In the past few years not many fields in medicine have been so profoundly transformed by seminal discoveries as the iron field. Key missing proteins in iron homeostasis have been characterised and their regulatory pathways or function dissected. Seemingly, the causative genes of the most important human diseases associated with deregulated iron metabolism and responsible for tissue iron overload have been identified. These giant steps forward have dramatically transformed the way we look at iron related diseases, their pathogenesis, diagnosis, and treatment. One of the best examples is the disorder known as haemochromatosis (HC) or hereditary haemochromatosis. This term was introduced to define the association of widespread tissue injury with massive tissue iron deposition1 and likely referred to a clinical entity named “bronze diabetes”2 and “cirrhose pigmentaire”3 first reported in France in the second half of the 19th century. After only one century the term was associated with an hereditary disease4 and linked to the major histocompatibility class I complex A3, on the short arm of chromosome 6.5,6 In 1996, the most prevalent HC gene, HFE, was cloned.7 However, once the HFE gene was identified, it appeared immediately clear that HFE mutations accounted for most but not all cases of HC.8 Since then, unprecedented progress in the field of iron genetics has led to the identification of new genes involved in iron metabolism whose mutations are responsible for cases of hereditary iron storage disorders.
The term haemochromatosis (HC) (synonymous for hereditary or idiopathic or primary haemochromatosis) defines an autosomal recessive disorder of iron metabolism characterised by tissue iron overload potentially leading to multiorgan disease, such as liver cirrhosis, endocrinopathy, and cardiomyopathy. The syndrome is the result of a genetically determined failure to stop iron from entering the circulatory pool when it is not needed. It is associated with pathogenic mutations of at least four HC genes (that is, HFE, TfR2, HJV, and HAMP) and it is likely due to a regulatory defect in iron homeostasis in the liver.9 Four basic features define HC and are characteristic of the classic disorder related to HFE C282Y homozygosity (the prototype for this subset and by far the most common form) and the rare disorders more recently attributed to loss of TfR2, HAMP, or HJV (table 1). These features include: hereditary, usually autosomal recessive, trait; iron overload initially involving the plasma compartment (reflected in increasing serum transferrin saturation); iron overload subsequently involving parenchymal cells (reflected in increasing serum ferritin) with the potential for organ damage and disease; unimpaired erythropoiesis and optimal response to phlebotomy.9 Established concepts regarding primary cause and molecular pathogenesis of HC have been more or less all challenged by recent new discoveries, particularly those pertaining to the genetic field. Based on this new information and on circumstantial evidence provided by recent human and animal studies, a unifying pathogenic model for HC is presented in this article.
Table 1 Haemochromatosis genes.
| Gene (symbol/location) | Gene product known or postulated function* | Pathogenic mutation(s)/ prevalence |
|---|---|---|
| Haemochromatosis gene (HFE/6p21.3) | Interaction with transferrin receptor 1 | One (C282Y) |
| Hepcidin regulator | Common | |
| Transferrin‐receptor 2 (TfR2/7q22) | Uptake of iron bound transferrin | Many |
| Hepcidin regulator | Rare | |
| Hepcidin antimicrobial peptide (HAMP /19q13.1) | Downregulation of iron efflux from macrophages, enterocytes, placenta | Many |
| Very rare | ||
| Hemojuvelin (HJV/1p21) | Hepcidin regulator | Many (G230V more prevalent) |
| Rare |
*There may be other as yet unknown functions related to iron overload while the known functions do not, at least at this time, always account for what is known about pathophysiology.
The pathogenic genes
HFE
HFE is a major histocompatibility class I‐like protein whose ancestral peptide binding groove is too narrow to allow classic antigen presentation10 while a possible non‐classic immunological activity has been recently proposed.11 It is incapable of binding iron.12 Interaction between HFE and the transferrin receptor, TfR1, which mediates transferrin bound iron uptake by most cells,12,13 has been fully documented although its biological effects are still uncertain. However, it is unclear whether the interaction of HFE with TfR1 is key for the pathogenesis of HC.14,15,16
The C282Y mutation (substitution of tyrosine for cysteine at position 282 due to a single base, 845G‐>A), the most common pathogenic mutation of HFE, is associated with disruption of a disulfide bond in HFE that is critical for its binding to β2 microglobulin.17 The latter interaction is necessary for the stabilisation (intracytoplasmic), transport, and expression of HFE on the cell surface and endosomal membranes where HFE interacts with TfR1. The H63D mutation, a common HFE mutation whose pathogenic significance is still uncertain, does not impair HFE‐TfR1 interaction. While the biological function of HFE is still unknown, circumstantial evidence indicate that it might be required for the synthesis of hepcidin, the iron hormone secreted by hepatocytes (see below).
Transferrin receptor 2 (TFR2)
In 1999, the gene for a second human transferrin receptor (TfR2) was cloned.18 Unlike TfR1, the new receptor was found to be highly expressed in the liver and it was not regulated by intracellular iron status.19 TfR2 mediates the uptake of transferrin bound iron by hepatocytes,18 possibly through the mechanism of receptor mediated endocytosis similar to that described for TfR1, but its in vitro affinity for transferrin is 25–30‐fold lower than that of TfR1.20 Yet, TfR2 mediated transferrin iron uptake may be of importance in hepatocytes, which express a low number of TfR1. The biological role and function of TFR2 remain unknown, but recent studies suggests a role for TfR2 in hepcidin synthesis in the liver (see below). In fact, its putative role in hepatocyte uptake of iron18 is difficult to reconcile with the HC phenotype observed in humans with pathogenic TfR2 mutations21 and in TfR2 knockout mice.22 TfR2 does not seem to interact with HFE,20 but its persistent hepatic expression during iron overload might conceivably reflect a contribution to the modulation of hepcidin synthesis in this setting (see below).
Hemojuvelin (HJV)
Hemojuvelin (also called HFE2, or repulsive guidance molecule C (RgmC)) is transcribed from a gene of 4265 bp into a full length transcript with five spliced isoforms.23 Analyses of hemojuvelin in human tissues detect substantial expression in adult and fetal liver, heart, and skeletal muscle.23 The putative full length protein is 426 amino acids with a large RGM motif, homologous to repulsive guidance molecules involved in neuronal cells migration; it contains a C terminal putative transmembrane domain characteristic of a glycosylphosphatidylinositol linked membrane anchor (GPI anchor). Removal of the GPI anchor or proteolysis would be expected to generate a soluble form of hemojuvelin, suggesting that it can be present in either a soluble or a cell associated form. The function of hemojuvelin is presently unknown. However, hepcidin levels are depressed in individuals with HJV mutations,23 and in HJV knockout mice,24 and a recent study in vitro indicated that HJV is a transcriptional regulator of hepcidin25 (see below). In this study, cellular hemojuvelin positively regulated hepcidin mRNA expression, and recombinant soluble hemojuvelin suppressed hepcidin mRNA expression in primary human hepatocytes in a log linear dose dependent manner, suggesting binding competition between soluble and cell associated hemojuvelin.
Hepcidin (HAMP)
Hepcidin, the long awaited iron hormone, is an antimicrobial peptide produced by hepatocytes in response to inflammatory stimuli and iron.26,27,28 It is the product of the HAMP gene, consisting of three exons and two introns located on chromosomes 7 and 19 in mouse and humans, respectively. Humans and rats have a single HAMP gene 28, whereas two functional genes, Hamp 1 and 2 are present in the mouse genome.29 Hepcidin mRNA is nearly confined to the liver. The transcript encodes a precursor protein of 84 amino acids, including a putative 24 amino acid leader peptide, while the circulating forms consist of only the C terminal portion (20 and 25 amino acid peptides). In solution, the small cysteine‐rich hepcidin peptides form a distorted beta sheet with an unusual vicinal disulphide bridge found at the turn of the hairpin, which is probably of functional significance.30 Due to significant antibacterial and antifungal activities of the C terminal peptide, hepcidin has been classified as a member of the cysteine‐rich, cationic, antimicrobial peptides, including the thionins and defensins.
Evidence from transgenic mouse models indicates that hepcidin is the principal downregulator of the transport of iron across the small intestine and the placenta, and its release from macrophages. Transgenic animals overexpressing hepcidin die perinatally due to severe iron deficiency anaemia occurring in the context of reticuloendothelial cell iron overload.29 In vivo injection of hepcidin into mice significantly reduced mucosal iron uptake and transfer to the carcass, independently of iron status or presence of HFE,31 or induces hypoferraemia in humans.32 The present view is that hepcidin downregulates iron efflux from the intestine and macrophages by interacting with the main iron export protein in mammals, ferroportin (FPN). In fact, it has been recently shown that hepcidin binds to FPN in cultured cells stably expressing FPN and, following complex internalisation, leads to FPN degradation.33 Moreover, hepcidin is highly concentrated in organs expressing FPN.32 This implies decreased FPN expression and reduced iron egress from cells such as enterocytes and macrophages whenever circulating hepcidin levels are high—namely, inflammation and iron overload. In fact, hepcidin functions as an antimicrobial peptide, and its production is readily stimulated by lipopolysaccharide 28 or turpentine34 both in vivo and in vitro. This stimulation is indirect and appears to be mainly mediated by the inflammatory cytokine interleukin 6.35,36,37 It is presently controversial whether HFE, the HC gene product, is required for hepcidin activation in response to inflammatory stimuli.37,38,39,40 Due to its sensitivity to inflammatory stimuli and owing to its effect on iron egress from macrophages and enterocytes, hepcidin is likely responsible, along with its cellular counterpart FPN, for iron perturbations occurring during chronic inflammatory disorders and know as anaemia of inflammation or anaemia of chronic disease.41
As to the regulatory role of iron on hepcidin synthesis, a correlation between hepcidin expression and hepatic iron stores or serum ferritin has been reported previously in humans.35,42,43 It may be that serum iron or transferrin saturation are the signals for hepcidin upregulation but the details of this stimulation are still obscure. In fact, exposure of cultured murine and human hepatocytes to ferric iron28 or iron saturated transferrin35 does not increase hepcidin mRNA and may even reduce it. This led to suggestions that in vivo iron sensing for hepcidin upregulation may take place in other cell types, possibly Kupffer cells,41 but recent studies have challenged this hypothesis.40,44 A study now points to Smad4, a transcription factor which might be directly responsible for hepcidin regulation in hepatocytes in response to iron and inflammatory stimuli and whose genetic disruption leads to systemic iron overload in mice.45
The fact that mice with genetic disruption of the transcription factors upstream stimulatory factor 2 (USF2) or C‐EBPa, both required for hepcidin transcriptional control, have an haemochromatotic phenotype46,47 and humans lacking hepcidin have a severe form of HC48 now places hepcidin at the centre of the pathogenesis of HC (see below).
A unifying pathogenic model for haemochromatosis
The first biochemical manifestation of HC is an increase in transferrin saturation which reflects an uncontrolled influx of iron into the bloodstream from enterocytes and macrophages. Duodenal transfer of iron to plasma is inappropriately high for body iron stores,49 suggesting downregulation failure in HC. Phlebotomy normally triggers sharp transient increases in absorption (from 1–2 mg/day to 5 mg/day), mainly to ensure bone marrow supplies, but in HC this response is exaggerated (8–10 mg/day) and the rate remains high for years.50 The end result is intestinal absorption of iron that generally exceeds loss by approximately 3 mg/day in HC.51 While the only way that total body iron can be increased is through increased intestinal iron absorption, macrophages (normally a much more important source of plasma iron than either enterocytes or hepatocytes)52 are also important in the pathogenesis of HC. They are invariably iron poor in HC and seem to release more iron or to retain less transferrin bound iron than their normal counterparts.9
Historically, the intestine has been seen as the primary site of the defect in HC and studies showing that HFE is normally expressed in intestinal crypts reinforced this idea leading to the development of a specific pathogenic model.9 This model attributed the relative iron deficiency of mature absorptive HC enterocytes and increased intestinal iron absorption to an abnormal interaction between TfR1 and mutant HFE in intestinal crypt cells. The presence in this model of C282Y mutant HFE which is unable to interact with TfR1, leads to iron deficient crypt cells, which give rise to iron deficient daughter cells. These cells are “programmed” to react to iron starvation by hyperactively and persistently absorbing iron from the intestinal lumen and transferring virtually all of it into the bloodstream, regardless of actual erythropoietic needs. More recently, however, this model has been challenged with the discovery of hepcidin and its central involvement in iron homeostasis. Consequently, attention has moved to the liver as the primary site of the defect in HC.
The progressive expansion of the plasma iron pool in HC, which occurs at a much faster rate in “juvenile” forms of the disorder (HJV and HAMP related HC) compared with late onset forms (HFE and TfR2‐related HC), is likely the result of increased transfer of iron to the blood compartment from enterocytes (that is, increased intestinal absorption) and from reticuloendothelial macrophages.9 As mentioned above, the main regulator of iron efflux from enterocytes and macrophages in humans is hepcidin. In HFE, TfR2, and HJV related HC, hepatic expression or serum/urine levels of this peptide are inappropriately low.42,43,53,54 Its expression in the liver is also significantly impaired in HFE, TfR2, and HJV knockout mice24,55,56,57 and hepatic deposition of iron in HFE‐KO animals can be prevented by hepcidin overexpression.58 These findings suggests a unifying pathogenic model for all forms of HC in which HFE, TfR2, and HJV are all independent but complimentary regulators of hepcidin synthesis in the liver (fig 1).
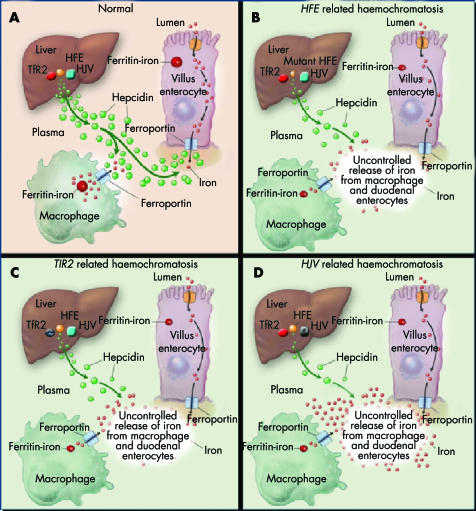
Figure 1 A unifying pathogenic model for hereditary haemochromatosis. In this model, HFE, TfR2, and HJV are considered independent but complementary modulators of hepatic synthesis of hepcidin, which downregulates the rate at which intestinal cells and macrophages release iron into the bloodstream. Loss of one of the regulatory proteins will result in an appreciable increase in iron influx into the bloodstream but residual hepcidin activity will be sustained by the other regulators. Specific genetic loss will have a different effect on circulatory iron overload (and on the phenotypic severity of the resulting syndrome) depending on the importance of the involved gene in hepcidin regulation. Therefore, the result is a mild “adult” haemochromatosis phenotype in HFE and TfR2 related haemochromatosis (B, C) and a more severe juvenile phenotype when the loss involves the “major” hepcidin regulator, HJV (D) (see text for details). Modified from Pietrangelo9 with permission.
Hypothetically, these three proteins may be important for sensing circulatory iron and turning on hepcidin gene transcription, albeit with different mechanisms and functional impact: HFE might have a role in endosomes and/or plasma membranes iron traffic even independently of TfR1; TfR2, highly expressed in hepatocytes even during iron overload, might signal transferrin bound or unbound iron; soluble and cell associated hemojuvelin might reciprocally regulate hepcidin expression in response to changes in extracellular iron concentration: soluble HJV might even signal the iron status of peripheral tissues, such as skeletal muscles where HJV also seems to be expressed. When all three proteins function correctly (and the HAMP gene that encodes hepcidin is normal), the amount of iron transferred into the blood will be appropriate to body needs, and excessive iron deposition in tissues will be avoided (fig 1A).
The relative contributions of the three genes to this modulatory process may be different, with a more substantial role assigned to HJV based on the more severe iron overload phenotype associated with HJV mutations. Loss of one of the minor regulatory proteins (HFE or TfR2 related HC) will result in an appreciable increase in iron influx into the bloodstream but residual hepcidin activity will be sustained by the second minor regulator and the major regulator, HJV gene (fig 1B, C). The result is a mild “adult” HC phenotype, with gradual plasma iron loading and gradual accumulation of iron in tissues. Loss of the “major” hepcidin regulator, HJV, will produce a more dramatic effect on iron into the bloodstream and result in a more severe, “juvenile”, HC. Combined loss of HFE and TfR2 (HFE+TfR2 related HC) would theoretically result in much more rapid and substantial increases in plasma iron and, consequently, greater iron overload in tissues: in short, a more severe juvenile phenotype, not unlike that produced by loss of HJV. In fact, a recent study has described patients with severe juvenile HC phenotype associated with combined mutations of HFE and TfR2.59 Finally, complete loss of hepcidin (HAMP related HH), in spite of normal HFE, TfR2, and HJV, will inevitably lead to massive uncontrolled release of iron into the circulation and severe HC phenotype.
Conclusions
For much of the twentieth century, HC was regarded as a clinically and genetically unique entity, albeit one with phenotypic variability. We now know that other iron gene mutations can be associated with more or less similar forms of iron overload, leading to tissue deposits of iron with different patterns and organ damaging potentials. Due to these advances, our concept of HC has been changing, evolving, stretching, and twisting to accommodate an increasingly rapid and rich succession of new discoveries, in particular those of the genetics era. This has also made some confusion. For instance, the OMIM database now lists the disease associated with FPN mutation, the ferroportin disease,60 a common cause of hereditary hyperferritinaemia, as type 4 HC.61 This is incorrect both on pathophysiological and clinical grounds, but it is also misleading for physicians, as diagnostic and therapeutic strategies may be different in the FPN disease compared with classic HC. The truth is that in FPN disease the basic pathological defect is the inappropriate retention of iron by macrophages and other cell type—namely, the defect lies at the other end of the hepcidin‐ferroportin axis.
The pathogenic model for HC presented in this article needs validation, and ongoing studies, particularly in transgenic animal models, will surely offer important clues. It clearly has the advantage of unifying within a unique syndromic entity a number of disorders that are, at first sight, distinct from a genetic point of view; the common pathogenic basis, instead, can explain common clinical manifestations, diagnostic strategy, and therapeutic approach. With the exception of HAMP, the precise functions of these genes remain to be defined, and their classic association with either juvenile or adult onset disease provides a valuable index of their importance in normal iron metabolism. Most notably, non‐HFE HC genes, while responsible for rarer cases of HC compared with HFE, are much more central than HFE in human iron homeostasis and understanding their function will greatly advance our comprehension of iron trafficking in health and disease.
Footnotes
Conflict of interest: None declared.
References
- 1.von Recklinghausen F D. Uber Haemochromatose. Taggeblatt der (62) Versammlung deutscher. Naturforscher and Aerzte in Heidelberg 1889324–325.
- 2.Trousseau A. Glycosurie; diabete sucre. Clinique Med de l'Hotel de Paris 18652663–698. [Google Scholar]
- 3.Troisier M. Diabète sucré. Bull Soc Anatomique Paris 187144231–235. [Google Scholar]
- 4.Sheldon J.Haemochromatosis. London: Oxford University Press, 1935
- 5.Simon M, Pawlotsky Y, Bourel M.et al Idiopathic haemochromatosis associated with HL‐A 3 tissular antigen] Nouv Presse Med 197541432. [PubMed] [Google Scholar]
- 6.Simon M, Le Mignon L, Fauchet R.et al A study of 609 HLA haplotypes marking for the hemochromatosis gene: (1) mapping of the gene near the HLA‐A locus and characters required to define a heterozygous population and (2) hypothesis concerning the underlying cause of hemochromatosis‐HLA association. Am J Hum Genet 19874189–105. [PMC free article] [PubMed] [Google Scholar]
- 7.Feder J N, Gnirke A, Thomas W.et al A novel MHC class I‐like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 199613399–408. [DOI] [PubMed] [Google Scholar]
- 8.Pietrangelo A. Hemochromatosis 1998: is one gene enough? J Hepatol 199829502–509. [DOI] [PubMed] [Google Scholar]
- 9.Pietrangelo A. Hereditary hemochromatosis—a new look at an old disease. N Engl J Med 20043502383–2397. [DOI] [PubMed] [Google Scholar]
- 10.Lebron J A, Bennett M J, Vaughn D E.et al Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell 199893111–123. [DOI] [PubMed] [Google Scholar]
- 11.Rohrlich P S, Fazilleau N, Ginhoux F.et al Direct recognition by alphabeta cytolytic T cells of Hfe, a MHC class Ib molecule without antigen‐presenting function. Proc Natl Acad Sci U S A 200510212855–12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feder J N, Penny D M, Irrinki A.et al The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci U S A 1998951472–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross C N, Irrinki A, Feder J N.et al Co‐trafficking of HFE, a nonclassical major histocompatibility complex class I protein, with the transferrin receptor implies a role in intracellular iron regulation. J Biol Chem 199827322068–22074. [DOI] [PubMed] [Google Scholar]
- 14.Zhang A S, Davies P S, Carlson H L.et al Mechanisms of HFE‐induced regulation of iron homeostasis: Insights from the W81A HFE mutation. Proc Natl Acad Sci U S A 20031009500–9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies P S, Zhang A S, Anderson E L.et al Evidence for the interaction of the hereditary haemochromatosis protein, HFE, with the transferrin receptor in endocytic compartments. Biochem J 2003373145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson H, Zhang A S, Fleming W H.et al The hereditary hemochromatosis protein, HFE, lowers intracellular iron levels independently of transferrin receptor 1 in TRVb cells. Blood 20051052564–2570. [DOI] [PubMed] [Google Scholar]
- 17.Waheed A, Parkkila S, Zhou X Y.et al Hereditary hemochromatosis: Effects of C282Y and H63D mutations on association with beta(2)‐microglobulin, intracellular processing, and cell surface expression of the HFE protein in COS‐7 cells. Proc Natl Acad Sci U S A 19979412384–12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawabata H, Yang R, Hirama T.et al Molecular cloning of transferrin receptor 2. A new member of the transferrin receptor‐like family. J Biol Chem 199927420826–20832. [DOI] [PubMed] [Google Scholar]
- 19.Kawabata H, Germain R S, Vuong P T.et al Transferrin receptor 2‐alpha supports cell growth both in iron‐chelated cultured cells and in vivo. J Biol Chem 200027516618–16625. [DOI] [PubMed] [Google Scholar]
- 20.West A P, Jr, Bennett M J, Sellers V M.et al Comparison of the interactions of transferrin receptor and transferrin receptor 2 with transferrin and the hereditary hemochromatosis protein HFE. J Biol Chem 200027538135–38138. [DOI] [PubMed] [Google Scholar]
- 21.Camaschella C, Roetto A, Cali A.et al The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet 20002514–15. [DOI] [PubMed] [Google Scholar]
- 22.Fleming R E, Ahmann J R, Migas M C.et al Targeted mutagenesis of the murine transferrin receptor‐2 gene produces hemochromatosis. Proc Natl Acad Sci U S A 20029910653–10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papanikolaou G, Samuels M E, Ludwig E H.et al Mutations in HFE2 cause iron overload in chromosome 1q‐linked juvenile hemochromatosis. Nat Genet 20043677–82. [DOI] [PubMed] [Google Scholar]
- 24.Huang F W, Pinkus J L, Pinkus G S.et al A mouse model of juvenile hemochromatosis. J Clin Invest 20051152187–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin L, Goldberg Y P, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell‐associated hemojuvelin. Blood 20051062884–2889. [DOI] [PubMed] [Google Scholar]
- 26.Krause A, Neitz S, Magert H J.et al LEAP‐1, a novel highly disulfide‐bonded human peptide, exhibits antimicrobial activity. FEBS Lett 2000480147–150. [DOI] [PubMed] [Google Scholar]
- 27.Park C H, Valore E V, Waring A J.et al Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem 20012767806–7810. [DOI] [PubMed] [Google Scholar]
- 28.Pigeon C, Ilyin G, Courselaud B.et al A new mouse liver‐specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem 20012767811–7819. [DOI] [PubMed] [Google Scholar]
- 29.Nicolas G, Bennoun M, Porteu A.et al Severe iron deficiency anaemia in transgenic mice expressing liver hepcidin. Proc Natl Acad Sci U S A 2002994596–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter H N, Fulton D B, Ganz T.et al The solution structure of human hepcidin, a peptide‐hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J Biol Chem 200227737597–37603. [DOI] [PubMed] [Google Scholar]
- 31.Laftah A H, Ramesh B, Simpson R J.et al Effect of hepcidin on intestinal iron absorption in mice. Blood 20041033940–3944. [DOI] [PubMed] [Google Scholar]
- 32.Rivera S, Nemeth E, Gabayan V.et al Synthetic hepcidin causes rapid dose‐dependent hypoferremia and is concentrated in ferroportin‐containing organs. Blood 20051062196–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemeth E, Tuttle M S, Powelson J.et al Hepcidin regulates iron efflux by binding to ferroportin and inducing its internalization. Science 20043062090–2093. [DOI] [PubMed] [Google Scholar]
- 34.Nicolas G, Chauvet C, Viatte L.et al The gene encoding the iron regulatory peptide hepcidin is regulated by anaemia, hypoxia, and inflammation. J Clin Invest 20021101037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemeth E, Valore E V, Territo M.et al Hepcidin, a putative mediator of anemia of inflammation, is a type II acute‐phase protein. Blood 20031012461–2463. [DOI] [PubMed] [Google Scholar]
- 36.Nemeth E, Rivera S, Gabayan V.et al IL‐6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest 20041131271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee P, Peng H, Gelbart T.et al The IL‐6‐ and lipopolysaccharide‐induced transcription of hepcidin in HFE‐, transferrin receptor 2‐, and beta2‐microglobulin‐deficient hepatocytes. Proc Natl Acad Sci U S A 20041019263–9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy C N, Custodio A O, de Graaf J.et al An Hfe‐dependent pathway mediates hyposideremia in response to lipopolysaccharide‐induced inflammation in mice. Nat Genet 200436481–485. [DOI] [PubMed] [Google Scholar]
- 39.Frazer D M, Wilkins S J, Millard K N.et al Increased hepcidin expression and hypoferraemia associated with an acute phase response are not affected by inactivation of HFE. Br J Haematol 2004126434–436. [DOI] [PubMed] [Google Scholar]
- 40.Montosi G, Corradini E, Garuti C.et al Kupffer cells and macrophages are not required for hepatic hepcidin activation during iron overload. Hepatology 200541545–552. [DOI] [PubMed] [Google Scholar]
- 41.Ganz T. Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood 2003102783–788. [DOI] [PubMed] [Google Scholar]
- 42.Bridle K R, Frazer D M, Wilkins S J.et al Disrupted hepcidin regulation in HFE‐associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet 2003361669–673. [DOI] [PubMed] [Google Scholar]
- 43.Gehrke S G, Kulaksiz H, Herrmann T.et al Expression of hepcidin in hereditary hemochromatosis: evidence for a regulation in response to serum transferrin saturation and non‐transferrin‐bound iron. Blood 2003102371–376. [DOI] [PubMed] [Google Scholar]
- 44.Lou D Q, Lesbordes J C, Nicolas G.et al Iron‐ and inflammation‐induced hepcidin gene expression in mice is not mediated by Kupffer cells in vivo. Hepatology 2005411056–1064. [DOI] [PubMed] [Google Scholar]
- 45.Wang R H, Li C.et al A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab 200522399–2409. [DOI] [PubMed] [Google Scholar]
- 46.Nicolas G, Bennoun M, Devaux I.et al Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc Natl Acad Sci U S A 2001988780–8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Courselaud B, Pigeon C, Inoue Y.et al C/EBPalpha regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Cross‐talk between C/EBP pathway and iron metabolism. J Biol Chem 200227741163–41170. [DOI] [PubMed] [Google Scholar]
- 48.Roetto A, Papanikolaou G, Politou M.et al Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet 20033321–22. [DOI] [PubMed] [Google Scholar]
- 49.Lynch S R, Skikne B S, Cook J D. Food iron absorption in idiopathic hemochromatosis. Blood 1989742187–2193. [PubMed] [Google Scholar]
- 50.Smith P M, Godfrey B E, Williams R. Iron absorption in idiopathic haemochromatosis and its measurement using a whole‐body counter. Clin Sci 196937519–531. [PubMed] [Google Scholar]
- 51.Crosby W H. The control of iron balance by the intestinal mucosa. Blood 196322441–449. [PubMed] [Google Scholar]
- 52.Bothwell T H. Overview and mechanisms of iron regulation. Nutr Rev 199553237–245. [DOI] [PubMed] [Google Scholar]
- 53.Nemeth E, Roetto A, Garozzo G.et al Hepcidin is decreased in TFR2‐hemochromatosis. Blood 20041051803–1806. [DOI] [PubMed] [Google Scholar]
- 54.Papanikolaou G, Tzilianos M, Christakis J I.et al Hepcidin in iron overload disorders. Blood 20051054103–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muckenthaler M, Roy C N, Custodio A O.et al Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat Genet 200334102–107. [DOI] [PubMed] [Google Scholar]
- 56.Kawabata H, Fleming R E, Gui D.et al Expression of hepcidin is down‐regulated in TfR2 mutant mice manifesting a phenotype of hereditary hemochromatosis. Blood 2005105376–381. [DOI] [PubMed] [Google Scholar]
- 57.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest 20051152180–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicolas G, Viatte L, Lou D Q.et al Constitutive hepcidin expression prevents iron overload in a mouse model of hemochromatosis. Nat Genet 20033497–101. [DOI] [PubMed] [Google Scholar]
- 59.Pietrangelo A, Caleffi A, Henrion J.et al Juvenile hemochromatosis associated with pathogenic mutations of adult hemochromatosis genes. Gastroenterology 2005128470–479. [DOI] [PubMed] [Google Scholar]
- 60.Pietrangelo A. The ferroportin disease. Blood Cells Mol Dis 200432131–138. [DOI] [PubMed] [Google Scholar]
- 61.OMIM Online Mendelian Inheritance in Man. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = OMIM (accessed 16 January 2006)