substrate-like segment may block substrates from entering the groove. However, based on the X-ray structure of DegP, the carboxylate-binding loop of PDZ2 (Fig. 1E, left panel) is highly flexible, and the possible reorientation of the substrate-like segment may allow binding of substrates.
In addition to the structural evidence presented previously, there is also experimental evidence demonstrating that other proteases bind substrates through their PDZ domains. For instance, the periplasmic E. coli tail-specific protein (Tsp) binds to the C-terminal end of substrates through its PDZ domain. This recruits the substrate to the catalytic site, which is located in a different domain, resulting in substrate cleavage at multiple sites (1, 16, 27, 30). Moreover, the X-ray structure of the E. coli DegS protein in complex with the C-terminal tail of the OmpC protein (35) shows that the DegS PDZ domain is used for substrate binding, also demonstrating how peptide binding to the PDZ domain modulates protease activity by inducing a series of reversible conformational changes that activate the protease (33, 35). Finally, a previous study (30) showed that the isolated PDZ domains of Salmonella enterica serovar Typhimurium DegP are able to bind the 11-amino-acid SsrA peptide. Interestingly, in this study E. coli DegP bound to the SsrA peptide, but the isolated PDZ domains of the E. coli protein were unable to bind to it. The SsrA peptide is usually appended to the C terminus of incompletely translated proteins, targeting them for proteolytic degradation and thus avoiding the buildup of ribosomes stalled on defective mRNA molecules (4-6, 15, 17). It therefore appears that substrate recognition by PDZ domains is a common mechanism in proteases.
In this work, we found that the PDZ1 domain of E. coli DegP is essential for protease activity and that both PDZ domains are dispensable for chaperone activity. Our results suggest that the two PDZ domains in DegP perform different functions; PDZ1 is involved in targeting substrates for proteolysis by recognizing the last three C-terminal residues of the substrate, whereas PDZ2 is required for maintenance of the hexameric cage of DegP. Additionally, we found that the loops between the protease and the PDZ1 domain and between the two PDZ domains are essential for protease activity in the hexameric DegP protein. The implications of these results for the protease-chaperone switch of DegP are discussed below.
MATERIALS AND METHODS
Plasmids and mutagenesis.
The pET21b-DegP, pET21b-ΔPDZ1 263-366 DegP, and pET21b-ΔPDZ2 DegP plasmids were obtained as described previously (10). The pET21b-ΔPDZ1+2 DegP plasmid was generated by PCR using the pET21b-DegP plasmid as the template, and it was subcloned into the expression vector pET21b. The pET21b-ΔPDZ1 263-354 DegP plasmid was obtained from pET21b-DegP by engineering NheI and XbaI restriction sites between residues 262 and 263 and between residues 354 and 355, respectively, using the QuikChange site-directed mutagenesis method (Stratagene). The plasmid was digested with NheI and XbaI, purified from the NheI-XbaI fragment by agarose gel electrophoresis, and ligated with T4 DNA ligase (Invitrogen). The QuikChange method was used to remove the remaining nucleotides. The pET21b plasmids used to express the Δ258-260 and Δ357-364 DegP mutants were obtained by the QuikChange method from the pET21b-DegP plasmid. The pET21b-Δ258-260+ΔPDZ2 plasmid was constructed from the pET21b-ΔPDZ2 DegP plasmid also by using the QuikChange method. The same method was used to construct all the proteolytically inactive variants (S210A) of the mutants.
The enhanced green fluorescent protein (EGFP) gene was amplified from the pEGFP-C2 vector (Clontech) by PCR and subcloned into the expression vector pPROEX-HTb (Invitrogen) using NcoI and XbaI sites. A stop codon was introduced after the XbaI site by the QuikChange method to obtain the pPROEX-HTb-EGFP expression plasmid. The pPROEX-HTb-EGFP-SsrA plasmid expressing EGFP-SsrA was constructed in several steps. First, two complementary oligonucleotides coding for the 11-residue SsrA tag followed by a stop codon and an NdeI site were synthesized with XbaI- and HindIII-compatible ends. Second, the pPROEX-HTb vector was digested with XbaI and HindIII and ligated with T4 DNA ligase to the annealed oligonucleotide coding for the SsrA tag to obtain the pPROEX-HTb-SsrA plasmid. Finally, the EGFP gene was obtained from the pPROEX-HTb-EGFP plasmid as an NcoI-XbaI insert and ligated into the pPROEX-HTb-SsrA plasmid digested with the NcoI and XbaI enzymes, producing the pPROEX-HTb-EGFP-SsrA plasmid that was used to express EGFP-SsrA.
The QuikChange method was used to obtain the pPROEX-HTb-EGFP-SFS plasmid from the pPROEX-HTb-EGFP plasmid by replacing nucleotides coding for the last three residues of EGFP (ISR). This plasmid was used to produce EGFP-SFS, and all the subsequent mutations in the SFS tag were generated by the QuikChange method from the pPROEX-HTb-EGFP-SFS plasmid.
Protein expression and purification.
Wild-type DegP and all the DegP mutants used in the assays were expressed as C-terminally His-tagged proteins. The procedure used to express and purify the proteins with a HiTrap metal chelating column (GE Healthcare Life Sciences) was performed as described previously (10).
To express EGFP, EGFP-SsrA, EGFP-SFS, and several variants of EGFP-SFS as N-terminally His-tagged proteins, the expression vector pPROEX-HTb containing one of the EGFPs was transformed into E. coli BL21(DE3) competent cells. The cells were grown in LB medium at 37°C to optical density at 600 nm of 0.5, and expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h at 25°C. Lysis was performed in 20 ml lysis buffer (50 mM Tris-HCl [pH 7.3], 0.1 M NaCl, 5% glycerol) by adding 256 μl of lysozyme (50 mg/ml) and incubating the preparation for 1 h at 4°C, followed by sonication on ice. The lysate was cleared by centrifugation at 39,000 × g for 40 min. NaCl was added to obtain a concentration to 0.5 M, and the lysate was filtered through a 0.45-μm filter and added to a HiTrap metal chelating column (GE Healthcare Life Sciences) equilibrated with 50 mM Tris-HCl (pH 7.3)-0.5 M NaCl-5% glycerol. Nonspecifically bound proteins were washed with increasing concentrations of imidazole up to 45 mM. EGFP (or one of the EGFP variants) was eluted with 240 mM imidazole. Fractions containing EGFP were pooled, diluted fivefold in buffer A (50 mM Tris-HCl [pH 7.3], 0.1 M NaCl), and loaded into a HiTrap Q HP column (GE Healthcare Life Sciences) equilibrated in buffer A. The column was washed with 50 ml of the same buffer, and the protein was eluted by increasing the concentration of NaCl to 200 or 400 mM depending on the EGFP mutant. Fractions containing the EGFP were pooled and loaded into a HiPrep 26/10 desalting column (GE Healthcare Life Sciences) equilibrated with 50 mM Tris-HCl (pH 7.3)-0.15 M NaCl. The eluted protein was concentrated to obtain a concentration of 8 mg/ml with a concentrator (Vivaspin 6; 10,000-molecular-weight cutoff; Vivascience), and glycerol was added to a final concentration of 20% (vol/vol).
Size exclusion chromatography.
A Superdex 200 10/300 GL column (GE Healthcare Life Sciences) equilibrated in 50 mM HEPES (pH 7.3) containing 100 mM NaCl at 4°C was used for the gel filtration chromatography experiments. Protein samples (100 μl) at concentrations between 0.5 and 3.5 mg/ml depending on the experiment were applied to the column. Samples were incubated at 4, 37, or 43°C before the protein was loaded into the column. A gel filtration calibration kit (high molecular weight; GE Healthcare Life Sciences) was used for column calibration.
Analytical centrifugation.
Sedimentation equilibrium and velocity experiments were carried out as described previously (10). Samples analyzed in the sedimentation equilibrium experiments were examined at three loading concentrations, as shown in Table 1. Data collected at different speeds and different loading concentrations were analyzed globally in terms of various species analysis models using SEDPHAT 4.0, (P. S. Schuck; http://www.analyticalultracentrifugation.com/sedphat/sedphat.htm). Solution densities were determined at 20°C with a Mettler-Toledo DE51 density meter and were corrected using values obtained at 4.0°C. Partial specific volumes were calculated based on the amino acid composition using SEDNTERP (J. Philo; http://www.jphilo.mailway.com/). Sedimentation coefficient distributions [given in the form c(s)] were obtained by analysis of the sedimentation velocity data with SEDFIT (25) (P. Schuck; http://www.analyticalultracentrifugation.com/default.htm). All sedimentation coefficients below are the values determined at 4.0°C in 50 mM HEPES (pH 7.3).
TABLE 1.
Properties of DegP mutants determined by analytical ultracentrifugationa
| Sample | Deletion positions | Monomer mass (Da) | Partial specific vol (cm3 g−1) | Sedimentation equilibrium at A280 |
|---|---|---|---|---|
| ΔPDZ1+2 DegPS210A | 257-446 | 27,003.4 | 0.7217 | 0.30, 0.65, 0.95 |
| Δ258-260 DegPS210A | 258-260 | 46,544.8 | 0.7279 | 0.25, 0.35, 0.52 |
| Δ357-364 DegPS210A | 357-364 | 45,983.3 | 0.7292 | 0.26, 0.58, 0.88 |
We assumed that the first 26 N-terminal residues of all DegP mutants were processed. Partial specific volumes were calculated at 4.0°C using SEDNTERP. The density determined was 1.00402 g cm−3 at 4.0°C.
Chaperone and protease activity assays.
Refolding assays using citrate synthase (Roche) as a substrate were performed as described previously (10). Similarly, protease assays with malate dehydrogenase (MDH) from porcine heart (Roche and Sigma), bovine milk β-casein (Sigma), and egg white lysozyme (Bioshop) were performed using previously described methods (10).
EGFP refolding assay.
Prior to the experiment EGFP and the EGFP-SsrA, EGFP-SFS, and EGFP-SFS variants were denatured by incubating 1 mg/ml of protein in 6 M guanidine-HCl containing 25 mM Tris-HCl (pH 7.3), 10 mM dithiothreitol, and 1 mM EDTA for 2 h at 25°C. To perform the assay, 200 pmol of the guanidine-HCl-denatured EGFP (or EGFP variant) was diluted into 500 μl of EGFP refolding buffer (50 mM Tris-HCl, pH 7.3) with or without 800 pmol of a specific DegPS210A mutant. Fluorescence was determined at 37°C with excitation at 489 nm and emission at 509 nm by using a fluorescence spectrophotometer (Cary Eclipse; Varian) (9, 28). The amount of refolded EGFP (or EGFP variant) was expressed as a percentage of the fluorescence intensity obtained for the same native EGFP at an identical concentration. Where indicated below, the EGFP refolding assay was done in the presence of the different DegP mutants. In these cases, a fluorescence value of 100% was considered the intensity of fluorescence produced by the same concentration of the native EGFP (or EGFP variant) in the presence of the specific DegP mutant at the concentration used in the assay. This control was necessary as we noticed a small increase in the fluorescence of the native EGFPs in the presence of DegP or DegP mutants.
RESULTS
PDZ domain requirements for protease and chaperone activities of DegP.
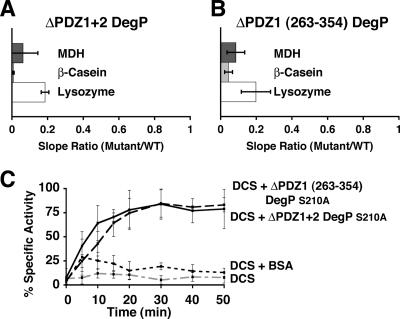
In agreement with previous reports (31), we showed that deletion of both PDZ domains (Fig. 2A) or the PDZ1 domain (10) resulted in a dramatic reduction in the protease activity of DegP with all three substrates tested (MDH, β-casein, and lysozyme). Conversely, we also showed (10) that the ΔPDZ2 DegP mutant was able to hydrolyze the substrates tested at rates that were similar to (MDH and β-casein) or higher than (lysozyme) the rates observed with wild-type DegP.
FIG. 2.
Protease and chaperone activities of the PDZ domain DegP mutants. (A and B) Protein substrates (MDH, β-casein, and lysozyme) were mixed with the ΔPDZ1+2 (A) or ΔPDZ1 263-354 DegP (B) mutant for the proteolysis reactions. Samples were removed during incubation, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and stained with Coomassie brilliant blue (MDH and β-casein assays) or deep purple total protein stain (lysozyme assays). The gels were scanned, and the substrate bands were quantified to obtain the hydrolysis curves. The hydrolysis rates for each mutant and substrate were calculated by determining the ratio of the slopes of the proteolysis curves for the mutant and wild-type (WT) DegP. The averages and standard deviations of the rates were calculated after each hydrolysis reaction was repeated three times. (C) The ΔPDZ1+2 or ΔPDZ1 263-354 DegPS210A mutant was mixed with unfolded citrate synthase in refolding buffer. At different times, samples were removed and citrate synthase activity was determined and expressed as a percentage of the highest specific activity obtained when the refolding reaction was performed in the presence of DegPS210A. Similar refolding reactions were performed in the presence of bovine serum albumin (BSA) and in the absence of any DegP protein. DCS, denatured citrate synthase.
Interestingly, none of the PDZ deletions had a significant effect on the chaperone activity of DegP (Fig. 2C) (10), as shown by analysis of the chaperone activity of the S210A proteolytically inactive variants of the mutants (in which serine 210 in the catalytic triad was changed to alanine).
We hypothesized that the lack of PDZ2 involvement in protease activity may be a consequence of residues 358 to 362 filling its binding cleft (Fig. 1E), as the ΔPDZ1 263-354 DegP mutant analyzed in the present study had very low proteolytic activity (Fig. 2B). In this mutant residues 358 to 362 were retained, but the PDZ1 domain (residues 263 to 354) was deleted. Consistent with previous results, the proteolytically inactive S210A variant of this mutant exhibited full chaperone activity (Fig. 2C). To investigate whether the substrate-like segment occupying the binding cleft blocks the access of potential substrates, we looked at our previously published data for a different ΔPDZ1 DegP mutant (ΔPDZ1 263-366) (10) in which both the PDZ1 domain and the substrate-like segment from the loop connecting the PDZ domains were removed. This should have facilitated access of the substrate to the C-terminal binding groove of PDZ2. Interestingly, the ΔPDZ1 263-366 DegP mutant exhibited no (MDH and β-casein) or very mild (lysozyme) protease activity. Therefore, the hypothesis that the substrate-like segment occupies the binding cleft and blocks the binding of potential substrates is an unlikely explanation for the dispensability of the PDZ2 domain for the protease activity of DegP. These results suggest that despite the flexible carboxylate-binding loop in PDZ2 that could allow reorientation of the substrate-like segment and entrance of substrates, this domain is not actually required for protease activity.
Together, these results indicate that neither of the PDZ domains is required for the chaperone activity of DegP and that only the PDZ1 domain is essential for its protease activity.
Both PDZ domains are essential for maintaining the hexameric cage of DegP.
In the “closed” conformation of the DegP hexamer, the PDZ2 domain of each monomer interacts with the PDZ1 and PDZ2 domains of the monomer at the opposite trimer in a zipper-like arrangement (Fig. 1C) (19). Our previous studies showed that residues 39 to 78 in the LA loops, as well as the PDZ2 domains, are essential for maintenance of the DegP hexameric cage (10). In our previous study, we characterized the oligomeric state of the ΔPDZ2 DegP mutant (and its S210A variant) by size exclusion chromatography and analytical ultracentrifugation and showed that this mutant is a trimer. Similarly, the ΔPDZ1 263-366 DegP mutant (and its S210A variant) (Fig. 1A and B) were shown to be trimers. We concluded that the PDZ2 domain is essential for the maintenance of the hexameric cage in DegP, as is the PDZ1 domain, which acts as a spacer allowing the PDZ2 domain to reach the PDZ1 and PDZ2 domains of the opposing monomer rather than the adjacent monomer (10).
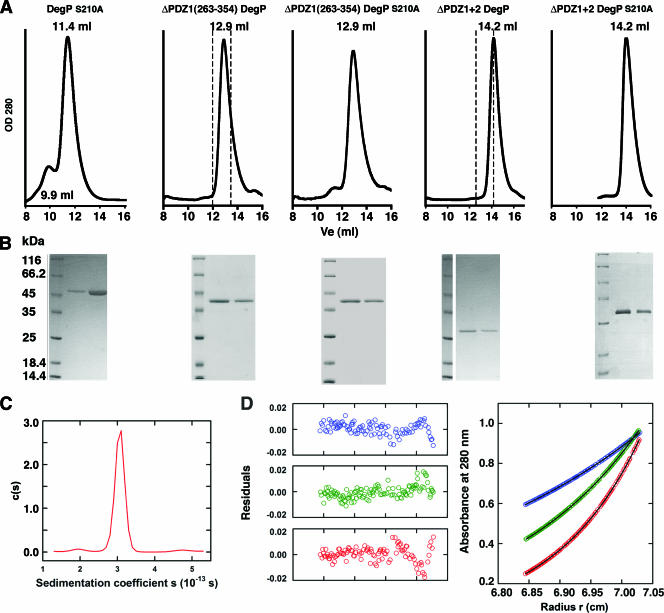
To rule out the possibility that the previously characterized ΔPDZ1 263-366 mutant (10) is a trimer because the PDZ2 domain is not adequately folded due to the absence of the substrate-like segment filling the binding cleft, we extended our analysis by characterizing the oligomeric state of the ΔPDZ1 263-354 mutant (Fig. 1A and B). This protein includes not only the entire PDZ2 domain but also residues 358 to 362 of the substrate-like segment (Fig. 1E). This mutant was incubated at 4, 37, or 43°C for 1 h before it was loaded onto a size exclusion chromatography column. In all cases this mutant and its proteolytically inactive variant (ΔPDZ1 263-354 DegPS210A) eluted at a position very close to the expected elution volume for the trimeric form of the protein (Fig. 3A and B).
FIG. 3.
PDZ domain deletion mutants of DegP are trimers. (A) Elution profiles of the PDZ domain mutants obtained with a size exclusion chromatography Superdex-200 column. Samples were incubated at 4°C for 1 h before they were loaded onto the column. The expected elution volumes (Ve) for the hexameric form (12 ml) and trimeric form (13.5 ml) of the ΔPDZ1 263-354 DegP mutant (dashed lines in the second plot from the left) and for the hexameric form (12.7 ml) and trimeric form (14.2 ml) of the ΔPDZ1+2 DegP mutant (dashed lines in the second plot from the right) are indicated. OD 280, optical density at 280 nm. (B) Two fractions from each peak were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. (C) Sedimentation coefficient distribution [c(s)] obtained for the ΔPDZ1+2 DegPS210A mutant by sedimentation velocity analysis. (D) Sedimentation equilibrium profiles expressed in terms of A280 versus the radius for the ΔPDZ1+2 DegPS210A mutant with a loading A280 of 0.95. Data were collected at 6,000 rpm (blue), 8,000 rpm (green), and 10,000 rpm (red) at 4.0°C and were analyzed in terms of a single ideal solute. Best fits are indicated by black lines through the experimental points in the plot on the right, and the corresponding distributions of the residuals are shown in the plots on the left.
Similar experiments with the ΔPDZ1+2 and ΔPDZ1+2 DegPS210A mutants yielded identical results. The ΔPDZ1+2 DegPS210A mutant was also characterized by analytical ultracentrifugation, and data consistent with the results of size exclusion chromatography were obtained. Sedimentation velocity data analyzed in terms of a continuous c(s) distribution resulted in excellent fits consistent with the presence of one well-resolved species having a sedimentation coefficient of 3.1 S (Fig. 3C). Sedimentation equilibrium data analyzed in terms of a single ideal solute also resulted in excellent fits; these data showed that the molecular mass was 78.9 ± 0.8 kDa (expected monomer mass, 27,003.4 Da; stoichiometry, n = 2.92 ± 0.03), which indicated that this DegP mutant is a monodisperse trimer in solution (Fig. 3D).
These results confirmed the essential role of the PDZ1 and PDZ2 domains in maintaining the hexameric cage. Furthermore, they showed the stability of the ΔPDZ1 263-366 and ΔPDZ1+2 mutants and their proteolytically inactive variants at the temperatures used in the protease and chaperone assays performed.
C-terminal tags enhance trapping of unfolded proteins by DegP.
We next wanted to study the mechanism of substrate recognition by DegP. The presence of two PDZ domains in this protein led us to investigate whether DegP recognizes substrates through their C-terminal residues. To this end, we used EGFP and constructed derivatives of EGFP with different C-terminal residues. EGFP was chosen because it fluoresces in its native state but not in nonnative conformations (9) and also because it has the ability to refold spontaneously into its native conformation upon incubation in refolding buffer.
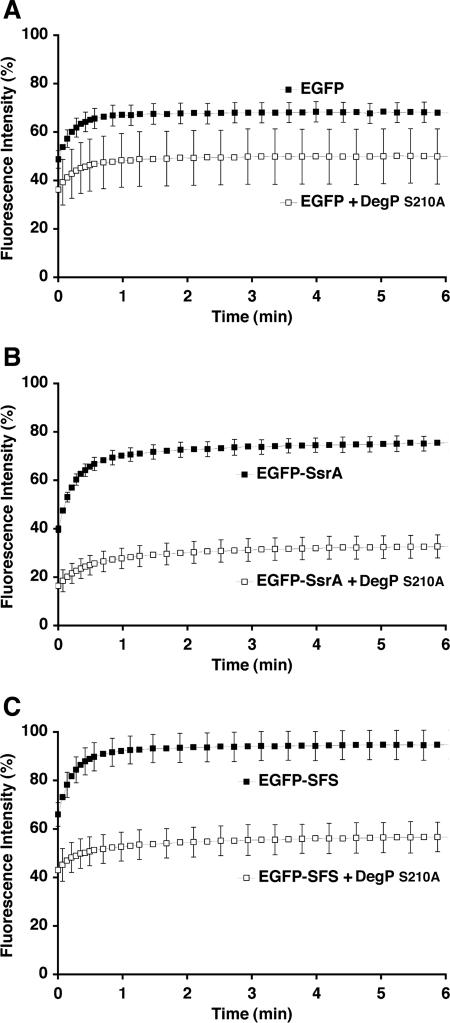
As a control, we first tested whether DegPS210A could trap guanidine-HCl-denatured EGFP in its nonnative form by chemically denaturing EGFP and then diluting it in the refolding reaction mixture with or without DegPS210A. Without DegPS210A, unfolded EGFP rapidly regained fluorescence, and 1 min after the reaction was initiated 68% of the EGFP reached a folded and fluorescent state. In contrast, only 50% of the initial fluorescence of EGFP was recovered after addition of DegPS210A to the refolding mixtures (Fig. 4A), indicating that DegPS210A trapped some of the unfolded EGFP before it refolded.
FIG. 4.
DegPS210A sequestration of unfolded EGFP with C-terminal tags. (A) Guanidine-HCl-denatured EGFP was diluted in EGFP refolding buffer in the absence (▪) or presence (□) of DegPS210A protein. The fluorescence intensity was determined at different times at 37°C and was expressed as a percentage of the fluorescence intensity of native EGFP at the same concentration. The experiment was done in triplicate, and averages and standard deviations of the percentages of the fluorescence intensities were calculated. The experiment was repeated with the EGFP-SsrA (B) and EGFP-SFS (C) substrates.
To test whether a C-terminal tag in EGFP influences sequestration of unfolded proteins by DegPS210A, we denatured EGFP-SsrA, which contained the 11-amino-acid SsrA protease recognition tag (AANNENYALAA) fused to the carboxyl terminus of EGFP. Dilution of the denatured protein in a refolding reaction mixture without DegPS210A resulted in recovery of about 76% of the original fluorescence, whereas in the presence of DegPS210A only 33% of the initial fluorescence was recovered (Fig. 4B), suggesting that the 11-residue SsrA tag enhances sequestration of unfolded EGFP by DegPS210A.
It has been shown that the last three residues of the C-terminal regions of substrates are the most critical residues for binding by PDZ domains (8). To determine whether this is the case with the PDZ domains of DegP, we obtained EGFP with its last three residues (ISR) changed to SFS (EGFP-SFS). Serine, phenylalanine, and serine are the three carboxyl-terminal amino acids of the PapG pilin, which is a natural substrate for DegP (11, 12). The refolding assay was performed, and we observed that only 57% of the unfolded EGFP-SFS was refolded in the presence of DegPS210A, compared to the 95% of the protein that was refolded in the absence of DegPS210A (Fig. 4C).
Altogether, these data suggest that C-terminal tags enhance trapping of unfolded substrates by DegP. Even though the tags are different lengths, the sequence of the last three carboxyl-terminal amino acids appears to be sufficient to enhance sequestration.
Residue at P−1 of the SFS C-terminal tag influences sequestration of unfolded EGFP by DegP.
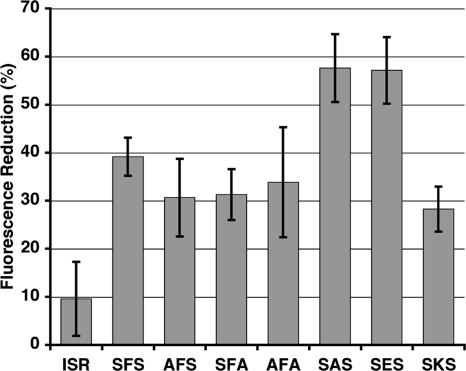
To determine which of the last three residues of the substrate C-terminal tag influences sequestration of unfolded EGFP by DegP, we generated point mutations in the original EGFP-SFS substrate. In particular, we were interested in determining whether changes at P−1 in the C-terminal tag alter substrate trapping by DegP. The X-ray structures of DegP (19) and other PDZ domain-containing proteins (3) indicated that the side chain of the P−1 residue points away from the surface of the binding cleft and is thus expected to have little influence on the binding specificity. We initially generated three EGFP-SFS mutants. The highly hydrophobic phenylalanine residue at P−1 was changed to a small nonpolar residue (alanine), a negatively charged residue (glutamate), or a positively charged residue (lysine). The change to a nonpolar residue (EGFP-SAS) or to a negatively charged residue (EGFP-SES) significantly increased the amount of EGFP sequestered in an unfolded state in the refolding assay with DegPS210A (Fig. 5). Conversely, the change to a positively charged lysine residue (EGFP-SKS) decreased the amount of EGFP sequestered, demonstrating that the residue at P−1 in the substrate molecule has a great influence on the trapping ability of DegP.
FIG. 5.
Importance of the residue at P−1 of the substrate C-terminal tag for sequestration by DegP. Guanidine-HCl-denatured EGFP variants with the C-terminal tags indicated (amino acids indicated by one-letter abbreviations) were refolded in the absence or presence of DegPS210A protein. For each EGFP, plots similar to those in Fig. 4 were generated, and then the difference between the percentage of protein refolded in the absence of DegPS210A and the percentage of protein refolded in the presence of DegPS210A protein at 8 min was calculated (at this time fluorescence had reached a plateau). Experiments were done in triplicate, and averages and standard deviations of the percentages of fluorescence reduction were calculated. Thus, the values reflect the percentage of each EGFP sequestered by DegPS210A protein in an unfolded state. ISR, results obtained for wild-type EGFP (isoleucine, serine, and arginine are the three C-terminal residues of EGFP).
Additionally, we changed the residues at P0 and P−2 in the C-terminal tag of EGFP-SFS from the polar amino acid serine to the nonpolar amino acid alanine either individually or simultaneously. The three mutants, EGFP-AFS, EGFP-SFA, and EGFP-AFA, showed a slight decrease in the amount of substrate sequestered by DegPS210A (Fig. 5). This result was surprising because in principle, the hydrophobic residues forming the pockets for P0 and P−2 in the PDZ1 domain of DegP are better suited to bind alanine than to bind serine. However, additional mutations in EGFP-SFS are required to understand the binding requirements at P0 and P−2.
The PDZ1 domain is responsible for sequestering the unfolded substrate through the C-terminal tag.
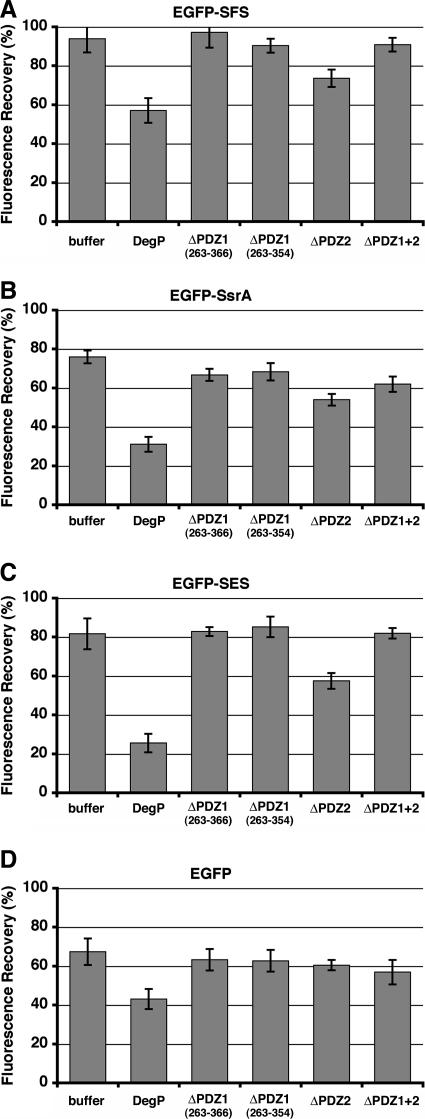
Having shown that the PDZ1 domain is essential for protease activity, we hypothesized that the PDZ1 domain is involved in sequestering the unfolded substrate through the C-terminal tag. To investigate this possibility, we examined the proteolytically inactive variants (DegPS210A) of our PDZ deletion mutants in an EGFP fluorescence assay using denatured EGFP-SFS as a substrate. The ΔPDZ1+2, ΔPDZ2, ΔPDZ1 263-354, ΔPDZ1 263-366 DegPS210A, and full-length DegPS210A mutants (Fig. 1A and B) were purified and used in the refolding reactions. We found that fluorescence recovery was not affected by the presence of the ΔPDZ1+2 DegPS210A mutant or either of the two ΔPDZ1 DegPS210A mutants. However, the level of fluorescence recovery significantly decreased upon incubation with ΔPDZ2 DegPS210A or DegPS210A, which was used as a control (Fig. 6A). This suggests that the amount of sequestered EGFP-SFS in an unfolded state increased and is consistent with the hypothesis that the PDZ1 domain is responsible for binding the three C-terminal SFS residues. Interestingly, ΔPDZ2 DegPS210A was not as efficient as at sequestering EGFP-SFS, full-length DegPS210A, suggesting that the PDZ2 domain plays a role in stabilizing the SFS C-terminal tag bound to the PDZ1 domain.
FIG. 6.
PDZ1 domain mediates sequestration of the unfolded substrate through the C-terminal tag. (A) Guanidine-HCl-denatured EGFP-SFS was allowed to refold either alone or in the presence of DegPS210A or one of the PDZ mutants. The intensity of fluorescence in each reaction was determined after 8 min of incubation at 37°C and was expressed as a percentage of the intensity of fluorescence of native EGFP-SFS at the same concentration in the presence of the DegPS210A mutant. The experiment was done in triplicate, and averages and standard deviations of the percentages of fluorescence intensities were calculated. Identical experiments were performed with EGFP-SsrA (B), EGFP-SES (C), and EGFP (D).
To confirm these results, we repeated the experiment using unfolded EGFP-SsrA (Fig. 6B) and unfolded EGFP-SES (Fig. 6C). Consistent with our previous results, neither the ΔPDZ1+2 DegPS210A mutant nor the two ΔPDZ1 DegPS210A mutants affected the refolding of EGFP-SsrA and EGFP-SES. Similarly, only the ΔPDZ2 DegPS210A and DegPS210A mutants showed significant sequestration of the unfolded substrates (Fig. 6B and C). As a negative control, we measured the effects of the mutants on the refolding of untagged EGFP. As expected, only the DegPS210A mutant exhibited some ability to sequester the unfolded form of EGFP. The differences between the fluorescence observed in the refolding reaction with EGFP in the presence of the mutants and the fluorescence observed in the reaction in the absence of DegP were not significant (Fig. 6D).
Loops connecting the protease and the PDZ1 domain and connecting the two PDZ domains are essential for protease activity.
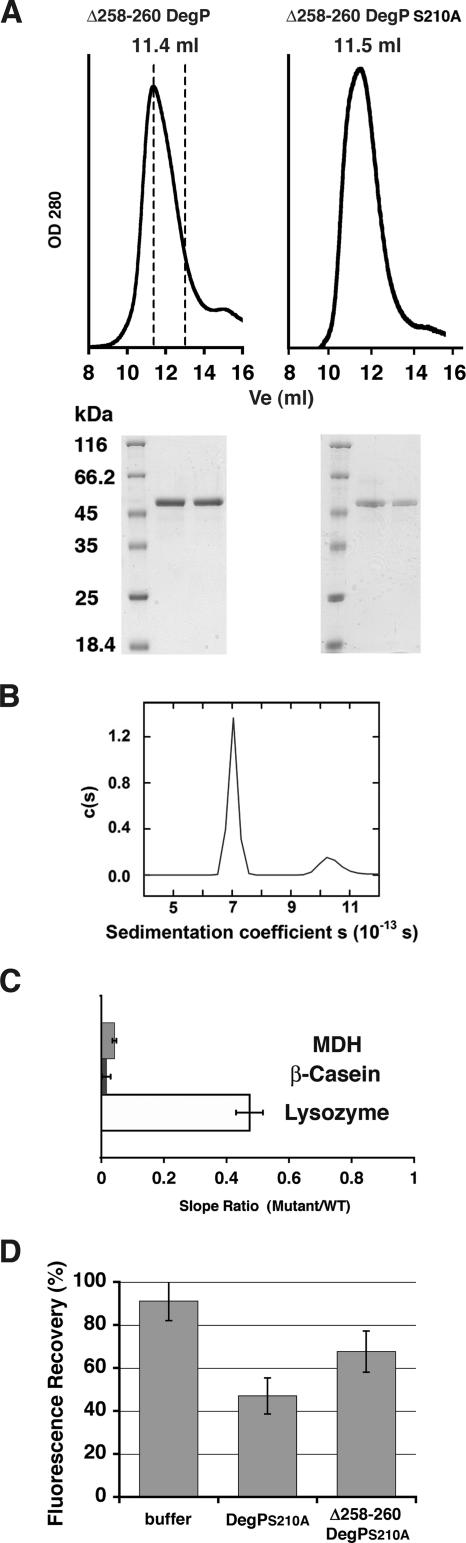
The PDZ1 domain is connected to the protease domain by a loop (Fig. 1A and B). To determine whether this loop is essential for DegP protease and chaperone activities, we generated the Δ258-260 DegP mutant and a proteolytically inactive variant of this mutant (Δ258-260 DegPS210A), which lacked most of the loop-forming residues. Both mutants eluted at the elution volume expected for a hexamer when they were loaded onto a size exclusion chromatography column after incubation at 4, 37, or 43°C for 1 h (Fig. 7A). Data obtained in sedimentation equilibrium experiments with the Δ258-260 DegPS210A mutant exhibited excellent fits when they were analyzed in terms of two noninteracting ideal solutes, returning molecular masses for predominantly hexamers of 281 ± 33 kDa (expected monomer mass, 46,544.8 Da; stoichiometry, n = 6.0 ± 0.7) and for some dodecamers of 549 ± 45 kDa (stoichiometry, n = 11.8 ± 1.0). The hexamer was the predominant species, an observation confirmed by the results of a sedimentation velocity analysis showing the presence of two well-resolved sedimenting species having uncorrected c(s) values of approximately 7 and 10.2 S. Integration of the c(s) distribution revealed the presence of approximately 10% dodecamers in this sample (Fig. 7B). Moreover, the c(s) distribution reproducibly returned a sedimentation coefficient identical to that observed for the DegPS210A mutant. This shows that the overall shapes of the two proteins are very similar and that the structural integrity of the Δ258-260 DegPS210A mutant is maintained. Thus, we concluded that both the Δ258-260 DegP and Δ258-260 DegPS210A mutants are stable hexamers similar to wild-type DegP.
FIG. 7.
Characterization of the Δ258-260 DegP mutant. (A) Analysis of the oligomeric state of the Δ258-260 DegP mutant and its proteolytically inactive S210A variant by size exclusion chromatography. The plots at the top show the elution profiles of the mutants. The dashed lines indicate the expected elution volumes (Ve) for a hexameric form (11.5 ml) and a trimeric form (13 ml) of the mutant. The gels below the plots contained samples from each peak resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. OD 280, optical density at 280 nm. (B) Sedimentation velocity data for the Δ258-260 DegPS210A mutant expressed as a continuous sedimentaion coefficient distribution [c(s)] obtained using the program SEDFIT. (C) Plot showing the averages and standard deviations for the hydrolysis rates for the Δ258-260 DegP mutant for three substrates (MDH, β-casein, and lysozyme). WT, wild type. (D) Plot showing the ability of the Δ258-260 DegPS210A mutant to sequester EGFP-SFS, generated as described in the legend to Fig. 6.
The chaperone activity of the Δ258-260 DegPS210A mutant was not affected (data not shown); however, the mutation had a dramatic effect on the protease activity with MDH and β-casein, which was completely eliminated. Hydrolysis of lysozyme was not completely eliminated, but it was significantly decreased compared with the activity of wild-type DegP (Fig. 7C).
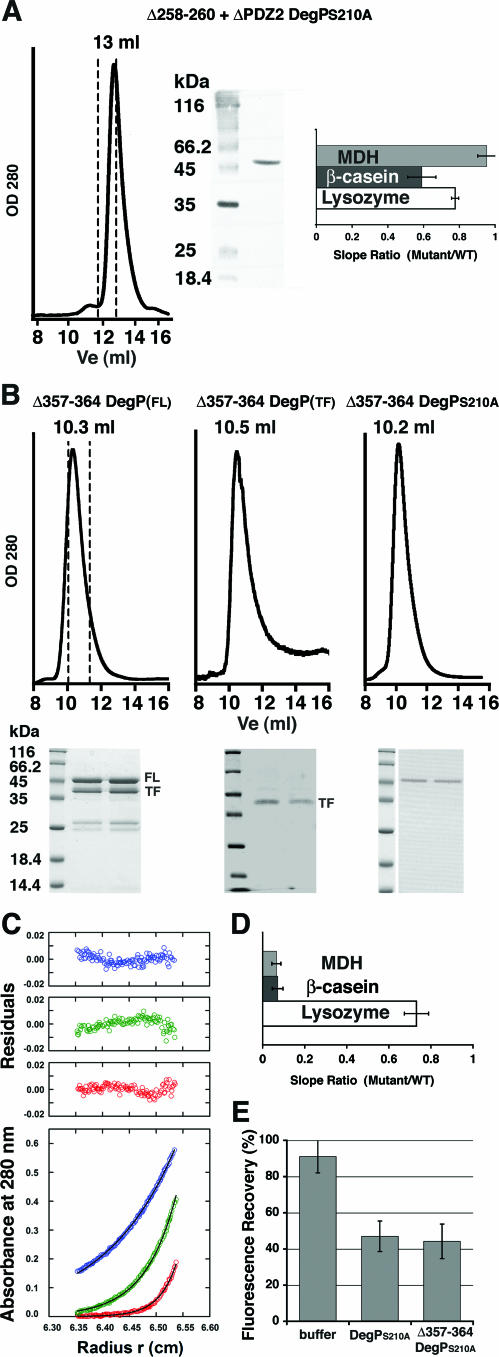
Interestingly, a mutant lacking the entire PDZ2 domain and the flexible loop connecting the protease and PDZ1 domains (Δ258-260+ΔPDZ2 DegP mutant) exhibited protease activity (Fig. 8A, right panel). This mutant eluted as a trimer in size exclusion chromatography (Fig. 8A, left and middle panels). Therefore, the loop appears to be essential for protease activity only when DegP forms a hexameric cage.
FIG. 8.
Characterization of the Δ258-260+ΔPDZ2 and Δ357-364 DegP mutants. (A) The plot on the left shows the elution profile of the Δ258-260+ΔPDZ2 DegPS210A mutant obtained with a Superdex-200 column after incubation at 4°C for 1 h. The dashed lines indicate the estimated elution volumes (Ve) for a hexameric form (11.8 ml) and a trimeric form (13.3 ml) of the mutant. A sample from the peak was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue (middle panel). The averages and standard deviations for the proteolysis rates for the Δ258-260+ΔPDZ2 DegP mutant with MDH, β-casein, and lysozyme are shown on the right. WT, wild type; OD 280, optical density at 280 nm. (B) Plots showing the elution profiles for full-length (FL) and truncated (TF) forms of the Δ357-364 DegP mutant. The elution profile for the proteolytically inactive variant of this mutant (S210A) is also shown. The dashed lines in the elution profile on the left indicate the estimated elution volumes for a dodecameric form (10 ml) and a hexameric form (11.5 ml) of the mutant. The gels below the plots contained fractions of each peak resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. (C) Sedimentation equilibrium profiles expressed in terms of A280 versus the radius for the Δ357-364 DegPS210A mutant at a loading A280 of 0.58. Data were collected at 4,000 rpm (blue), 6,000 rpm (green), and 8,000 rpm (red) at 4.0°C and were analyzed in terms of a single ideal solute. Best fits are indicated by black lines through the experimental points, and the corresponding distributions of the residuals are shown. (D) Graph showing the averages and standard deviations for the proteolysis rates of the Δ357-364 DegP mutant with different substrates. (E) Plot showing the percentages of unfolded EGFP-SFS refolded in the absence and presence of the Δ357-364 DegPS210A mutant compared to the results obtained for the DegPS210A mutant. The plot was generated as described in the legend to Fig. 6.
We hypothesized that the loop between the protease and the PDZ1 domain is essential only in the context of a hexameric DegP because the flexibility that this loop may confer allows exposure of the binding cleft of PDZ1 to substrate molecules, which may otherwise be buried within the hexameric cage. However, the hexameric Δ258-260 DegPS210A mutant was able to trap unfolded EGFP-SFS (Fig. 7D) and EGFP-SES (data not shown) in the fluorescence assay, suggesting that the PDZ1 binding cleft in this mutant is readily accessible to the C-terminal residues of the substrate molecules.
We next examined whether the flexible loop between the PDZ1 and PDZ2 domains is required for DegP activities by generating the Δ357-364 DegP mutant (Fig. 1A and B) and its inactive S210A variant. Analysis of the oligomeric state showed that these two mutants were stable dodecamers as determined by size exclusion chromatography (Fig. 8B, left and right panels) and sedimentation equilibrium analysis (Fig. 8C) (experimental molecular mass, 566 ± 38 kDa, corresponding to a stoichiometry of 12.3 ± 0.8). Two hexameric cages presumably associate to form the dodecamers observed. Consistent with our previous results, the mutation did not alter the chaperone activity of the protein (data not shown). However, the Δ357-364 DegP mutant was unable to hydrolyze MDH and β-casein (Fig. 8D). As observed for the mutant lacking the loop between the protease and PDZ1 domains, the PDZ1 binding cleft in the Δ357-364 DegP mutant was also accessible to the substrate molecules, as shown by the EGFP fluorescence assay (Fig. 8E), indicating that the loop between the PDZ domains in DegP is also essential for proteolysis but is not required for substrate access to the PDZ1 binding cleft.
Surprisingly, the Δ357-364 DegP mutant hydrolyzed lysozyme at rates similar to the rates observed for the wild type. These data, together with the slow lysozyme hydrolysis observed with the DegP protein lacking the loop between the protease and PDZ1 domains and the ΔPDZ1 DegP mutants, suggest that lysozyme may be able to access the catalytic triad of DegP in the protease domain without assistance from the PDZ1 domain. Consistently, we noticed that in our experimental conditions degradation of β-casein or MDH occurred with incubation times on the order of minutes, while the incubation times in the lysozyme hydrolysis assays had to be extended to 4 h in order to observe significant degradation (10). This observation also suggests that the targeting of lysozyme for degradation by DegP may be different from the targeting of β-casein and MDH.
Interestingly, the Δ357-364 DegP mutant showed partial cleavage during expression and purification, and an approximately 43-kDa truncated polypeptide was formed (Fig. 8B, left panel). A sample of this mutant containing mainly the truncated form of the protein was obtained by inducing expression for 3 h in the presence of 7 mM dithiothreitol. This mutant eluted as a dodecamer when it was loaded on a size exclusion chromatography column (Fig. 8B, middle panel), and it hydrolyzed lysozyme at rates similar to those observed for the predominantly nontruncated protein (data not shown). Therefore, it appears that truncation occurred without affecting the oligomeric structure of the protein, and we concluded that the lack of protease activity exhibited by this mutant in the presence of MDH and β-casein was due to the absence of the loop between the PDZ1 and PDZ2 domains rather than to the presence of ∼40% of the protein in the truncated form.
DISCUSSION
Our data showing that the PDZ1 domain of E. coli DegP is essential for protease activity and that the last three C-terminal residues of the substrate molecules enhance sequestration of unfolded proteins provide new insights into the mechanism of substrate recognition for proteolytic degradation by DegP. More generally, because our study showed that both PDZ domains are dispensable for the chaperone activity of DegP, the data imply that DegP recognizes substrate molecules targeted for degradation and substrate molecules targeted for refolding in different ways. Thus, we hypothesized that the mechanism through which DegP recognizes the substrate may play a role in the protease-chaperone switch, dictating whether the substrate is degraded or refolded.
Our results are consistent with a mechanism in which the DegP protease first recognizes its substrates by binding to exposed C-terminal residues, much like other PDZ-containing proteins (e.g., Tsp [1, 16]). By doing this, it recruits a substrate to the catalytic site and subsequently cleaves it at multiple sites. In this context, the C-terminal residues allow DegP to tether the substrate and attack the primary cleavage sites (12). It has been shown that in other proteases, such as DegS, binding of the C terminus of a substrate to the PDZ domain induces a conformational change that activates the catalytic triad (35). Whether binding of the C-terminal residues of the substrate to the PDZ1 domain of DegP also causes such allosteric activation of the proteolytic domain is currently unknown. In this study, we found that the loop connecting the protease domain and the PDZ1 domain is essential for protease activity. In a manner similar to that in DegS, this loop in DegP may be required to transmit the conformational change that makes the catalytic site active. However, based on the protease activity observed with the Δ258-260+ΔPDZ2 DegP mutant that was also missing this loop but was proteolytically active, we concluded that this is probably not the case. Alternatively, we hypothesized that residues 258 to 260 between the protease and the PDZ1 domain and residues 357 to 364 between the two PDZ domains (Fig. 1A and B) are flexible loops required to expose the binding cleft of PDZ1 and allow substrate binding, but the EGFP fluorescence assay showed that the PDZ1 binding cleft in these mutants seems to be readily accessible to the C-terminal residues of the substrate molecules. Interestingly, the loop between the protease and the PDZ1 domain was found to be dispensable for the protease activity in a trimeric mutant of DegP. Additional experiments are necessary to determine why these loops are essential for the protease activity of hexameric DegP but dispensable in a trimeric mutant of this protein.
The X-ray structure of DegP suggests that the hydrophobic binding pockets for P0 and P−2 in the binding cleft in the PDZ1 domain of DegP confer the binding specificity. Several crystal structures, including the structure of PSD-95 PDZ3 bound to its peptide ligand, initially suggested that the residue at P−1 points away from the interaction surface of the binding cleft, which correlates with the relatively low specificity of this PDZ domain with respect to substrate residues at P−1 (3). However, subsequent studies indicated that substitutions at this site could affect the binding preference for PSD-95 PDZ3 (22) and other PDZ domains, including PDZ1 in the NHERF (13, 14, 34) and InaD (18) proteins. Our results indicate that point mutations in the residue at P−1 of the SFS C-terminal tag greatly influenced the ability of DegP to sequester the unfolded substrate. In fact, changes in a negatively charged or small nonpolar residue, such as alanine, dramatically increased trapping by PDZ1, which correlated well with the presence of the positively charged residue arginine 325 in the vicinity of the −1 binding pocket in the PDZ1 domain (Fig. 1D).
Our results showed that the C-terminal SsrA tag and other C-terminal tags increased the sequestration of unfolded EGFP. Although the EGFP refolding assay used in this study does not measure binding directly, the increased sequestration of unfolded substrate is consistent with changes in the C-terminal tag binding to the E. coli PDZ1 DegP domain. Moreover, the results obtained with the SsrA tag are in agreement with the results of a previous report showing the ability of the PDZ domains of S. enterica serovar Typhimurium DegP to bind the SsrA peptide (30). Interestingly, in this previous report, isolated E. coli DegP PDZ domains were unable to bind the SsrA peptide.
Previous studies have shown that PDZ domains are highly modular domains and that their structure is usually not compromised by mutations within the PDZ domain itself (32) or in neighboring domains. Many of our results confirmed that the mutants in this study were properly folded, that all of the mutants tested showed chaperone activity, and that the oligomeric structures were stable even upon incubation for 1 h at 43°C. Further proof that the structural integrity of these mutants was maintained was obtained from sedimentation velocity analyses; mutants with small deletions (e.g., Δ258-260 DegPS210A mutant) had a c(s) distribution very similar to that of the corresponding full-length DegP, whereas mutants with larger deletions had sedimentation values that appeared to correlate with their masses. It is therefore unlikely that the results obtained with these mutants reflected incorrect folding or protein instability.
Acknowledgments
We thank Alba Guarne and Cecelia Trainor for insightful comments and critical reading of the manuscript. We thank Mochan Li for preliminary work on the Δ357-364 DegP mutant and Saeed Darvish-Kazem for constructing the EGFP-SFS mutants.
This work was supported by grants from the Canadian Institutes of Health Research, the Canada Foundation for Innovation, and the Ontario Innovation Trust. J.O. was a recipient of a Canadian Institutes of Health Research salary award. R.G. was supported by the Intramural Research Program of the NIH National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Beebe, K. D., J. Shin, J. Peng, C. Chaudhury, J. Khera, and D. Pei. 2000. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry 39:3149-3155. [DOI] [PubMed] [Google Scholar]
- 2.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 3.Doyle, D. A., A. Lee, J. Lewis, E. Kim, M. Sheng, and R. MacKinnon. 1996. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 85:1067-1076. [DOI] [PubMed] [Google Scholar]
- 4.Farrell, C. M., A. D. Grossman, and R. T. Sauer. 2005. Cytoplasmic degradation of ssrA-tagged proteins. Mol. Microbiol. 57:1750-1761. [DOI] [PubMed] [Google Scholar]
- 5.Flynn, J. M., I. Levchenko, M. Seidel, S. H. Wickner, R. T. Sauer, and T. A. Baker. 2001. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. USA 98:10584-10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris, B. Z., and W. A. Lim. 2001. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114:3219-3231. [DOI] [PubMed] [Google Scholar]
- 8.Harrison, S. C. 1996. Peptide-surface association: the case of PDZ and PTB domains. Cell 86:341-343. [DOI] [PubMed] [Google Scholar]
- 9.Hoskins, J. R., S. K. Singh, M. R. Maurizi, and S. Wickner. 2000. Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc. Natl. Acad. Sci. USA 97:8892-8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jomaa, A., D. Damjanovic, V. Leong, R. Ghirlando, J. Iwanczyk, and J. Ortega. 2007. The inner cavity of Escherichia coli DegP protein is not essential for molecular chaperone and proteolytic activity. J. Bacteriol. 189:706-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, C. H., P. N. Danese, J. S. Pinkner, T. J. Silhavy, and S. J. Hultgren. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16:6394-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, C. H., P. Dexter, A. K. Evans, C. Liu, S. J. Hultgren, and D. E. Hruby. 2002. Escherichia coli DegP protease cleaves between paired hydrophobic residues in a natural substrate: the PapA pilin. J. Bacteriol. 184:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karthikeyan, S., T. Leung, G. Birrane, G. Webster, and J. A. Ladias. 2001. Crystal structure of the PDZ1 domain of human Na+/H+ exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains. J. Mol. Biol. 308:963-973. [DOI] [PubMed] [Google Scholar]
- 14.Karthikeyan, S., T. Leung, and J. A. Ladias. 2001. Structural basis of the Na+/H+ exchanger regulatory factor PDZ1 interaction with the carboxyl-terminal region of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 276:19683-19686. [DOI] [PubMed] [Google Scholar]
- 15.Karzai, A. W., E. D. Roche, and R. T. Sauer. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7:449-455. [DOI] [PubMed] [Google Scholar]
- 16.Keiler, K. C., and R. T. Sauer. 1996. Sequence determinants of C-terminal substrate recognition by the Tsp protease. J. Biol. Chem. 271:2589-2593. [DOI] [PubMed] [Google Scholar]
- 17.Keiler, K. C., P. R. Waller, and R. T. Sauer. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990-993. [DOI] [PubMed] [Google Scholar]
- 18.Kimple, M. E., D. P. Siderovski, and J. Sondek. 2001. Functional relevance of the disulfide-linked complex of the N-terminal PDZ domain of InaD with NorpA. EMBO J. 20:4414-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. [DOI] [PubMed] [Google Scholar]
- 20.Liao, D. I., J. Qian, D. A. Chisholm, D. B. Jordan, and B. A. Diner. 2000. Crystal structures of the photosystem II D1 C-terminal processing protease. Nat. Struct. Biol. 7:749-753. [DOI] [PubMed] [Google Scholar]
- 21.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 171:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niethammer, M., J. G. Valtschanoff, T. M. Kapoor, D. W. Allison, T. M. Weinberg, A. M. Craig, and M. Sheng. 1998. CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90. Neuron 20:693-707. [DOI] [PubMed] [Google Scholar]
- 23.Pallen, M. J., and C. P. Ponting. 1997. PDZ domains in bacterial proteins. Mol. Microbiol. 26:411-413. [DOI] [PubMed] [Google Scholar]
- 24.Ponting, C. P. 1997. Evidence for PDZ domains in bacteria, yeast, and plants. Protein Sci. 6:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78:1606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz, J., T. Doerks, C. P. Ponting, R. R. Copley, and P. Bork. 2000. More than 1,000 putative new human signalling proteins revealed by EST data mining. Nat. Genet. 25:201-204. [DOI] [PubMed] [Google Scholar]
- 27.Silber, K. R., K. C. Keiler, and R. T. Sauer. 1992. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc. Natl. Acad. Sci. USA 89:295-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh, S. K., R. Grimaud, J. R. Hoskins, S. Wickner, and M. R. Maurizi. 2000. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc. Natl. Acad. Sci. USA 97:8898-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Songyang, Z., A. S. Fanning, C. Fu, J. Xu, S. M. Marfatia, A. H. Chishti, A. Crompton, A. C. Chan, J. M. Anderson, and L. C. Cantley. 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275:73-77. [DOI] [PubMed] [Google Scholar]
- 30.Spiers, A., H. K. Lamb, S. Cocklin, K. A. Wheeler, J. Budworth, A. L. Dodds, M. J. Pallen, D. J. Maskell, I. G. Charles, and A. R. Hawkins. 2002. PDZ domains facilitate binding of high temperature requirement protease A (HtrA) and tail-specific protease (Tsp) to heterologous substrates through recognition of the small stable RNA A (ssrA)-encoded peptide. J. Biol. Chem. 277:39443-39449. [DOI] [PubMed] [Google Scholar]
- 31.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 32.Stricker, N. L., K. S. Christopherson, B. A. Yi, P. J. Schatz, R. W. Raab, G. Dawes, D. E. Bassett, Jr., D. S. Bredt, and M. Li. 1997. PDZ domain of neuronal nitric oxide synthase recognizes novel C-terminal peptide sequences. Nat. Biotechnol. 15:336-342. [DOI] [PubMed] [Google Scholar]
- 33.Walsh, N. P., B. M. Alba, B. Bose, C. A. Gross, and R. T. Sauer. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61-71. [DOI] [PubMed] [Google Scholar]
- 34.Webster, G., T. Leung, S. Karthikeyan, G. Birrane, and J. A. Ladias. 2001. Crystallographic characterization of the PDZ1 domain of the human Na+/H+ exchanger regulatory factor. Acta Crystallogr. D Biol. Crystallogr. 57:714-716. [DOI] [PubMed] [Google Scholar]
- 35.Wilken, C., K. Kitzing, R. Kurzbauer, M. Ehrmann, and T. Clausen. 2004. Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell 117:483-494. [DOI] [PubMed] [Google Scholar]