Abstract
Objectives
To perform a case–control study of a functional M196R polymorphism of tumour necrosis factor receptor type 2 (TNF‐RII) in a Japanese population and a meta‐analysis of all published reports on the polymorphism to investigate the association of the M196R polymorphism of TNF‐RII with systemic lupus erythematosus (SLE).
Methods
The functional M196R polymorphism of TNF‐RII was genotyped by using polymerase chain reaction combined with the subsequent single‐strand conformation polymorphism (PCR—SSCP) analysis for screening, followed by nucleotide sequencing for confirmation. A total of 331 patients and 359 controls were subjected to a case–control study. A meta‐analysis of the available case–control studies including all published data as well as our own data was performed to investigate the association of the functional M196R polymorphism of TNF‐RII with SLE.
Results
Our case–control study did not show any significant association of a functional M196R polymorphism of TNF‐RII with SLE, although there was a trend towards association. A meta‐analysis of seven case–control studies in eight different ethnic populations including our own showed that 196M/R and 196R/R genotypes combined was significantly associated with an increased risk of SLE (odds ratio (OR) 1.29, 95% confidence interval (CI) 1.04 to 1.60; p = 0.02). Stratification by ethnicity showed a more significant association in Asians, including Japanese, Korean and Vietnamese (OR 1.40, 95% CI 1.10 to 1.78; p = 0.006). The effect of the 196R allele on SLE was not clear in Caucasians.
Conclusions
The 196R allele of the functional M196R polymorphism of TNF‐RII is a risk factor for SLE, especially in the Asian population.
Systemic lupus erythematosus (SLE) is a systemic autoimmune disorder characterised by the production of a broad variety of autoantibodies and subsequent immune complex deposition, which results in chronic inflammation in multiple organ systems. Although the pathogenesis of SLE is still unclarified, a substantial contribution of genetic factors has been implicated.1
Accumulating evidence suggests that tumour necrosis factor α (TNFα) and its receptors are actively involved in the pathogenesis and development of SLE. TNFα is a pleiotropic cytokine that plays a pivotal role in health and disease. The biological effects of TNFα are mediated through two distinct cell surface receptors, TNF‐RI (TNFRSF1A) and TNF‐RII (TNFRSF1B).2,3 A number of polymorphisms in the TNFα gene and in the nearby TNFβ gene have been shown to be associated with SLE.4,5,6,7 One of the polymorphisms, TNF‐α‐308A, located in the promoter region of the TNFα gene, was shown to be associated with increased production of TNFα.3 In a mouse model of SLE, TNFα administration modulates its disease severity, depending on the dose and time of its administration.8,9 In human SLE, TNFα is highly expressed in glomeruli in the kidney and correlates well with renal inflammatory activity.10 These findings suggest that TNFα may be involved in the pathogenesis of SLE.
The association of TNF receptors with SLE has been implicated as well. Recent genome‐wide linkage studies in multiplex SLE families have identified chromosome 1p36 as one of the candidate loci for SLE.11,12 The gene for TNF‐RII is located on the chromosome 1p36.3–p36.2. Komata et al first demonstrated that a methionine to arginine substitution at position 196 (196R) of TNF‐RII was significantly increased in SLE patients.13 Our previous study14 coincided with their findings in Japanese SLE patients, but others did not.15,16,17,18 We have demonstrated that increased production of interleukin‐6 and cell death occur in 196R TNF‐RII‐transfected HeLa cells compared with those transfected with wild‐type TNF‐RII.14 This finding that 196R allele was associated with functional alteration has recently been confirmed by others.19 This polymorphism was reported to be associated with susceptibility to chronic inflammatory disorders other than SLE, such as familial rheumatoid arthritis and ulcerative colitis.20,21,22 Recently, the molecular basis of a subset of the hereditary periodic fever syndromes has been shown to reside in defects of the TNFRSF1A.23 Its clinical symptoms, such as recurrent episodes of fever, skin rash, arthritis and serositis, mimic those of SLE. In fact, we and others have reported that novel polymorphisms in the TNFRSF1A were associated with SLE.24,25 These lines of evidence prompted us to perform a case–control study in newly enrolled Japanese patients with SLE and controls (331 cases and 359 controls), which did not overlap with the cases and controls in our previous report.14 Furthermore, in the present study, a meta‐analysis of data from case–control studies, including our present case–control study, on the M196R polymorphism of TNF‐RII and SLE was performed.
Materials and methods
Patients and DNA
We studied a total of 331 patients with SLE who are followed up at the rheumatology clinics of Kyushu University Hospital, Fukuoka, Japan, and its affiliated hospitals. All patients fulfilled the American College of Rheumatology 1997 revised criteria for SLE.26,27 A total of 359 healthy individuals living in the Kyushu area were enrolled in the association study. All individuals were Japanese. The study protocol was approved by our institutional review board, and all participants provided written informed consent. DNA was purified from peripheral blood mononuclear cells by using a QIAamp DNA Blood Maxi Kit (Qiagen, Hilden, Germany) or according to other standard protocols. All subjects were newly enrolled in this study and did not overlap with the patients and controls in our previous study.14
Determination of M196R polymorphism of TNF‐RII
The primers for detecting the M196R polymorphism in exon 6 of TNF‐RII were synthesised on the basis of the genomic DNA sequence as described previously.14 Polymerase chain reaction (PCR) and the subsequent single‐strand conformation polymorphism (SSCP) analysis were performed to determine M196R polymorphism as described previously.14 Briefly, PCR was performed in a total volume of 10 μl containing 50 ng of template DNA, 0.25 μM each primer, 2 mM MgCl2, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 0.2 mM each nucleotide (Amersham Biosciences, Piscataway, New Jersey, USA) and 0.25 U of AmpliTaq DNA polymerase (Applied Biosystems, Foster City, California, USA). The thermal cycling profile was conducted for 40 cycles of 30 s at 94°C for denaturation, 1 min at 62°C for annealing and 1 min at 72°C for extension.
PCR products were separated on 12% polyacrylamide gels at 25°C with 5% glycerol. DNA fragments were visualised by silver staining. Sequencing analysis was performed for PCR products of interest to confirm the PCR–SSCP data.
Meta‐analysis
We conducted Medline, Current Contents and Web of Science searches using “TNF‐RII”, “TNF‐R2”, “TNFRSF1B”, “systemic lupus erythematosus” and “polymorphism” for papers published before April 2006. Additional articles were identified through the references cited in the first series of articles selected. Articles included in the meta‐analysis were in any language, with human subjects, published in the primary literature, and had no obvious overlap of subjects with other studies. We excluded studies with the same data or overlapping data by the same authors. Using the Medline database, we identified six case–control studies that provided information on SLE associated with the M196R polymorphism of TNF‐RII. For studies including subjects of different ethnic groups, data were extracted separately for each ethnic group whenever possible. We assessed the Hardy–Weinberg equilibrium via a goodness‐of‐fit test (Pearson) to compare the observed and expected genotype frequencies among controls.
Data were combined using both fixed effects (Mantel–Haenszel) and random effects (DerSimonian and Laird method) models.28 The random effects model is more appropriate when heterogeneity is present.28 Thus, estimated values were basically calculated on the basis of the random effects model. Heterogeneity, evaluated by the Cochrane Q test among the studies, was considered significant at a level of p<0.10.29,30 To test for publication bias, both Begg's31 and Egger's32 tests were used to assess whether smaller studies reported greater associations than larger studies. Publication bias was considered significant at a level of p<0.10. All the calculations were performed with the computer program STATA V.8.2 (Stata Corporation).
Results
A case–control study for M196R polymorphism
For the case–control study, 331 patients with SLE and 359 healthy individuals were enrolled. Table 1 shows the estimated frequencies of the M196R polymorphism. In all, 10 (3.0%) were homozygous and 87 (26.3%) were heterozygous for the 196R allele in the 331 patients with SLE, whereas out of the 359 healthy individuals, 5 (1.4%) were homozygous and 85 (23.7%) were heterozygous. When a combined frequency of the 196R/R and 196M/R genotypes was compared with that of the 196M/M genotype, there was no significant association between the M196R polymorphism and SLE (p = 0.224). The number of 196R alleles was 107 out of a total of 662 (16.2%) alleles in patients with SLE, whereas that in healthy individuals was 95 out of 718 (13.5%) alleles, which showed no significant difference in allele frequency either (p = 0.124).
Table 1 Association study for TNF‐RII polymorphism at codon 196 with our patients with SLE.
| SLE | Control | p Value | |
|---|---|---|---|
| Genotype | |||
| 196R/R | 10 (3) | 5 (1.4) | 0.224 |
| 196R/M | 87 (26.3) | 85 (23.7) | |
| 196M/M | 234 (70.7) | 269 (74.9) | |
| Total | 331 (100) | 359 (100) | |
| Allele | |||
| 196R | 107 (16.2) | 95 (13.5) | 0.124 |
| 196M | 555 (83.8) | 623 (86.5) | |
| Total | 662 (100) | 718 (100) |
SLE, systemic lupus erythematosus.
Values are number (%).
Individual and summary frequencies of the case–control studies
As the minor R allele is related to increased production of IL‐6,14 subjects with at least one minor allele may be associated with an increased risk of SLE. As the 196R/R genotype has not been separated because of a low prevalence of the minor 196R allele in several studies (table 2), we combined the 196M/R genotype with the 196R/R genotype.
Table 2 Summary of the reported case–control studies for the meta‐analysis.
| Reports | Ethnicity | Number of | Genotype frequency (%) | p Value* | Frequency of the 196 R allele | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SLE | Control | |||||||||||||
| SLE | Control | M/M | M/R | R/R | M/R+R/R | M/M | M/R | R/R | M/R+R/R | SLE | Control | |||
| Komata et al, 199913 | Japanese | 81 | 207 | 63.0 | 35.8 | 1.2 | 37.0 | 81.2 | 17.9 | 0.9 | 18.8 | 0.003 | 0.191 | 0.99 |
| Al‐Ansari et al, 200015 | Spanish | 128 | 141 | 56.3 | 39.1 | 4.7 | 43.8 | 58.9 | 35.5 | 5.7 | 41.1 | 0.80 | 0.242 | 0.234 |
| UK | 74 | 90 | 52.7 | 35.1 | 12.2 | 47.3 | 47.8 | 47.8 | 4.4 | 52.2 | 0.11 | 0.297 | 0.283 | |
| Morita et al, 200114 | Japanese | 105 | 99 | 62.9 | 33.3 | 3.8 | 37.1 | 74.7 | 25.3 | 0 | 25.3 | 0.0498 | 0.205 | 0.126 |
| Takahashi et al, 200116 | Japanese | 42 | 238 | 75.4 | 23.9 | 0.1 | 24.6 | 77.3 | 21.8 | 0.8 | 22.7 | 0.87 | 0.127 | 0.118 |
| Lee et al, 200117 | Korean | 139 | 137 | 65.5 | 30.9 | 3.6 | 34.5 | 69.3 | 26.3 | 4.4 | 30.7 | 0.68 | 0.191 | 0.175 |
| Khoa et al, 200518 | Vietnamese | 44 | 42 | 61.4 | 34.1 | 4.5 | 38.6 | 66.7 | 29.5 | 2.4 | 33.3 | 0.93 | 0.216 | 0.179 |
| Present study | Japanese | 331 | 359 | 70.7 | 26.3 | 0.3 | 29.3 | 74.9 | 23.7 | 1.4 | 25.0 | 0.22 | 0.162 | 0.132 |
| Summary* | Frequency (%) of the M/R and R/R genotypes combined | Frequency of the R allele | ||||||||||||
| Japanese only (4 populations) | 22.8 (19.8 to 25.8) | 0.119 (0.104 to 0.134) | ||||||||||||
| Asian populations (6 populations) | 24.5 (20.9 to 28.0) | 0.130 (0.108 to 0.151) | ||||||||||||
| Caucasian (2 populations) | 46.2 (35.4 to 57.0) | 0.254 (0.206 to 0.301) | ||||||||||||
SLE, systemic lupus erythematosus.
*Differences in distribution of three genotypes of TNF‐RII M196R polymorphism between cases and controls.
Among six case–control studies,13,14,15,16,17,18 one study included two different Caucasian groups.15 Data from our own population were added to the meta‐analysis (table 2). Six studies encompassed Asian populations including Japanese, Korean and Vietnamese, and two studied Caucasian populations. No case–control studies were done for African populations. Of the eight studies, two Japanese studies revealed a significant association between the M196R polymorphism of TNF‐RII and SLE.13,14 The remaining six studies did not show any statistically significant difference; however, there was a tendency towards increased 196R allele frequency in patients with SLE among all of the four studies from Asian groups. This tendency was not observed in the two studies dealing with Caucasian populations. In healthy individuals, there seems to be an ethnic difference in the 196R allele frequency. For a total of 1082 healthy Asian individuals enrolled, the summary frequency of the 196R allele was 0.130 (95% CI = 0.108 to 0.151), whereas that of Caucasians was 0.254 (95% CI = 0.206 to 0.301) in 231 control individuals (table 2). The frequency was somewhat higher among Caucasians than among Asians.
Individual and summary ORs for the combined genotype
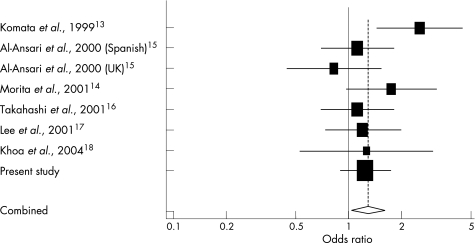
As shown in table 3 and fig 1, only one study found that the combined genotype was significantly associated with an increased risk of SLE (OR = 2.53, 95% CI = 1.37 to 4.65).13 All the eight studies were in Hardy–Weinberg equilibrium in controls (p>0.05), and were thus included in the meta‐analysis for the association study of the M196R polymorphism with SLE. Heterogeneity and publication bias were absent in the analysis. When all of the eight studies were combined and subjected to a meta‐analysis, 196R carriers (196M/R and 196R/R genotypes combined) were at a significantly increased risk of SLE (OR = 1.29, 95% CI = 1.04 to 1.60; p = 0.02). When meta‐analyses were performed for the studies of Asian and Japanese populations, the OR of the combined genotype for SLE was 1.40 (95% CI = 1.10 to 1.78; p = 0.006) and 1.26 (95% CI = 1.01 to 1.58; p = 0.043), respectively. There was no statistically significant association for SLE in the homozygotes for the 196R allele, probably because of the limited sample size. The meta‐analysis of the two case–control studies dealing with Caucasians could not detect any association between M196R polymorphism and SLE (OR = 0.99, 95% CI = 0.68 to 1.45; p = 0.96).
Table 3 Studies of M196R TNF‐RII polymorphism and risk of SLE.
| OR (95% CI) for the M/R and R/R genotypes combined | Method of genotyping | Source of control | ||||
|---|---|---|---|---|---|---|
| Komata et al, 199913 | 2.53 (1.37 to 4.65) | PCR–SSCP | Community control | |||
| Al‐Ansari et al, 200015 | ||||||
| Spanish | 1.11 (0.67 to 1.86) | PCR–RFLP | Community control | |||
| UK | 0.82 (0.42 to 1.59) | |||||
| Morita et al, 200114 | 1.75 (0.92 to 3.35) | PCR–SSCP | Community control | |||
| Takahashi et al, 200116 | 1.11 (0.66to 1.86) | PCR–SSCP | Community control | |||
| Lee et al, 200117 | 1.19 (0.70 to 2.04) | PCR–SSCP | Community control (blood donor) | |||
| Khoa et al, 200518 | 1.26 (0.48 to 3.35) | PCR–RFLP | Community control (blood donor) | |||
| Present study | 1.84 (0.92 to 3.64) | PCR–SSCP | Community control | |||
| Summary | OR for the M/R and R/R genotypes combined | Cochran Q test for heterogeneity | Test for publication bias | |||
| Random effects model | Fixed effects model | Begg's test | Egger's test | |||
| All | 1.29 (1.04 to 1.60) | 1.28 (1.07 to 1.53) | p = 0.23 | p = 0.71 | p = 0.74 | |
| Japanese only | 1.26 (1.01 to 1.58) | 1.26 (1.01 to 1.58) | p = 0.70 | p = 0.74 | p = 0.73 | |
| Asian populations | 1.40 (1.10 to 1.78) | 1.38 (1.13 to 1.69) | p = 0.26 | p = 0.26 | p = 0.48 | |
| Caucasians | 0.99 (0.68 to 1.45) | 0.99 (0.68 to 1.45) | p = 0.45 | p = 1.00 | Not calculable | |
PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; SSCP, single‐stranded conformation polymorphism.
Figure 1 Meta‐analysis of seven case–control studies in eight different study populations of systemic lupus esythematosus (SLE) and the 196M/R and 196R/R genotypes combined. The centre of a box and the horizontal line (logarithm) indicate the odds ratio (OR) and the 95% confidence interval (CI) in each study, with the areas of the boxes representing the weight of each study. The summary OR on the basis of random effects model is represented by the middle of a diamond the width of which indicates the 95% CI. The summary OR is shown by the dotted vertical line. Statistical heterogeneity between studies was assessed using Cochran's Q test (Q = 9.3, p = 0.23). Summary: OR, 1.29 (1.04 to 1.60).
Discussion
Accumulating evidence suggests that TNF‐RII is involved in the pathophysiology of inflammatory disorders. Although TNF‐RII was initially considered to play a supporting role to TNF‐R1 in TNFα‐mediated signalling, TNF‐RII itself is now considered to mediate signals distinct from those by TNF‐RI. A critical role for TNF‐RII in systemic inflammation and tissue damage has been shown in a murine model.33 It has been demonstrated that TNF‐RII is a main receptor for transmembrane TNFα.34 A number of polymorphisms have been identified within TNF‐RII that consists of ten exons and spans 42 kb. A polymorphism at codon 196 of TNF‐RII exon 6 changes the amino acid from methionine to arginine. In the cells carrying the 196R allele, we have demonstrated increased cytokine production and apoptosis after TNFα stimulation.14 The difference in intracellular signalling event between the 196M allele and 196R allele has recently been shown by another group,19 which supports the functional alteration of the M196R polymorphism. Association of this functional M196R polymorphism with a number of human inflammatory diseases other than SLE, such as familial rheumatoid arthritis20,21 and inflammatory bowel disease, has been reported.22
Findings of association studies between the 196R polymorphism and SLE have been controversial.13,14,15,16,17,18 In the present study, we therefore performed genotyping of a set of Japanese patients and controls, followed by meta‐analysis of eight reported case–control studies including our own study. Meta‐analysis is the statistical combination of a number of studies to produce a single estimate of the magnitude of the effect of the intervention under investigation.32 On increasing the sample size and statistical power, meta‐analysis is considered to be a powerful tool to summarise conflicting results from different studies. In our case–control study, a total of 331 patients with SLE and 359 healthy individuals of Japanese ancestry were studied, which includes the largest number of cases and controls compared with the previously reported M196R genotyping studies (table 2). Although there was a tendency for association, there was no statistically significant association between the M196R polymorphism and an increased risk of SLE. It is suggested that the effect size of M196R polymorphism is not large enough to be detected in a single association study. In contrast, the meta‐analysis of all the seven case–control studies in five different ethnic populations showed that the 196R allele of the M196R polymorphism is a significant risk factor for SLE (OR = 1.29, 95% CI 1.04 to 1.60; p = 0.02; table 3). Even a more significant association between the M196R polymorphism and SLE was observed in the Asian population including Japanese, Korean and Vietnamese (OR = 1.40, 95% CI 1.10 to 1.78; p = 0.006). As no case–control studies for Africans have been reported, further accumulation of samples would be needed for this population, whose risk of developing SLE is higher than those of Caucasians and Asians.35 In Caucasians, the M196R polymorphism was not associated with an increased risk of SLE, although the sample size was small. It is noteworthy that two transmission disequilibrium tests have been performed to avoid the possible population admixture. One study enrolled 91 Caucasian families with SLE in southern California,36 and the other studied 456 families with SLE in the UK,37 most of which were Caucasians. These two transmission disequilibrium tests demonstrated no evidence of association, suggesting that the 196R allele of the M196R polymorphism is not likely to be a risk factor for SLE in the Caucasian population.
Successful introduction of anti‐TNFα agents in the treatment of diseases such as rheumatoid arthritis and Crohn's disease strongly suggests that TNFα is actively involved in the pathogenesis of chronic inflammatory disorders.38 In the case of SLE, the application of anti‐TNFα agents has been stopped for the following two main reasons.39 Firstly, treatment with TNF blockers sometimes leads to the development of anti‐nuclear antibodies and antibodies to double‐stranded DNA, although this condition is reversible when anti‐TNF therapy is stopped and only rarely results in definite SLE. Secondly, TNFα is protective against the development of SLE‐like conditions in some lupus model mice. However, a recent open‐label study showing that the anti‐TNFα antibody, infliximab, may be effective in improving the inflammatory manifestations of SLE suggests the important role of TNFα in the pathogenesis of human SLE.40 There is now substantial evidence that TNFα plays an important role in the organ inflammation secondary to immune complex formation and resultant monocyte or macrophage activation.41 The association of a more active polymorphism of TNF‐RII (M196R) with SLE fits well with this evidence. It is of interest that transmembrane TNFα, a main ligand for TNF‐RII, is markedly upregulated in CD8 T cells from patients with SLE.42 Clinical phenotypes, such as arthritis and nephritis, of our patients carrying the M196R polymorphism did not differ from those of patients who were non‐carriers (data not shown), which was consistent with our previous findings.14 Inflammatory tissue damage was not different between the M196R carriers and non‐carriers.
In conclusion, the 196R allele of the functional polymorphism M196R of TNF‐RII was significantly associated with an increased risk of SLE as shown by the meta‐analysis of a total of seven case–control studies in eight different study populations (table 3, fig 1). Stratification by ethnicity showed that the 196R allele is likely to be a risk factor for SLE in the Asian population. Further study of Caucasian and African descendars is needed before making conclusions about the association of the M196R polymorphism with SLE.
Abbreviations
PCR - polymerase chain reaction
SSCP - single‐strand conformation polymorphism
SLE - systemic lupus erythematosus
TNF‐RII - tumour necrosis factor receptor type 2
References
- 1.Tsao B P. The genetics of human systemic lupus erythematosus. Trends Immunol 200324595–602. [DOI] [PubMed] [Google Scholar]
- 2.Vandenabeele P, Declerq W, Beyaert R, Fiers W Two tumor necrosis factor receptors: structure and function Trends Cell Biol. 1995;5:392–399. doi: 10.1016/s0962-8924(00)89088-1. [DOI] [PubMed] [Google Scholar]
- 3.Aggarwal B B. Signaling pathways of the TNF superfamily: a double‐edged sword. Nat Rev Immunol 20033745–756. [DOI] [PubMed] [Google Scholar]
- 4.Rudwaleit M, Tikly M, Khamashta M, Gibson K, Klinke J, Hughes G, Interethnic differences in the association of tumor necrosis factor promoter polymorphisms with systemic lupus erythematosus J Rheumatol. 1996;23:1725–1728. [PubMed] [Google Scholar]
- 5.Wilson A G, Gordon C, di Giovine F S, de Vries N, van de Putte L B, Emery P, A genetic association between systemic lupus erythematosus and tumor necrosis factor alpha Eur J Immunol. 1994;24:191–195. doi: 10.1002/eji.1830240130. [DOI] [PubMed] [Google Scholar]
- 6.Hajeer A H, Worthington J, Davies E J, Hillarby M C, Poulton K, Ollier W E. TNF microsatellite a2, b3 and d2 alleles are associated with systemic lupus erythematosus. Tissue Antigens 199749222–227. [DOI] [PubMed] [Google Scholar]
- 7.D'Alfonso S, Colombo G, Della Bella S, Scorza R, Momigliano‐Richiardi P. Association between polymorphisms in the TNF region and systemic lupus erythematosus in the Italian population. Tissue Antigens 199647551–555. [DOI] [PubMed] [Google Scholar]
- 8.Jacob C O, McDevitt H O. Tumor necrosis factor‐α in murine autoimmune ‘lupus' nephritis. Nature 1988331356–358. [DOI] [PubMed] [Google Scholar]
- 9.Theofilopoulos A N, Lawson B R. Tumor necrosis factor and other cytokines in murine lupus. Ann Rheum Dis 199958149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aringer M, Zimmermann C, Graninger W B, Petera P, Steiner G, Ulrich W. TNF is an essential mediator in lupus nephritis. Arthritis Rheum 2002463418–3419. [Google Scholar]
- 11.Gaffney P, Kearns G M, Shark K B, Ortmann W A, Selby S A, Malmgren M L. A genome‐wide search for susceptibility genes in human lupus erythematosus sib‐pair families. Proc Natl Acad Sci USA 19989514875–14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shai R, Quismorio F P, Jr, Li L, Kwon O, Morrison J, Wallace D J. Genome‐wide screen for systemic lupus erythematosus susceptibility genes in multiplex families. Hum Mol Genet 19998639–644. [DOI] [PubMed] [Google Scholar]
- 13.Komata T, Tsuchiya N, Matsushita M, Hagiwara K, Tokunaga K. Association of tumor necrosis factor receptor 2 (TNFR2) polymorphism with susceptibility to systemic lupus erythematosus. Tissue Antigens 199953527–533. [DOI] [PubMed] [Google Scholar]
- 14.Morita C, Horiuchi T, Tsukamoto H, Hatta N, Kikuchi Y, Arinobu Y. Association of tumor necrosis factor receptor type II polymorphism 196R with systemic lupus erythematosus in the Japanese. Molecular and functional analysis. Arthritis Rheum 2001442819–2827. [DOI] [PubMed] [Google Scholar]
- 15.Al‐Ansari A S, Ollier W E R, Villarreal J, Ordi J, The L ‐ S, Hajeer A H. Tumor necrosis factor receptor II (TNFRII) exon 6 polymorphism in systemic lupus erythematosus. Tissue Antigens 20005597–99. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi M, Hashimoto H, Akizuki M, Sasazuki T, Nishikimi N, Ouchi H. Lack of association between the Met196Arg polymorphism in the TNFR2 gene and autoimmune diseases accompanied by vasculitis including SLE in Japanese. Tissue Antigens 20015766–69. [DOI] [PubMed] [Google Scholar]
- 17.Lee E B, Yoo J E, Lee YJ Choi Y J, Park K S, Song Y W. Tumor necrosis factor receptor 2 polymorphism in systemic lupus erythematosus: no association with disease. Hum Immunol 2001621148–1152. [DOI] [PubMed] [Google Scholar]
- 18.Khoa P D, Sugiyama T, Yokochi T. Polymorphism of IL‐10 promoter and tumor necrosis factor receptor II in Vietnamese patients with systemic lupus erythematosus. Clin Rheumatol 20052411–13. [DOI] [PubMed] [Google Scholar]
- 19.Till A, Rosenstiel P, Krippner‐Heidenreich A, Mascheretti‐Croucher S, Croucher P J P, Schafer H. The Met‐196>Arg variation of human tumor necrosis factor receptor 2 (TNFR2) affects TNF‐α‐induced apoptosis by impaired NF‐κB signaling and target gene expression. J Biol Chem 20052805994–6004. [DOI] [PubMed] [Google Scholar]
- 20.Barton A, John S, Ollier W E, Silman A, Worthington J. Association between rheumatoid arthritis and polymorphism of tumor necrosis factor receptor II, but not tumor necrosis factor receptor I, in Caucasians. Arthritis Rheum 20014461–65. [DOI] [PubMed] [Google Scholar]
- 21.Dieude P, Petit E, Cailleau‐Moindrault S, Osorio J, Pierlot C, Martinez M. Association between tumor necrosis factor receptor II and familial, but not sporadic, rheumatoid arthritis: evidence for genetic heterogeneity. Arthritis Rheum 2002462039–2044. [DOI] [PubMed] [Google Scholar]
- 22.Pierik M, Vermeire S, Steen K V, Joossens S, Claessens G, Vlietinck R. Tumour necrosis factor‐alpha receptor 1 and 2 polymorphisms in inflammatory bowel disease and their association with response to infliximab. Aliment Pharmacol Ther 200420303–310. [DOI] [PubMed] [Google Scholar]
- 23.McDermott M F, Aksentijevich I, Galon J, McDermott E M, Ogunkolade B W, Centola M. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell 199997133–144. [DOI] [PubMed] [Google Scholar]
- 24.Horiuchi T, Tsukamoto H, Mitoma H, Miyagawa H, Tamimoto Y, Yoshizawa Sl. Novel mutations in TNFRSF1A in patients with typical tumor necrosis factor receptor‐associated periodic syndrome and with systemic lupus erythematosus in Japanese. Int J Mol Med 200414813–818. [PubMed] [Google Scholar]
- 25.Ida H, Kawasaki E, Miyashita T, Tanaka F, Kamachi M, Izumi Y. A novel mutation (T61I) in the gene encoding tumor necrosis factor receptor superfamily 1A (TNFRSF1A) in a Japanese patient with tumor necrosis factor receptor‐associated periodic syndrome (TRAPS) associated with systemic lupus erythematosus. Rheumatology 2004431292–1299. [DOI] [PubMed] [Google Scholar]
- 26.Tan E M, Cohen A S, Fries J F, Masi A T, McShane D J, Rothfield N F. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1982251272–1277. [DOI] [PubMed] [Google Scholar]
- 27.Hochberg M C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997401725. [DOI] [PubMed] [Google Scholar]
- 28.DerSimonian R, Laird N. Meta‐analysis in clinical trials. Control Clin Trials 19867177–188. [DOI] [PubMed] [Google Scholar]
- 29.Cochran W G. The combination of estimates from different experiments. Biometrics 195410101–129. [Google Scholar]
- 30.Whitehead A, Whitehead J. A general parametric approach to the meta‐analysis of randomized clinical trials. Stat Med 1991101665–1667. [DOI] [PubMed] [Google Scholar]
- 31.Begg C B, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994501088–1101. [PubMed] [Google Scholar]
- 32.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta‐analysis detected by a simple graphical test. BMJ 1997315629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douni E, Kollias G. A critical role of the p75 tumor necrosis factor receptor (p75 TNF‐R) in organ inflammation independent of TNF, lymphotoxin α or the p55 TNF‐R. J Exp Med 19981881343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grell M, Douni E, Wajant H, Lohden M, Clauss M, Maxeiner B. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 199583793–802. [DOI] [PubMed] [Google Scholar]
- 35.Molokhia M, McKeigue P. Risk for rheumatic disease in relation to ethnicity and admixture. Arthritis Res 20002115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuchiya N, Kawasaki A, Tsao B P, Komata T, Grossman J M, Tokunaga K. Analysis of the association of HLA‐DRB1, TNFalpha promoter and TNFR2 (TNFRSF1B) polymorphisms with SLE using transmission disequilibrium test. Genes Immun 20012317–322. [DOI] [PubMed] [Google Scholar]
- 37.Chadha S, Miller K, Farwell L Sacks S, Daly M J, Rioux J D. Haplotype analysis of tumour necrosis factor receptor genes in 1p36: no evidence for association with systemic lupus erythematosus. Eur J Hum Genet 20061469–78. [DOI] [PubMed] [Google Scholar]
- 38.Feldmann M, Maini R N. Anti‐TNF alpha therapy of rheumatoid arthritis: what have we learned? Annu Rev Immunol 200119163–196. [DOI] [PubMed] [Google Scholar]
- 39.Aringer M, Smolen J S. Complex cytokine effects in a complex autoimmune disease: tumor necrosis factor in systemic lupus erythematosus. Arthritis Res Ther 20035172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aringer M, Graninger W B, Steiner G, Smolen J S. Safety and efficacy of tumor necrosis factor α blockade in systemic lupus erythematosus. An open‐label study. Arthritis Rheum 2004503161–3169. [DOI] [PubMed] [Google Scholar]
- 41.Aringer M, Smolen J S. Tumour necrosis factor and other proinflammatory cytokines in systemic lupus erythematosus: a rationale for therapeutic intervention. Lupus 200413344–347. [DOI] [PubMed] [Google Scholar]
- 42.Horiuchi T, Morita C, Tsukamoto H, Mitoma H, Sawabe T, Harashima S. Increased expression of membrane TNF‐α on activated peripheral CD8+ T cells in systemic lupus erythematosus. Int J Mol Med 200617875–879. [PubMed] [Google Scholar]