Recent advances in basic and clinical science have driven epigenetics to the forefront of cancer research. Together with genetic changes, the disruption of epigenetic mechanisms is now established as a hallmark of cancer in humans. Colorectal cancer, long a classic model for the genetic basis of cancer, is now providing researchers with the opportunity to view epigenetic events in the context of neoplasia in humans. Knowledge of the heritable changes in gene expression that result from epigenetic events is of increasing relevance to clinical practice, particularly in terms of diagnostic and prognostic molecular markers, as well as novel therapeutic targets.
Background
Colorectal cancer, for many years a prototypic model for the genetic basis of cancer, is now increasingly cited as an exemplar of the role of epigenetic changes in tumorigenesis. In part, this is because colorectal neoplasia provides a wide range of accessible lesions, from aberrant crypt foci to carcinoma. It also serves as a poster child for epigenetic change because of the possible role that DNA methylation has in the initiation and progression of this disease. For both these reasons, it provides an excellent opportunity to understand how epigenetics and genetics collude to produce malignancy.
This review will provide a broad overview of common epigenetic processes as they occur in the normal cell and in the cancer cell, and will highlight recent findings in the epigenetics of colorectal neoplasia. It will briefly discuss the clinical implications of epigenetic changes, in terms of both the identification of disease predisposition and the therapeutic opportunities that a better understanding of these changes may provide. The term epigenetics, while variously defined,1 will be used in this review to describe those heritable changes in gene function that do not entail a change in DNA sequence.2 Table 1 shows the key historical milestones in the research on cancer epigenetics.
Table 1 Milestones in cancer genetics and epigenetics, in relation to the clinical management of colorectal cancer.
| Decade | Genetics3 | Epigenetics4 | Clinical management |
|---|---|---|---|
| 1940 | Proposed existence of cancer stem cells | C H Waddington coins the terms epigenetics and epigenome | Dukes' staging 1932 |
| 1950 | Two‐hit hypotheses | No‐touch technique for colon surgery | |
| 1960 | Chromosomal translocations | Description of X chromosome inactivation | Flexible sigmoidoscopy and colonoscopy |
| 1970 | First human oncogene | 5‐Methylcytosine as a mechanism of gene control in mammals | Therapeutic polypectomy |
| Tumour‐suppressor genes | |||
| 1980 | Oncogene cooperation | Global hypomethylation of cancer cells | Total mesorectal excision for rectal cancer |
| Hypermethylation of the RB gene | |||
| Chromatin modification linked to DNA methylation | |||
| 1990 | Genetic basis for cancer predisposition | Invention of bisulphite technique | Adjuvant chemotherapy introduced |
| First imprinted genes identified | |||
| DNA methylation involved in genomic imprinting | |||
| Loss of imprinting in cancer | |||
| 2000 | Expression profiling of cancer genes | Drug trials on humans target the epigenetic modifications in DNA | Biological and targeted treatments |
Epigenetic events in normal human cells
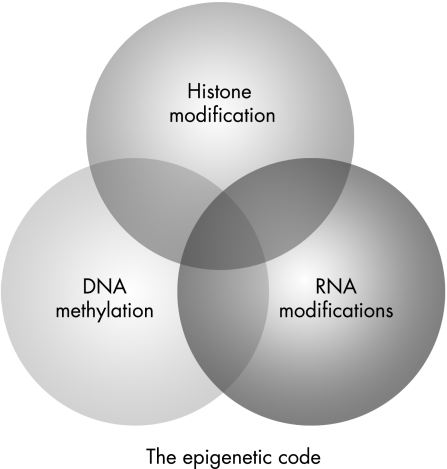
Although the nucleotide sequence of the human genome has long been recognised as the blueprint from which all macromolecular structures are derived, it has also been apparent that other factors within the material of the cell nucleus can also determine gene expression, and hence the structure and function of the cells and tissues that they form. As these factors are heritable, in that they can be passed from cell to cell, they have been referred to as the “epigenetic code”. Ingeniously, this code marks the DNA sequence in ways that do not involve modification of the DNA sequence itself. The repertoire of epigenetic marks includes modifications to histone proteins, methylation of DNA and the phenomenon of RNA interference as described in plants5 and fungi,6 and possibly in mammalian cells7 (fig 1). Whereas the genetic code provides the blueprint for all cellular elements, the epigenetic code controls elaboration of that blueprint, including the particular suites of “luxury” proteins that set apart one differentiated cell from the next.8 In effect, this means that individuals have a single genome but many “epigenomes”.
Figure 1 Schematic of the inter‐related cellular processes that constitute the epigenetic code. RNA modification includes the roles of RNA interference and microRNA in changing gene expression.
Histone modifications and the histone code
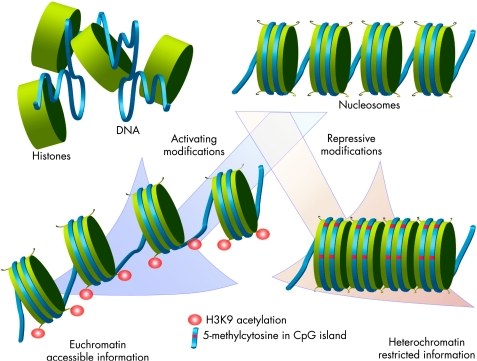
Much of the epigenetic code is carried through chemical modifications of individual amino acids on the tails of proteins called histones. The basic unit of human chromatin, the nucleosome, consists of a 146‐bp loop of DNA wrapped over an octamer of core histones (H2A/H2B dimers and H3/H4 tetramer). Covalent modifications of histone proteins can change densely compacted, inactive heterochromatin to the open and active configuration of euchromatin, and vice versa (fig 2). These modifications, which include acetylation, methylation, phosphorylation and ubiquitinylation, are reversible events that occur at the N‐terminal and C‐terminal domains of all core histones. Each of these modifications can be subjected to further variations that can change its function. For instance, methylation of arginine can involve the addition of 1, 2 or 3 methyl groups, each conferring subtly different functional consequences. The histone modifications are made possible by several families of enzymes, including histone acetyltransferases (HATs), histone deacetylases (HDACs) and histone methyltransferases. The balanced activity of these enzymes and related proteins is pivotal to normal cellular function, and any change in their function causes diverse and often profound disorders.9
Figure 2 A model of epigenetic modifications and their effect on transcription. The nucleosome is assembled from DNA and histones, and the chemical modification of histone tails induces conformational changes that can cause either activation or repression of transcription. Repressive modifications include methylation of lysine 9 residue of histone 3 (H3K9), lysine 27 residue of histone 3 (H3K27) and lysine 20 residue of histone 4 (H4K20) in association with DNA methylation. Changes such as acetylation at H3K9 (shown) are associated with open chromatin formation (euchromatin).
Generally, the active chromatin structure corresponding to increased transcriptional activity is associated with increased histone acetylation (fig 2). HATs such as P300 and cellobiose phosphorylase (CBP) catalyse acetylation of lysine (lys) residues on histones H3 and H4.10 Acting antagonistically to HATs, HDACs produce transcriptional repression by a complicated mechanism that involves interaction with de novo methyltransferases11,12 and methyl‐CpG‐binding proteins.13,14 Likewise, methylation and phosphorylation of histones are also involved in regulation of the activation state of chromatin.9 Methylation at lys 4 and lys 14 as well as phosphorylation of serine 10 on histone H3 have all been linked to gene activation, whereas methylation of lys 9 on histone H3 has been associated with gene silencing.15 Taken together, this is the pattern of histone modifications that constitutes the “histone code” and that complements the primary DNA sequence in defining transcription states.16,17
DNA methylation
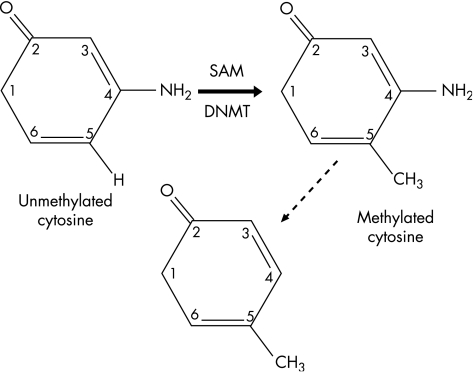
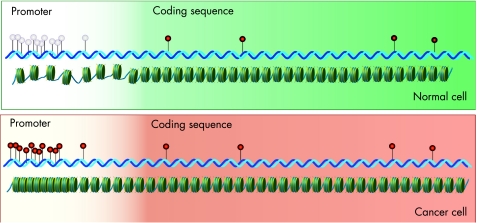
Among all mechanisms of epigenetic modification, enzymatic modification of cytosine bases in DNA to form 5‐methylcytosine is perhaps the most widely studied and the best understood (fig 3). In the mammalian genome, methylation of cytosine residues occurs most commonly at the 5′‐CG‐3′ dinucleotides (also termed CpG dinucleotides) and occasionally at 5′‐CA‐3′ or 5′‐CT‐3′ residues.18 The resultant base, 5‐methylcytosine, is relatively unstable, and prone to spontaneous deamination to form thymine (fig 3); in this way, DNA methylation can be seen as an endogenous mutagen. About 70% of all CpG dinucleotides in the human genome are heavily methylated,19 and the remainder are typically seen in CpG‐rich regions of ⩾200 bp that span the promoters and sometimes the first exons of genes. These regions, known as CpG islands,20 are found in association with about 60% of all human genes. Apparently, the configuration of CpG methylation in the genome produces a recognisable pattern of non‐methylated CpG islands scattered on a background of DNA that is methylated at low density (fig 4). These genomic patterns of CpG methylation are reprogrammed in the early embryo, but maintained with considerable fidelity thereafter, and are of great functional relevance in normal cells. Patterns of methylation cooperate in the regulation of the differential expression of genes, such as the silencing of genes on the inactive X chromosome, and the production of age‐related and tissue‐specific gene expression.21
Figure 3 Methylation of cytosine residues and its consequences. De novo methyltransferase (DNMT) catalyses the methylation at position 5 of cytosine, using S‐adenosylmethionine (SAM) as the methyl donor. Spontaneous deamination of 5‐methylcytosine results in its conversion to thymine, an event which is in itself mutagenic, and which has caused progressive depletion of cytosine bases from the eukaryotic genome throughout evolution.
Figure 4 Organisation and consequences of CpG methylation in normal and cancer cells. The upper panel shows a normal cell, in which a cluster of CG dinucleotides (CpG island) remains unmethylated (pale pins), where as scattered cytosines elsewhere are methylated (red pins). In the absence of methylation of this CpG island, DNA in the promoter region remains accessible to transcription factors, and the gene is expressed. In the lower panel, a cancer cell shows characteristic CpG island methylation, with concomitant compact chromatin structure in the promoter region, causing silencing of gene expression.
Genomic imprinting is a variant of the process of DNA methylation that allows monoallelic gene expression in a parent‐of‐origin‐specific manner. About 80 imprinted loci have now been described, and they are typically characterised by tissue‐specific and stage‐specific patterns of expression.22 This is clearly a key epigenetic process, and one which has been extensively reviewed previously.21,23
Patterns of genomic methylation are of vital importance in both health and disease, and an understanding of the mechanism by which methylation leads to transcriptional silencing is developing rapidly.24 However, less is known about the factors that determine the positioning and the de novo development of these epigenetic marks within the genome. There has been considerable interest in the role of transposable elements in inducing methylation events, but limited data support this contention in higher organisms.25 De novo development of DNA methylation is suggested to be the result of loss of transcription from the gene itself.26
Epigenetic events and mechanisms in colorectal carcinogenesis
Given the powerful role of epigenetic changes in changing gene expression, as well as their close relationship with development, it is not surprising that cancer cells show a marked change in the configuration of epigenetic marks on their genome.8,27,28 Historically, much work has focused on the changes in DNA methylation patterns seen in cancer cells both in terms of global hypomethylation and focal hypermethylation at CpG islands. More recently, work has begun to elucidate the changes to chromatin structure seen in this disease. Both of these will be reviewed briefly.
DNA hypomethylation
In the late 1970s, it was shown that the genome of tumour cells showed a progressive and global decrease in the number of cytosine bases that had been methylated to form 5‐methylcytosine.28,29,30,31 This phenomenon, usually referred to as DNA hypomethylation, is a typical finding in all neoplasms, both benign and malignant.4 In the particular case of colorectal neoplasia, global hypomethylation has been found in lesions across the neoplastic spectrum, from adenomatous polyps to carcinomas,32 as well as in hyperplastic polyps.33
Hypomethylation has been linked to several mechanisms that could drive neoplastic progression. In contrast with normal cells, hypomethylation in tumour cells typically occurs at the repetitive sequences residing in satellite or pericentromeric regions. This pattern of hypomethylation may make chromosomes more susceptible to breakage, and therefore lead directly to genomic instability.34,35 Hypomethylation can also result in the reactivation of previously silenced retrotransposons, leading to the disruption of normal gene structure and function.36,37 Further, the activity of transposable elements may govern the methylation state of their neighbouring genes through the phenomenon of transcriptional interference, which has been observed in maize and wheat,38,39 but not to date in animals. DNA hypomethylation can also lead to the activation of oncogenes, an event that has been documented with the S100A4 metastasis‐associated gene in colorectal carcinoma, and in the cyclin D240 and maspin41 genes in gastric carcinoma. Finally, decreased methylation of DNA can lead to loss of imprinting (LOI), and this can drive cellular proliferation in cancer. The clearest example of this phenomenon is the LOI at the IGF2/H19 region as a result of hypomethylation at the differentially methylated region of IGF2,42 an event seen in about 40% of colorectal cancer tissue.43
Hypermethylation
In concert with global hypomethylation, focal hypermethylation at CpG islands is also regarded as a critical event in cancer development44,45,46 (fig 4). Not surprisingly, research in this discipline has focused on tumour‐suppressor genes, as promoter silencing by hypermethylation provides a mechanism other than sequence mutation for the inactivation of these key genes. Since the demonstration of methylation‐induced silencing of the RB gene in cancer,47 many more tumour‐suppressor genes have been identified as targets for this process, including p16INK4A, VHL, APC, CDH1 (E‐cadherin) and MLH1.4 Yet, silencing of tumour‐suppressor genes is not the only mechanism by which hypermethylation can favour the development of cancer. Hypermethylation can also lead to LOI in cancer. In Wilms' tumour, for example, hypermethylation at the IGF2 differentially methylated region causes LOI of the normally silenced maternal allele of IGF2.48,49 Similar events are seen with the p73 gene in haematological malignancies,50 and the ARH1 gene in follicular carcinoma of the thyroid.51
CpG island methylation is a common epigenetic event in colorectal neoplasia, with MLH1 promoter methylation representing a classic example of this phenomenon. A long list of hypermethylated genes is associated with colorectal neoplasia, including tumour‐suppressor, mismatch‐repair and cell‐cycle‐regulatory genes (table 2). This list is likely to grow as methods for the discovery of methylation targets are improved. Importantly, and as recently summarised by Baylin and Ohm,78, these genes have been drawn from many of the key functional groups that define the cancer phenotype, including Wnt signalling (SRFP genes), mismatch repair (MLH1), cell‐cycle regulation (CDKN2A), epithelial differentiation (GATA4,5), p53‐mediated damage responses (HIC1) and cell–matrix interactions (TIMP3).
Table 2 Some of the genes silenced by promoter methylation in colorectal neoplasia.
| Gene | Function | Frequency (%) | References |
|---|---|---|---|
| APC | Signal transduction, beta‐catenin regulation | 10–50 | 52–57 |
| CDH13 | Cell signalling (cell recognition and adhesion) | 30–40 | 58 |
| CDKN2A | Cell‐cycle regulation | 15–30 | 55, 59, 60 |
| CHFR | Mitotic stress checkpoint | 30–40 | 61, 62 |
| HIC1 | Regulation of DNA damage responses | ∼80 | 63, 64 |
| HPP1 | Transmembrane transforming growth factor (TGF)‐β antagonist | ∼80 | 65 |
| LKB1 | Cell signalling, cell polarity | 5–10 | 66 |
| MGMT | Repair of DNA guanosine methyl adduct | 30–40 | 55, 56, 67–69 |
| MLH1 | Mismatch repair | 10–20 | 56, 70–72 |
| p14ARF | Cell‐cycle regulation | 20–30 | 55, 73, 74 |
| RASSF1A | DNA repair, cell‐cycle regulation | >50 | 55, 75, 76 |
| SOCS1 | Cell signalling | 5–10 | 58 |
| THBS1 | Angiogenesis | 10–20 | 55, 56 |
| TIMP3 | Matrix remodelling, tissue invasion | 10–30 | 55, 77 |
Interesting recent observations have challenged the dogma that hypermethylation is confined to discrete CpG islands. Frigola et al79 showed that many colorectal cancers exhibited epigenetic silencing of an entire 4 Mb band of chromosome 2. This finding shows that epigenetic silencing can be a regional phenomenon with an effect on the expression of multiple rather than single genes.
Dysregulation of histone modification
In comparison with DNA methylation, current knowledge regarding dysregulation of the histone code in cancer is less advanced. At a simplistic level, this involves replacement of histones with variants, or changes in the decorations on the histone tails through chemical modifications of individual amino acids. Certainly, aberrant methylation of tumour‐suppressor genes is accompanied by two key modifications in the histone code—namely, deacetylation and methylation of the lysine (K) 9 residue of histone H3. These two moieties are mutually exclusive as they affect the same position. Acetylation of lysine (K) 9 residue of histone H3 correlates with gene expression, whereas methylation of this residue is associated with gene silencing and acts by recruiting heterochromatin‐associated proteins.80 Changes of these types are well documented in colorectal cancer.81,82,83 More recently, a pattern of changes to the core histone H4, characterised by the loss of both monoacetylation from lys 16 and trimethylation from lys 20, has been proposed as a universal marker for malignant transformation.84 Overexpression of a putative histone methyltransferase SMYD3 that methylates the lysine 4 residue of histone H3 in colorectal cancer has been reported.85 As methylation of the lysine 9 residue of histone H3 has been associated with gene activation,4 this suggests that the increased activity of SMYD3 can potentially promote transcription of oncogenes, homeobox genes and cell‐cycle‐regulatory genes. These types of changes in histone modification are characteristic of many human tumours.86 Individual histones may be replaced by histone variants such as H3.3 for the canonical H3 histone87 or the H2A.Z variant for H2A. The H2A.Z variant has a crucial role in embryogenesis88; and also by depositing it at the 5′ end of genes, it can retain the boundaries that prevent the spread of heterochromatin into euchromatic regions.89 Inappropriate inclusion of histone variants possibly disturb the boundaries between euchromatin and heterochromatin. The disturbances in the epigenetic machinery that induce these changes are the focus of current research,84,86 as are the consequences of such changes.
Relationship between epigenetic events, genetic change and pathology in colorectal neoplasia
Epigenetics of microsatellite and chromosomal instability
There is apparently a close interplay between genetic mutations and epigenetic modifications within the neoplastic cell. For example, by silencing one allele of a tumour‐suppressor gene, methylation can work in concert with a sequence mutation of the other allele to fulfil Knudson's two‐hit hypothesis. Yet, although it is possible to consider the epigenetic events seen in colorectal cancer in isolation, it is perhaps more informative to see these changes within the existing framework of established pathways for the development and progression of colorectal neoplasms.
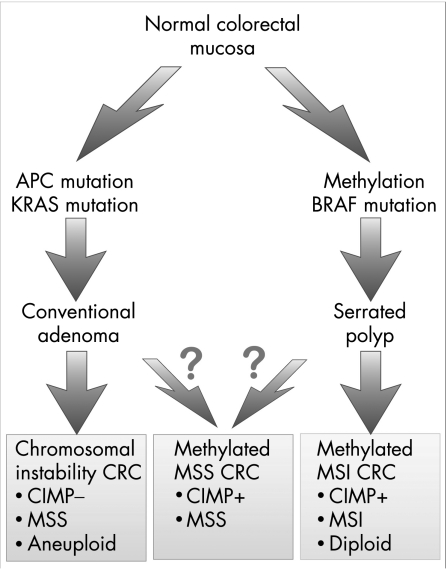
Current paradigms of colorectal cancer progression suggest at least two distinct pathways for progression, the traditional chromosomal instability pathway and the more recently elucidated microsatellite instability (MSI) pathway.90 These pathways represent divergent patterns, in terms of underlying genetics and tumour biology, including precursor lesions and morphology91 (fig 5). Epigenetic events are clearly involved in the chromosomal instability pathway, with hypomethylation establishing opportunities for chromosomal instability, and for activation of oncogenes such as c‐myc.92 However, the MSI pathway, characterised by early loss of mismatch‐repair activity within the tumour clone, and thus by the accumulation of errors at microsatellite loci, serves as an exemplar of epigenetic carcinogenesis.
Figure 5 Proposed pathways for colorectal tumorigenesis and their relationship with the CpG island methylator phenotype (CIMP). A working model of the dichotomy between chromosomal instability and microsatellite instability (MSI) pathways in colorectal carcinogenesis (CRC), and the common morphological and genetic changes that accompany each subtype. A subgroup of tumours, which are characterised by CpG island methylation (CIMP positive) and microsatellite stability (MSS), is shown. It is not clear whether these tumours arise from either or both of the main pathways, or whether they develop separately. MSI, microsatellite instability; serrated polyp, hyperplastic polyp or serrated adenoma.
The MSI pathway was recognised largely because of its occurrence in the cancer predisposition syndrome of hereditary non‐polyposis colorectal cancer (HNPCC), and it was only several years later that CpG island methylation was recognised as being critical to the development of the 15% of sporadic cancers that also followed this pathway.93 It is now well established that biallelic methylation of MHL1 followed by transcriptional inactivation of the gene is seen in nearly all sporadic MSI cancers. Like HNPCC tumours, sporadic MSI colorectal carcinomas have distinct clinicopathological features, including poor differentiation, intraepithelial lymphocytic infiltrates and location in the proximal colon.94 Curiously, however, they occur predominantly in elderly women, and a recent systematic review has confirmed that they have a considerably better outcome than those with microsatellite‐stable cancers of similar stage and grade.95
Although MLH1 methylation is the hallmark of the MSI pathway for colorectal cancer and epigenetic silencing of other genes is common (table 2), it is noteworthy that these tumours also show particular types of genetic change. For instance, activating mutations of the BRAF gene are very common in sporadic MSI cancers,96,97,98,99 although this gene is rarely if ever mutated in cancers arising in patients with HNPCC. Likewise, an interdependence has been reported between MGMT hypermethylation and TP53 mutations.100 The precise inter‐relationship between genetics and epigenetics in the MSI pathway, including the chronology of key events, remains to be elucidated.
The CpG island methylator phenotype
In 1999, Toyota et al101 identified a set of CpG islands that could be methylated in tumours but were not methylated in normal epithelial cells. They were able to show that many of these loci were heavily methylated in a subset of colorectal cancers, and they coined the acronym CIMP to describe tumours characterised by multiple, concordant methylation events.101 Subsequent population‐based studies on patients with colorectal cancer have suggested that CIMP tumours are clinically, pathologically and genetically distinct. They are characterised by many of the features typical of MSI tumours, such as right‐sidedness, high grade, mucinous type and high incidences in elderly people and in females.102,103,104 However, about half of the tumours that display widespread CpG island methylation are microsatellite stable (fig 5). Evidence also suggests that they may be unique in terms of behaviour. Our group has reported difference in outcome between subgroups of the CIMP tumours depending on microsatellite status,105 and highlighted the poor prognosis of patients with CIMP‐positive, microsatellite‐stable tumours.
The CIMP concept has not been accepted by all researchers in this field, and over the past few years there has been much debate as to whether the CIMP tumours represent a biologically distinct group of colorectal cancers or are an artificially selected group from a continuum of tumours showing different degrees of methylation at particular loci.106 Underpinning this debate is the important issue of whether the cells that give rise to the CIMP tumours have a definable change in their machinery of methylation that produces what Issa107 has referred to as “epigenetic instability”, and whether this is integral to tumour initiation and progression. This is an important question; if this were true, then a better understanding of the CIMP tumours would shed more light on the mechanisms that control CpG island methylation, and potentially on the appropriate management of this type of cancer. An affirmative answer would also support the concept that predisposition to the CIMP tumour may partly be hereditary, an observation initially suggested108 but not confirmed in larger studies.104,109 At present, issues regarding the operational definition of the CIMP are clearly limiting the attainment of consensus on these important matters,107 and the biological basis of the CIMP remains uncertain.
Chronology of genetic and epigenetic events in colorectal cancer
Research over the past decade has consistently shown that epigenetic changes such as promoter hypermethylation67,110,111 and LOI43 can occur in histologically normal colonic epithelium, and that these changes are more likely in patients with CIMP or MSI cancers.112 The early occurrence of these epigenetic events, and their relevance to the emerging science of stem cell biology, serves to highlight their theoretical significance in neoplastic development. Baylin and Ohm78 have recently advocated the primacy of epigenetic events in colorectal neoplasia, arguing that such epigenetic changes in stem cells may predetermine the nature of subsequent genetic events. Such a concept, if true, would help to explain the distinctive pattern of genetic changes in colorectal carcinogenesis made famous by Vogelstein et al.113 Feinberg et al114 have also recently highlighted the early role of epigenetic changes in neoplastic progression, suggesting that epigenetic modifications within stem cells and their progeny are responsible for forming a polyclonal cellular milieu from which neoplastic clones can develop.
Causes of epigenetic changes in colorectal neoplasia
Clearly, if epigenetic events are present at the earliest stages of colorectal tumorigenesis, then this holds important implications for both the recognition of cancer predisposition and, possibly, the chemoprevention of this disease. At a minimum, it is important to understand the factors that may induce epigenetic changes.
Environmental factors influencing epigenetic changes in colorectal neoplasia
The influence of environmental factors on the epigenetic state of cells (epimutagens) is a rapidly expanding field, and will be discussed only briefly in this review. With regard to dietary factors, folate is perhaps the best studied link to colorectal neoplasia. As an essential donor of one‐carbon units, folate is important in methylation reactions as well as in DNA synthesis and repair. Both epidemiological and experimental studies have shown that dietary folate correlates inversely with risk of colorectal neoplasia,115,116,117 but the effect of folate intake on tumorigenesis remains complex, and may depend in part on the stage of tumour development.118,119 From an epigenetic viewpoint, increased methylation secondary to dietary folate supplementation may have contradictory effects, from the beneficial restoration of gene hypomethylation to the disadvantageous silencing of genes. The complexity of this situation is compounded by related dietary factors, such as alcohol consumption, which may abrogate the protective role of folate.120,121 Finally, in considering dietary factors, it must be recognised that early maternal nutrition markedly affects the epigenetic patterning in the fetus, and it has been hypothesised that this in turn influences adult phenotypes, through the persistence of epigenetic changes at susceptible loci.122
Advancing age also correlates closely with epigenetic changes in normal colorectal mucosa. In these tissues, methylation of many genes, including the ESR1,112,123MLH1,70HIC1 and IGF2,124 has been shown to increase progressively with age. For at least some of these genes, this process seems to be accelerated in patients with colorectal cancer.101,125 These epigenetic changes may reflect the clinical truism that colorectal carcinoma is a disease in elderly people.
Inherited factors in the epigenetics of colorectal cancer
Given that epigenetic changes are stable and potentially heritable through meiosis, some of the ways in which inheritance may influence epigenetic changes associated with colorectal neoplasia should be considered. As discussed earlier, there has been considerable interest, albeit scant supporting evidence, for the proposition that the changes that underpin CIMP, whether genetic or epigenetic, may be inherited. Perhaps a clearer example of inherited epigenetic risk is seen in the case of LOI at IGF2. Certainly, patients with widespread LOI for this gene have an increased risk of developing colorectal cancer43 and are also more likely to have a family history of colorectal neoplasia.126 However, it is unclear whether LOI is a germline or somatic event.
Our group has recently found germline epimutations in MLH1, which predispose individuals to young‐onset MSI tumours in the large bowel and at extracolonic sites.127,128 These germline epimutations manifest by somawide uniparental methylation of the MLH1 promoter in the absence of an intragenic sequence mutation,127 and cause transcriptional silencing of the affected allele.128 These observations indicate that germline epigenetic changes can mimic hereditary cancer syndromes, and may be inherited.127,128,129,130,131 To date, such somawide epimutations have not been found in other genes such as APC,131 and further research on the family members of individuals with this abnormality is required to understand this phenomenon better.
Clinical importance of epigenetic changes in colorectal neoplasia
Given the increasing recognition of epigenetic changes in the histologically normal colorectal mucosa and in precursor lesions such as aberrant crypt foci, adenomas and serrated polyps, these changes can clearly serve as a marker for those at risk of colorectal cancer.112 Epigenetic markers are also increasingly being used in screening tests for colorectal neoplasia,110 yet much work remains to be carried out before such observations can be meaningfully translated into routine clinical practice.
The epigenetic events in colorectal cancer may soon influence treatment decisions. For instance, although still controversial,132 growing evidence from retrospective analyses suggests that MSI tumours respond differently to traditional chemotherapeutic agents,133,134 and that outcomes for some patients with these cancers may be worse with standard treatments.135 Such observations may reflect fundamental differences in drug responsiveness that are driven not by MSI but rather by underlying and as yet unrecognised epigenetic mechanisms.136,137
Not surprisingly, a better understanding of the epigenetic events in carcinogenesis, and the recognition that these events are potentially reversible, has brought with it a plethora of potential “epigenetic” therapies. Currently, there are two broad classes of epigenetic drugs designed to inhibit either DNA methylation or histone deacetylation. At least some of these drugs are in current clinical practice, and many more are in the clinical trials pipeline. Although only transient,138 inhibitors of DNA methylation such as 5‐azacytidine139 inactivate DNA methyltransferases, and can thus revert methylation‐induced silencing.140 Inhibitors of HDAC, such as suberoylanilide hydroxamic acid (SAHA) and newer derivatives,9 have been slower to emerge, and given the interdependence of these epigenetic processes, combination therapy approaches may also be beneficial.9 Whether these treatments will have sufficient specificity in practice to provide a useful therapeutic window awaits the outcome of current and future trials. Nevertheless, the experience gained in this process is likely to inform the mechanism of action of these drugs, and indeed the significance of epigenetic events in colorectal carcinogenesis.
Conclusions
Although our knowledge of the molecular genetics of colorectal neoplasia has developed rapidly over the past several decades, it is only in recent years that we have begun to understand the epigenetic events that underpin neoplastic initiation and progression in the large bowel. Currently, epigenetics of colorectal cancer is a bourgeoning field, and as was the case with the genetics of cancer, the lessons learnt from colorectal neoplasia are serving to throw light on the epigenetics of other common cancers. It is difficult to predict the extent to which knowledge of epigenetics gained over the next decade will transform our understanding of the disease and its precursors, but it clearly has the potential to entirely rework our current paradigms of cancer development, if not management.
Acknowledgements
We thank Megan Hitchins for helpful discussions regarding the manuscript.
Abbreviations
CIMP - CpG island methylator phenotype
HAT - histone acetyl transferase
HDAC - histone deacetylase
HNPCC - hereditary non‐polyposis colorectal cancer
LOI - loss of imprinting
lys - lysine
MSI - microsatellite instability
Footnotes
Funding: This work was supported in part by grants from the National Health and Medical Research Council of Australia and The Cancer Council of New South Wales. JJLW is the recipient of an International Postgraduate Research Scholarship.
Competing interests: None.
References
- 1.Holliday R. Epigenetics comes of age in the twentyfirst century. J Genet 2002811–4. [DOI] [PubMed] [Google Scholar]
- 2.Wu C T, Morris J R. Genes, genetics, and epigenetics: a correspondence. Science 20012931103–1105. [DOI] [PubMed] [Google Scholar]
- 3. Nature Milestones [cited 2006 20 April]; Available from: www.nature.com/milestones/milecancer/timeline.html S7–23 (accessed 27 Oct 2006)
- 4.Feinberg A P, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 20044143–153. [DOI] [PubMed] [Google Scholar]
- 5.Zilberman D, Cao X, Jacobsen S E. ARGONAUTE4 control of locus‐specific siRNA accumulation and DNA and histone methylation. Science 2003299716–719. [DOI] [PubMed] [Google Scholar]
- 6.Volpe T A, Kidner C, Hall I M.et al Regulation of heterochromatic silencing and histone H3 lysine‐9 methylation by RNAi. Science 20022971833–1837. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 2004431211–217. [DOI] [PubMed] [Google Scholar]
- 8.Holliday R. DNA methylation and epigenetic defects in carcinogenesis. Mutat Res 1987181215–217. [DOI] [PubMed] [Google Scholar]
- 9.Egger G, Liang G, Aparicio A.et al Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004429457–463. [DOI] [PubMed] [Google Scholar]
- 10.Ogryzko V V, Schiltz R L, Russanova V.et al The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 199687953–959. [DOI] [PubMed] [Google Scholar]
- 11.Fuks F, Burgers W A, Brehm A.et al DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet 20002488–91. [DOI] [PubMed] [Google Scholar]
- 12.Rountree M R, Bachman K E, Baylin S B. DNMT1 binds HDAC2 and a new co‐repressor, DMAP1, to form a complex at replication foci. Nat Genet 200025269–277. [DOI] [PubMed] [Google Scholar]
- 13.Ng H H, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol 2000201394–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito M, Ishikawa F. The mCpG‐binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J Biol Chem 200227735434–35439. [DOI] [PubMed] [Google Scholar]
- 15.Kondo Y, Issa J ‐ P J. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev 20042329–39. [DOI] [PubMed] [Google Scholar]
- 16.Jenuwein T, Allis C D. Translating the histone code. Science 20012931074–1080. [DOI] [PubMed] [Google Scholar]
- 17.Lieb J D, Clarke N D. Control of transcription through intragenic patterns of nucleosome composition. Cell 20051231187–1190. [DOI] [PubMed] [Google Scholar]
- 18.Ramsahoye B H, Biniszkiewicz D, Lyko F.et al Non‐CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA 2000975237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehrlich M, Gama‐Sosa M A, Huang L H.et al Amount and distribution of 5‐methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res 1982102709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA 19939011995–11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plass C, Soloway P D. DNA methylation, imprinting and cancer. Eur J Hum Genet 2002106–16. [DOI] [PubMed] [Google Scholar]
- 22.Robertson K D. DNA methylation and human disease. Nat Rev Genet 20056597–610. [DOI] [PubMed] [Google Scholar]
- 23.Wilkins J F. Genomic imprinting and methylation: epigenetic canalization and conflict. Trends Genet 200521356–365. [DOI] [PubMed] [Google Scholar]
- 24.Klose R J, Bird A P. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 20063189–97. [DOI] [PubMed] [Google Scholar]
- 25.Simmen M W, Leitgeb S, Charlton J.et al Nonmethylated transposable elements and methylated genes in a chordate genome. Science 19992831164–1167. [DOI] [PubMed] [Google Scholar]
- 26.Macleod D, Charlton J, Mullins J.et al Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev 199482282–2292. [DOI] [PubMed] [Google Scholar]
- 27.Riggs A D, Jones P A. 5‐Methylcytosine, gene regulation, and cancer. Adv Cancer Res 1983401–30. [DOI] [PubMed] [Google Scholar]
- 28.Feinberg A P, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 198330189–92. [DOI] [PubMed] [Google Scholar]
- 29.Lapeyre J ‐ N, Becker F F. 5‐Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun 197987698–705. [DOI] [PubMed] [Google Scholar]
- 30.Flatau E, Bogenmann E, Jones P A. Variable 5‐methylcytosine levels in human tumor cell lines and fresh pediatric tumor explants. Cancer Res 1983434901–4905. [PubMed] [Google Scholar]
- 31.Gama‐Sosa M A, Slagel V A, Trewyn R W.et al The 5‐methylcytosine content of DNA from human tumors. Nucleic Acids Res 1983116883–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goelz S E, Vogelstein B, Hamilton S R.et al Hypomethylation of DNA from benign and malignant human colon neoplasms. Science 1985228187–190. [DOI] [PubMed] [Google Scholar]
- 33.Bariol C, Suter C, Cheong K.et al The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol 20031621361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji W, Hernandez R, Zhang X Y.et al DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat Res 199737933–41. [DOI] [PubMed] [Google Scholar]
- 35.Qu G Z, Grundy P E, Narayan A.et al Frequent hypomethylation in Wilms tumors of pericentromeric DNA in chromosomes 1 and 16. Cancer Genet Cytogenet 199910934–39. [DOI] [PubMed] [Google Scholar]
- 36.Esteller M, Herman J G. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol 20021961–7. [DOI] [PubMed] [Google Scholar]
- 37.Suter C M, Martin D I, Ward R L. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis 20041995–101. [DOI] [PubMed] [Google Scholar]
- 38.Fincham J R, Sastry G R. Controlling elements in maize. Annu Rev Genet 1974815–50. [DOI] [PubMed] [Google Scholar]
- 39.Kashkush K, Feldman M, Levy A A. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 200333102–106. [DOI] [PubMed] [Google Scholar]
- 40.Oshimo Y, Nakayama H, Ito R.et al Promoter methylation of cyclin D2 gene in gastric carcinoma. Int J Oncol 2003231663–1670. [PubMed] [Google Scholar]
- 41.Akiyama Y, Maesawa C, Ogasawara S.et al Cell‐type‐specific repression of the maspin gene is disrupted frequently by demethylation at the promoter region in gastric intestinal metaplasia and cancer cells. Am J Pathol 20031631911–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui H, Onyango P, Brandenburg S.et al Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res 2002626442–6446. [PubMed] [Google Scholar]
- 43.Cui H, Horon I L, Ohlsson R.et al Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat Med 199841276–1280. [DOI] [PubMed] [Google Scholar]
- 44.Baylin S B, Hoppener J W, de Bustros A.et al DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer Res 1986462917–2922. [PubMed] [Google Scholar]
- 45.Jones P A, Laird P W. Cancer epigenetics comes of age. Nat Genet 199921163–167. [DOI] [PubMed] [Google Scholar]
- 46.Herman J G, Baylin S B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 20033492042–2054. [DOI] [PubMed] [Google Scholar]
- 47.Greger V, Passarge E, Hopping W.et al Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet 198983155–158. [DOI] [PubMed] [Google Scholar]
- 48.Rainier S, Johnson L A, Dobry C J.et al Relaxation of imprinted genes in human cancer. Nature 1993362747–749. [DOI] [PubMed] [Google Scholar]
- 49.Ogawa O, Eccles M R, Szeto J.et al Relaxation of insulin‐like growth factor II gene imprinting implicated in Wilms' tumour. Nature 1993362749–751. [DOI] [PubMed] [Google Scholar]
- 50.Corn P G, Kuerbitz S J, van Noesel M.et al Transcriptional silencing of the p73 gene in acute lymphoblastic leukemia and Burkitt's lymphoma is associated with 5′ CpG island methylation. Cancer Res 1999593352–3356. [PubMed] [Google Scholar]
- 51.Weber F, Aldred M A, Morrison C D.et al Silencing of the maternally imprinted tumor suppressor ARHI contributes to follicular thyroid carcinogenesis. J Clin Endocrinol Metab 2005901149–1155. [DOI] [PubMed] [Google Scholar]
- 52.Esteller M, Sparks A, Toyota M.et al Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res 2000604366–4371. [PubMed] [Google Scholar]
- 53.Kim J C, Koo K H, Roh S A.et al Genetic and epigenetic changes in the APC gene in sporadic colorectal carcinoma with synchronous adenoma. Int J Colorectal Dis 200318203–209. [DOI] [PubMed] [Google Scholar]
- 54.Bai A H C, Tong J H M, To K ‐ F.et al Promoter hypermethylation of tumor‐related genes in the progression of colorectal neoplasia. Int J Cancer 2004112846–853. [DOI] [PubMed] [Google Scholar]
- 55.Lee S, Hwang K S, Lee H J.et al Aberrant CpG island hypermethylation of multiple genes in colorectal neoplasia. Lab Invest 200484884–893. [DOI] [PubMed] [Google Scholar]
- 56.Kim H C, Roh S A, Ga I H.et al CpG island methylation as an early event during adenoma progression in carcinogenesis of sporadic colorectal cancer. J Gastroenterol Hepatol 2005201920–1926. [DOI] [PubMed] [Google Scholar]
- 57.Ebert M P A, Mooney S H, Tonnes‐Priddy L.et al Hypermethylation of the TPEF/HPP1 gene in primary and metastatic colorectal cancers. Neoplasia 20057771–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hibi K, Kodera Y, Ito K.et al Aberrant methylation of HLTF, SOCS‐1, and CDH13 genes is shown in colorectal cancers without lymph node metastasis. Dis Colon Rectum 2005481282–1286. [DOI] [PubMed] [Google Scholar]
- 59.Merlo A, Herman J G, Mao L.et al 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 19951686–692. [DOI] [PubMed] [Google Scholar]
- 60.Hawkins N J, Ward R L. Sporadic colorectal cancers with microsatellite instability and their possible origin in hyperplastic polyps and serrated adenomas. J Natl Cancer Inst 2001931307–1313. [DOI] [PubMed] [Google Scholar]
- 61.Corn P G, Summers M K, Fogt F.et al Frequent hypermethylation of the 5′ CpG island of the mitotic stress checkpoint gene Chfr in colorectal and non‐small cell lung cancer. Carcinogenesis 20032447–51. [DOI] [PubMed] [Google Scholar]
- 62.Toyota M, Sasaki Y, Satoh A.et al Epigenetic inactivation of CHFR in human tumors. Proc Natl Acad Sci USA 20031007818–7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maekawa M, Sugano K, Ushiama M.et al Heterogeneity of DNA methylation status analyzed by bisulfite‐PCR‐SSCP and correlation with clinico‐pathological characteristics in colorectal cancer. Clin Chem Lab Med 200139121–128. [DOI] [PubMed] [Google Scholar]
- 64.Chen W Y, Wang D H, Yen R C.et al Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53‐Dependent DNA‐damage responses. Cell 2005123437–448. [DOI] [PubMed] [Google Scholar]
- 65.Young J, Biden K G, Simms L A.et al HPP1: a transmembrane protein‐encoding gene commonly methylated in colorectal polyps and cancers. Proc Natl Acad Sci USA 200198265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Esteller M, Avizienyte E, Corn P G.et al Epigenetic inactivation of LKB1 in primary tumors associated with the Peutz‐Jeghers syndrome. Oncogene 200019164–168. [DOI] [PubMed] [Google Scholar]
- 67.Shen L, Kondo Y, Rosner G L.et al MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst 2005971330–1338. [DOI] [PubMed] [Google Scholar]
- 68.Fox E J, Leahy D T, Geraghty R.et al Mutually exclusive promoter hypermethylation patterns of hMLH1 and O6‐methylguanine DNA methyltransferase in colorectal cancer. J Mol Diagn 2006868–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wynter C V A, Kambara T, Walsh M D.et al DNA methylation patterns in adenomas from FAP, multiple adenoma and sporadic colorectal carcinoma patients. Int J Cancer 2006118907–915. [DOI] [PubMed] [Google Scholar]
- 70.Nakagawa H, Nuovo G J, Zervos E E.et al Age‐related hypermethylation of the 5′ region of MLH1 in normal colonic mucosa is associated with microsatellite‐unstable colorectal cancer development. Cancer Res 2001616991–6995. [PubMed] [Google Scholar]
- 71.Herman J G, Umar A, Polyak K.et al Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 1998956870–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miyakura Y, Sugano K, Konishi F.et al Extensive methylation of hMLH1 promoter region predominates in proximal colon cancer with microsatellite instability. Gastroenterology 20011211300–1309. [DOI] [PubMed] [Google Scholar]
- 73.Esteller M, Cordon‐Cardo C, Corn P G.et al p14ARF silencing by promoter hypermethylation mediates abnormal intracellular localization of MDM2. Cancer Res 2001612816–2821. [PubMed] [Google Scholar]
- 74.Shen L, Kondo Y, Hamilton S R.et al p14 methylation in human colon cancer is associated with microsatellite instability and wild‐type p53. Gastroenterology 2003124626–633. [DOI] [PubMed] [Google Scholar]
- 75.Sakamoto N, Terai T, Ajioka Y.et al Frequent hypermethylation of RASSF1A in early flat‐type colorectal tumors. Oncogene 2004238900–8907. [DOI] [PubMed] [Google Scholar]
- 76.Oliveira C, Velho S, Domingo E.et al Concomitant RASSF1A hypermethylation and KRAS//BRAF mutations occur preferentially in MSI sporadic colorectal cancer. Oncogene 2005247630–7634. [DOI] [PubMed] [Google Scholar]
- 77.Brueckl W M, Grombach J, Wein A.et al Alterations in the tissue inhibitor of metalloproteinase‐3 (TIMP‐3) are found frequently in human colorectal tumours displaying either microsatellite stability (MSS) or instability (MSI). Cancer Lett 2005223137–142. [DOI] [PubMed] [Google Scholar]
- 78.Baylin S B, Ohm J E. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 20066107–116. [DOI] [PubMed] [Google Scholar]
- 79.Frigola J, Song J, Stirzaker C.et al Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet 200638540–549. [DOI] [PubMed] [Google Scholar]
- 80.Peters A H F M, Mermoud J E, O'Carroll D.et al Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet 20023077–80. [DOI] [PubMed] [Google Scholar]
- 81.Fahrner J A, Eguchi S, Herman J G.et al Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res 2002627213–7218. [PubMed] [Google Scholar]
- 82.Kondo Y, Shen L, Issa J ‐ P J. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol 200323206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bachman K E, Park B H, Rhee I.et al Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell 2003389–95. [DOI] [PubMed] [Google Scholar]
- 84.Fraga M F, Ballestar E, Villar‐Garea A.et al Loss of acetylation at lys16 and trimethylation at lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 200537391–400. [DOI] [PubMed] [Google Scholar]
- 85.Hamamoto R, Furukawa Y, Morita M.et al SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol 20046731–740. [DOI] [PubMed] [Google Scholar]
- 86.Esteller M. Epigenetics provides a new generation of oncogenes and tumour‐suppressor genes. Br J Cancer 200694179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ahmad K, Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci USA 200299(Suppl 4)16477–16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Faast R, Thonglairoam V, Schulz T C.et al Histone variant H2A.Z is required for early mammalian development. Curr Biol 2001111183–1187. [DOI] [PubMed] [Google Scholar]
- 89.Raisner R M, Hartley P D, Meneghini M D.et al Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 2005123233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lengauer C, Kinzler K W, Vogelstein B. Genetic instabilities in human cancers. Nature 1998396643–649. [DOI] [PubMed] [Google Scholar]
- 91.Jass J R. Serrated adenoma of the colorectum and the DNA‐methylator phenotype. Nat Clin Pract Oncol 20052398–405. [DOI] [PubMed] [Google Scholar]
- 92.Sharrard R M, Royds J A, Rogers S.et al Patterns of methylation of the c‐myc gene in human colorectal cancer progression. Br J Cancer 199265667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Veigl M L, Kasturi L, Olechnowicz J.et al Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci USA 1998958698–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jass J R, Do K A, Simms L A.et al Morphology of sporadic colorectal cancer with DNA replication errors. Gut 199842673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Popat S, Hubner R, Houlston R S. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 200523609–618. [DOI] [PubMed] [Google Scholar]
- 96.Oliveira C, Pinto M, Duval A.et al BRAF mutations characterize colon but not gastric cancer with mismatch repair deficiency. Oncogene 2003229192–9196. [DOI] [PubMed] [Google Scholar]
- 97.Wang L, Cunningham J M, Winters J L.et al BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res 2003635209–5212. [PubMed] [Google Scholar]
- 98.Deng G, Bell I, Crawley S.et al BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res 200410191–195. [DOI] [PubMed] [Google Scholar]
- 99.Kambara T, Simms L A, Whitehall V L.et al BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 2004531137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Esteller M, Risques R A, Toyota M.et al Promoter hypermethylation of the DNA repair gene O(6)‐methylguanine‐DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res 2001614689–4692. [PubMed] [Google Scholar]
- 101.Toyota M, Ahuja N, Ohe‐Toyota M.et al CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999968681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hawkins N, Norrie M, Cheong K.et al CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology 20021221376–1387. [DOI] [PubMed] [Google Scholar]
- 103.van Rijnsoever M, Grieu F, Elsaleh H.et al Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut 200251797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Samowitz W S, Albertsen H, Herrick J.et al Evaluation of a large, population‐based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 2005129837–845. [DOI] [PubMed] [Google Scholar]
- 105.Ward R L, Cheong K, Ku S ‐ L.et al Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol 2003213729–3736. [DOI] [PubMed] [Google Scholar]
- 106.Yamashita K, Dai T, Dai Y.et al Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell 20034121–131. [DOI] [PubMed] [Google Scholar]
- 107.Issa J ‐ P. CpG island methylator phenotype in cancer. Nat Rev Cancer 20044988–993. [DOI] [PubMed] [Google Scholar]
- 108.Frazier M L, Xi L, Zong J.et al Association of the CpG island methylator phenotype with family history of cancer in patients with colorectal cancer. Cancer Res 2003634805–4808. [PubMed] [Google Scholar]
- 109.Ward R L, Williams R, Law M.et al The CpG island methylator phenotype is not associated with a personal or family history of cancer. Cancer Res 2004647618–7621. [DOI] [PubMed] [Google Scholar]
- 110.Petko Z, Ghiassi M, Shuber A.et al Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clin Cancer Res 2005111203–1209. [PubMed] [Google Scholar]
- 111.Nuovo G J, Nakagawa H, Sotamaa K.et al Hypermethylation of the MLH1 promoter with concomitant absence of transcript and protein occurs in small patches of crypt cells in unaffected mucosa from sporadic colorectal carcinoma. Diagn Mol Pathol 20061517–23. [DOI] [PubMed] [Google Scholar]
- 112.Kawakami K, Ruszkiewicz A, Bennett G.et al DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer. Br J Cancer 200694593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vogelstein B, Fearon E R, Hamilton S R.et al Genetic alterations during colorectal‐tumor development. N Engl J Med 1988319525–532. [DOI] [PubMed] [Google Scholar]
- 114.Feinberg A P, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006721–33. [DOI] [PubMed] [Google Scholar]
- 115.Giovannucci E, Stampfer M J, Colditz G A.et al Folate, methionine, and alcohol intake and risk of colorectal adenoma. J Natl Cancer Inst 199385875–884. [DOI] [PubMed] [Google Scholar]
- 116.Kim Y ‐ I. Folate, colorectal carcinogenesis, and DNA methylation: lessons from animal studies. Environ Mol Mutagen 20044410–25. [DOI] [PubMed] [Google Scholar]
- 117.Ulrich C M. Nutrigenetics in cancer research—folate metabolism and colorectal cancer. J Nutr 20051352698–2702. [DOI] [PubMed] [Google Scholar]
- 118.Pufulete M, Emery P W, Sanders T A B. Folate, DNA methylation and colo‐rectal cancer. Proc Nutr Soc 200362437–445. [PubMed] [Google Scholar]
- 119.Song J, Medline A, Mason J B.et al Effects of dietary folate on intestinal tumorigenesis in the apcMin mouse. Cancer Res 2000605434–5440. [PubMed] [Google Scholar]
- 120.Giovannucci E, Rimm E B, Ascherio A.et al Alcohol, low‐methionine–low‐folate diets, and risk of colon cancer in men. J Natl Cancer Inst 199587265–273. [DOI] [PubMed] [Google Scholar]
- 121.van Engeland M, Weijenberg M P, Roemen G M J M.et al Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res 2003633133–3137. [PubMed] [Google Scholar]
- 122.Waterland R A, Jirtle R L. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003235293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ahuja N, Li Q, Mohan A L.et al Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res 1998585489–5494. [PubMed] [Google Scholar]
- 124.Yuasa Y. DNA methylation in cancer and ageing. Mech Ageing Dev 20021231649–1654. [DOI] [PubMed] [Google Scholar]
- 125.Feinberg A P. The epigenetics of cancer etiology. Semin Cancer Biol 200414427–432. [DOI] [PubMed] [Google Scholar]
- 126.Cui H, Cruz‐Correa M, Giardiello F M.et al Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science 20032991753–1755. [DOI] [PubMed] [Google Scholar]
- 127.Suter C M, Martin D I K, Ward R L. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet 200436497–501. [DOI] [PubMed] [Google Scholar]
- 128.Hitchins M, Williams R, Cheong K.et al MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology 20051291392–1399. [DOI] [PubMed] [Google Scholar]
- 129.Gazzoli I, Loda M, Garber J.et al A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability‐high tumor. Cancer Res 2002623925–3928. [PubMed] [Google Scholar]
- 130.Miyakura Y, Sugano K, Akasu T.et al Extensive but hemiallelic methylation of the hMLH1 promoter region in early‐onset sporadic colon cancers with microsatellite instability. Clin Gastroenterol Hepatol 20042147–156. [DOI] [PubMed] [Google Scholar]
- 131.Hitchins M, Suter C, Wong J.et al Germline epimutations of APC are not associated with inherited colorectal polyposis. Gut 200655586–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Watanabe T, Wu T T, Catalano P J.et al Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med 20013441196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Carethers J M, Smith E J, Behling C A.et al Use of 5‐fluorouracil and survival in patients with microsatellite‐unstable colorectal cancer [see comments]. Gastroenterology 2004126394–401. [DOI] [PubMed] [Google Scholar]
- 134.Benatti P, Gafa R, Barana D.et al Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res 2005118332–8340. [DOI] [PubMed] [Google Scholar]
- 135.Ribic C M, Sargent D J, Moore M J.et al Tumor microsatellite‐instability status as a predictor of benefit from fluorouracil‐based adjuvant chemotherapy for colon cancer. N Engl J Med 2003349247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van Rijnsoever M, Elsaleh H, Joseph D.et al CpG island methylator phenotype is an independent predictor of survival benefit from 5‐fluorouracil in stage III colorectal cancer. Clin Cancer Res 200392898–2903. [PubMed] [Google Scholar]
- 137.Glasspool R M, Teodoridis J M, Brown R. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br J Cancer 2006941087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McGarvey K M, Fahrner J A, Greene E.et al Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res 2006663541–3549. [DOI] [PubMed] [Google Scholar]
- 139.Constantinides P G, Jones P A, Gevers W. Functional striated muscle cells from non‐myoblast precursors following 5‐azacytidine treatment. Nature 1977267364–366. [DOI] [PubMed] [Google Scholar]
- 140.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst 2005971498–1506. [DOI] [PubMed] [Google Scholar]