Abstract
FeMo cofactor (FeMoco) biosynthesis is one of the most complicated processes in metalloprotein biochemistry. Here we show that Mo and homocitrate are incorporated into the Fe/S core of the FeMoco precursor while it is bound to NifEN and that the resulting fully complemented, FeMoco-like cluster is transformed into a mature FeMoco upon transfer from NifEN to MoFe protein through direct protein–protein interaction. Our findings not only clarify the process of FeMoco maturation, but also provide useful insights into the other facets of nitrogenase chemistry.
Keywords: biosynthesis, nitrogenase
Nitrogenase is the key player in nature's ingenious scheme to convert the inert atmospheric dinitrogen to the bioavailable form of ammonia (for recent reviews, see refs. 1–8). The Mo-nitrogenase of Azotobacter vinelandii is composed of the iron (Fe) protein and the molybdenum–iron (MoFe) protein. The homodimeric Fe protein has one nucleotide-binding site per subunit and a single [4Fe-4S] cluster bridged between the two subunits. The α2β2-tetrameric MoFe protein contains two unique metal clusters per αβ-subunit: the [8Fe-7S] P-cluster (9), which is located at the αβ-interface and ligated to six protein residues; and the [Mo-7Fe-9S-X-homocitrate] (the identity of X is unknown but is considered to be C, O, or N; ref. 10) FeMo cofactor (FeMoco), which is situated within the α-subunit and bound to only two protein residues and an exogenous homocitrate ligand. Both P-cluster and FeMoco are composed of smaller substructures: the P-cluster comprises two [4Fe-4S] subclusters that share a μ6-sulfide (9) and FeMoco consists of [Mo-3Fe-3S] and [4Fe-3S] subcubanes that are bridged by three μ2-sulfides and share a central μ6-light atom (10). These metal clusters are essential for nitrogenase reaction, a process that involves ATP-dependent electron transfer from the [4Fe-4S] cluster of the Fe protein to the P-cluster of the MoFe protein and finally to FeMoco where substrate reduction takes place, and consequently become the major subjects in the vigorous studies of nitrogenase catalysis (1–8). Meanwhile, there is an emerging understanding of nitrogenase biosynthesis, in particular, P-cluster and FeMoco assembly in A. vinelandii, that is poised to further clarify the structure and function of these important clusters while also serving as a paradigm for the field of complex metalloprotein biosynthesis (11, 12).
The P cluster is a classical example of high-nuclearity clusters containing only Fe and S, and it is likely assembled at its targeted location (“in situ” assembly). Using a biochemical/spectroscopic approach, we identified the presence of two pairs of [4Fe-4S]-like clusters that likely represent P-cluster precursors in a FeMoco-deficient MoFe protein purified from a nifH-deletion strain (13, 14). This protein becomes catalytically active upon incubation with the deleted gene product, indicating that it might represent a physiologically relevant intermediate during P-cluster assembly (15). These results suggest that the P-cluster is formed through the fusion of its substructural units, a reaction mechanism that is well known in synthetic inorganic chemistry (16) and particularly appropriate considering the “modular” composition of the P cluster.
Unlike the P cluster, FeMoco, which contains additional heterometal (Mo) and organic moiety (homocitrate), is first assembled on a scaffold protein (17, 18) and then inserted into its destined location in MoFe protein (“ex situ” assembly). Biosynthesis of FeMoco presumably starts with the production of the Fe/S core by NifB (encoded by nifB) (19, 20), which is then transferred to, and further processed on, the α2β2 tetrameric NifEN protein (17, 21). Sequence similarity between the respective subunit-encoding genes led to the proposal that NifE and NifN form a structurally homologous complex to the MoFe protein (22, 23) and that, by analogy, NifEN also contains two types of metal cluster sites, one corresponding to the P-cluster site and the other to the FeMoco site of the MoFe protein (17, 21). The P-cluster analog was identified as a [4Fe-4S] cluster likely coordinated by conserved Cys residues at the NifE–NifN interface (21). The FeMoco analog had not been captured on NifEN until an efficient one-step purification procedure, which minimized the degradation of proteins and the consequent loss of metal clusters, was applied to a His-tagged form of the NifEN protein (17). In a so-called FeMoco maturation assay comprising NifEN, molybdate, homocitrate, Fe protein, MgATP and ΔnifB MoFe protein, the FeMoco analog on NifEN was proven to be a FeMoco precursor by its ability to activate FeMoco-deficient ΔnifB MoFe protein. Extended x-ray absorption fine structure (EXAFS) analysis of the NifEN-bound precursor showed that it was structurally similar to FeMoco except for the notable absence of Mo (18). Therefore, FeMoco cannot be formed through condensation of [Mo-3Fe-3S] and [4Fe-3S] partial cubanes; rather, the Fe/S structure of FeMoco is formed first, possibly through condensation of smaller Fe/S subclusters in a fashion similar to that proposed for P-cluster assembly and, then, Mo and homocitrate are added to complete the synthesis. Although instrumental in clarifying the trajectory of FeMoco biosynthesis, our original study left such unanswered questions as: when Mo and homocitrate are inserted into the cluster; how FeMoco is transferred from NifEN to the MoFe protein; and what role the Fe protein and MgATP play in FeMoco maturation.
The current study addresses these remaining questions by following the final steps of FeMoco assembly in A. vinelandii using a similar biochemical/spectroscopic strategy to that used previously (17, 18). Through this approach, we show that Mo and homocitrate are incorporated into the Fe/S core of the FeMoco precursor while it is bound to NifEN and that the fully complemented cluster is subsequently transferred from NifEN to MoFe protein through direct protein–protein interaction. The function of Fe protein and MgATP in FeMoco maturation is described in a companion paper (24).
Results
The FeMoco maturation assay, which was designed to test the capability of NifEN in FeMoco biosynthesis, comprises (i) NifEN, the source of FeMoco precursor; (ii) molybdate and homocitrate, the constituents of FeMoco absent from the precursor; (iii) Fe protein and MgATP, the factors assisting the maturation of the precursor in an unknown fashion; and (iv) FeMoco-deficient ΔnifB MoFe protein, the “receptor” for fully converted FeMoco (17). Based on this assay, we developed a new strategy to test the extent of FeMoco maturation on NifEN that involves (i) repurification of NifEN after incubation with all of the components of the FeMoco maturation assay except ΔnifB MoFe protein and (ii) subsequent analysis of re-purified NifEN (designated NifENcomplete). The analysis includes metal quantitation, EPR, visible region absorption and x-ray spectroscopies, and activity assays that measure the capacity to reconstitute ΔnifB MoFe protein. By systematically altering the composition of the incubation mixture, a suite of repurified NifEN proteins is produced, the analysis of which establishes the required components for FeMoco maturation on NifEN. ΔnifB NifENcomplete, which is treated identically to NifENcomplete except for the replacement of NifEN by precursor-free ΔnifB NifEN, serves as a negative control in this NifEN-specific FeMoco maturation assay. The complete set of repurified NifEN proteins are herein categorically designated NifEN′ and distinguished by different superscripts (see Materials and Methods for the complete list of NifEN′).
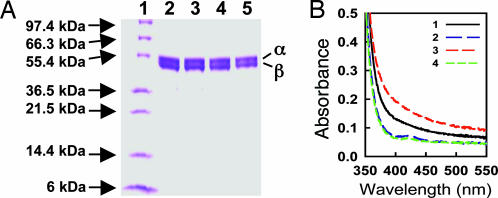
As shown in Fig. 1A, NifENcomplete and ΔnifB NifENcomplete, like their respective unprocessed counterparts NifEN and ΔnifB NifEN, are composed of α (≈52 kDa) and β (≈49 kDa) subunits (17). The molecular masses of both proteins are ≈200 kDa based on their elution profiles on gel filtration Sephacryl S-200 HR column (data not shown), indicating that both proteins are α2β2 tetramers. Metal analysis shows that unprocessed NifEN contains 16.1 ± 1.1 mol Fe and no Mo per mol of protein (Table 1), allowing for the assignment of two permanent [4Fe-4S] clusters (21) and one Mo-free precursor per protein molecule (17, 18). NifENcomplete, in contrast, contains 15.8 ± 0.6 mol Fe and 1.2 ± 0.1 mol Mo per mol of protein (Table 1), which is consistent with the presence of one cluster having the same metal composition as FeMoco in addition to the two permanent clusters per molecule of NifEN. The absence of the nifB gene should prevent the formation of a FeMoco precursor and, accordingly, both ΔnifB NifEN and ΔnifB NifENcomplete are shown to have sufficient Fe to form the permanent clusters alone: ≈8 mol Fe and no Mo per mol of protein (Table 1). Consistent with the metal analysis results, the visible region absorption spectra of these proteins reveal that the NifENcomplete spectrum (Fig. 1B, 3) is significantly more intense than that of NifEN (Fig. 1B, 1) at the same protein concentration, indicating that the precursor on NifENcomplete is modified from that on NifEN; whereas the intensity of the ΔnifB NifENcomplete spectrum (Fig. 1B, 4) is almost identical to that of the ΔnifB NifEN (Fig. 1B, 2), indicating that the permanent clusters are unchanged.
Fig. 1.
Protein purification and visible region absorption spectroscopy. (A) Coomassie blue-stained 10–20% gradient SDS/PAGE of NifEN, ΔnifB NifEN, NifENcomplete and ΔnifB NifENcomplete. Lane 1, 10 μg protein standard; lane 2, 15 μg purified NifEN; lane 3, 15 μg of purified ΔnifB NifEN; lane 4, 15 μg of purified NifENcomplete; lane 5, 15 μg of purified ΔnifB NifENcomplete. (B) Visible region spectra of the same protein samples in A. Spectra of dithionite-reduced NifEN (1), ΔnifB NifEN (2), NifENcomplete (3), and ΔnifB NifENcomplete (4) are shown between 350 and 550 nm. The samples were prepared at a concentration of 5 mg/ml, as described in Supporting Text, which is published as supporting information on the PNAS web site.
Table 1.
Metal contents of NifEN′
| Protein | Metal | |
|---|---|---|
| Mo | Fe | |
| NifEN | <0.01 | 16.1 ± 1.1 |
| NifENcomplete | 1.2 ± 0.1 | 15.8 ± 0.6 |
| ΔnifB NifEN | <0.01 | 8.2 ± 1.0 |
| ΔnifB NifENcomplete | <0.01 | 7.7 ± 0.1 |
| NifENminus Mo/homocitrate | <0.01 | 14.9 ± 0.1 |
| NifENminus homocitrate | 0.3 ± 0.1 | 15.2 ± 0.1 |
| NifENminus Mo | <0.01 | 15.7 ± 0.2 |
| NifENminus MgATP | <0.01 | 15.6 ± 0.2 |
| NifENminus Fe protein | <0.01 | 15.3 ± 0.1 |
| NifENapo Fe protein | <0.01 | 14.8 ± 0.2 |
| NifENA157S Fe protein | 0.2 ± 0.1 | 15.4 ± 0.1 |
| NifENM156C Fe protein | 0.2 ± 0.1 | 14.6 ± 0.4 |
| NifENA157G Fe protein | <0.01 | 16.1 ± 1.0 |
| NifENMgADP | <0.01 | 16.4 ± 1.1 |
| NifENATPγS | <0.01 | 15.3 ± 0.1 |
| NifENAMPPNP | <0.01 | 16.5 ± 1.0 |
Data are expressed as moles of metal per mole of protein.
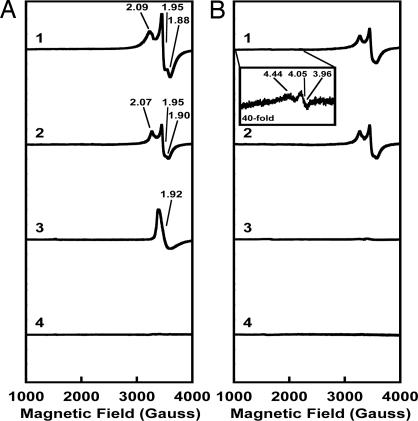
EPR analyses provide additional, more detailed evidence that the precursor on NifENcomplete is further processed (Fig. 2). In the dithionite-reduced state, NifEN exhibits an S = 1/2 signal in the g = 2 region, which has a rhombic line shape with a distinct feature between g values of 1.95 and 1.88 (Fig. 2A, 1). As established previously, this signal arises from both the precursor and the permanent [4Fe-4S] clusters on NifEN (17). Compared to unprocessed NifEN, NifENcomplete shows an S = 1/2 signal with a slightly rhombic line shape (Fig. 2B, 1) of significantly lower intensity. The decreased signal intensity of NifENcomplete is not caused by the loss of clusters, as shown by the metal analyses (Table 1); rather, it reflects the disappearance of the signal arising from the precursor upon further processing. As a result, the signal of NifENcomplete, like those of the ΔnifB NifEN proteins, should arise solely from the permanent [4Fe-4S] clusters in the protein. Indeed, the S = 1/2 signal of NifENcomplete is identical to that of ΔnifB NifEN (Fig. 2A, 2), which, as the spectrum of ΔnifB NifENcomplete shows (Fig. 2B, 2), remains unchanged upon processing. Consistent with these results, the unique signal of NifEN in the indigo disulfonate (IDS)-oxidized state with a g value of 1.92 (Fig. 2A, 3) is not observed in NifENcomplete (Fig. 2B, 3), indicating the disappearance of this Mo-free precursor-associated signal (17) upon further processing. As expected, this unique signal is also missing from the spectra of ΔnifB NifEN (Fig. 2A, 4) and ΔnifB NifENcomplete (Fig. 2B, 4) due to the absence of precursors in these proteins (17). Additionally, in the dithionite-reduced state, NifENcomplete shows a very minor EPR signal that has a slightly rhombic line shape with g values of 4.44, 4.05 and 3.96 (Fig. 2B, 1, Inset). This signal could arise from the processed precursor or some intermediates associated with its assembly (24). It is important to note that despite the presence of stoichiometric Mo for FeMoco formation, the characteristic S = 3/2 signal associated with MoFe protein-bound FeMoco (1) is not observed in NifENcomplete. These results suggest that the processed cluster in NifENcomplete has a different spin state than both the unprocessed NifEN-bound precursor and the mature MoFe protein-bound FeMoco.
Fig. 2.
EPR spectra of unprocessed (A) and processed (B) NifEN and ΔnifB NifEN. (A) EPR Spectra of dithionite-reduced NifEN (1), dithionite-reduced ΔnifB NifEN (2), IDS-oxidized NifEN (3), and IDS-oxidized ΔnifB NifEN (4). (B) EPR Spectra of dithionite-reduced NifENcomplete (1), dithionite-reduced ΔnifB NifENcomplete (2), IDS-oxidized NifENcomplete (3), and IDS-oxidized ΔnifB NifENcomplete (4). All spectra were measured at a protein concentration of 15 mg/ml, as described in the Supporting Text. The g values are indicated. (Inset) The spectrum of dithionite-reduced NifENcomplete between 1,000 and 2,000 G at a magnification of 40-fold.
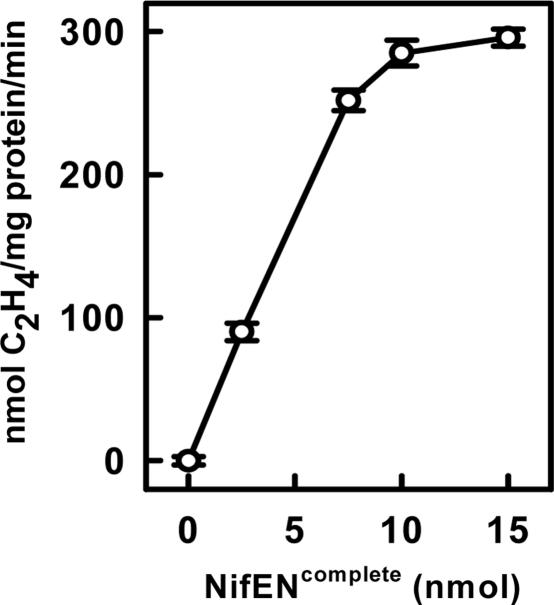
NifENcomplete alone does not show any substrate reducing activities, as expected (Table 2). Nevertheless, NifENcomplete can activate the FeMoco-deficient ΔnifB MoFe protein to a maximum activity of ≈300 nmol of C2H4 formation per mg of ΔnifB MoFe protein per min (Fig. 3). Upon activation by NifENcomplete, MoFe protein shows not only C2H4-formation but also H2-formation and N2-fixation activities that are comparable with those resulting from the complete FeMoco maturation assay (Table 2). Although activities in these maturation assays are lower than those resulting from activation of ΔnifB MoFe protein with isolated FeMoco (13), the activities for all substrates are proportionally reduced, which would not be expected if the cofactor were structurally perturbed or homocitrate was incorrectly bound (25). Thus, these activity results indicate that the precursor on NifENcomplete has all components necessary to become a mature FeMoco upon delivery from NifEN to the FeMoco binding site in MoFe protein. Furthermore, these data suggest that the transfer of the precursor from NifEN to MoFe protein can occur through direct protein–protein interaction, excluding the absolute requirement for a specific cluster carrier in this process.
Table 2.
Reconstitution of MoFe protein with NifEN′
| Assay condition | Activities* | |||
|---|---|---|---|---|
| C2H4 formation under C2H2/Ar |
H2 formation under Ar |
NH3 formation under N2 |
H2 formation under N2 |
|
| FeMoco maturation assay | ||||
| Complete | 290 ± 26 (100) | 350 ± 50 (100) | 111 ± 20 (100) | 65 ± 7 (100) |
| MoFe protein reconstitution with NifEN′† | ||||
| NifENcomplete | 284 ± 17 (98) | 375 ± 12 (107) | 142 ± 4 (127) | 70 ± 7 (108) |
| ΔnifB NifENcomplete | 0 (0) | 0 (0) | 0 (0) | 1 ± 1 (<1) |
| NifENminus Mo/homocitrate | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| NifENminus homocitrate‡ | 6 ± 1 (2) | 0 (0) | 0 (0) | 0 (0) |
| NifENminus Mo | 1 ± 1 (<1) | 0 (0) | 0 (0) | 0 (0) |
| NifENminus MgATP | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| NifENminus Fe protein | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| NifENapo Fe protein | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| NifENA157S Fe protein§ | 33 ± 1 (11) | 29 ± 2 (8) | 8 ± 1 (7) | 8 ± 1 (12) |
| NifENM156C Fe protein§ | 23 ± 1 (8) | 31 ± 1 (9) | 5 ± 1 (5) | 8 ± 1 (12) |
| NifENA157G Fe protein | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| NifENMgADP, NifENATPγS or NifENAMPPNP | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Activity assays with NifEN′ alone¶ | ||||
| NifENcomplete or ΔnifB NifENcomplete | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
Data are expressed as nanomoles per minute per milligram of protein. Percentages are given in parentheses.
*The lower detection limits were 0.01, 0.02, 0.001, and 0.02 nmol per min per mg of protein for C2H4 formation under C2H2/Ar, H2 formation under Ar, NH3 formation under N2 and H2 formation under N2, respectively.
†Except for ΔnifB NifENcomplete, all NifEN′ proteins contain normal amounts of Mo-free precursor based on their activities in FeMoco maturation assays, which are not considerably lower compared with NifEN.
‡NifENminus homocitrate can be activated to ≈10% upon incubation with all components of FeMoco maturation assay except molybdate, indicating the accumulation of a lower amount of molybdenum on the NifEN-bound precursor in the absence of homocitrate.
§A157S and M156C Fe proteins show 23% and 16% of MgATP hydrolysis activities, respectively, compared with the wild-type Fe protein (data not shown).
¶Assays were performed as described earlier (35) except that MoFe protein was replaced by NifENcomplete or ΔnifB NifENcomplete.
Fig. 3.
MoFe protein reconstitution with NifENcomplete. Assays were performed as described in Materials and Methods except that the amounts of NifENcomplete were varied between 0 and 15 nmol. The data presented here are the average of three independent experiments. The error bars are indicated.
Formation of the processed, activatable precursor on NifENcomplete requires molybdate, homocitrate, Fe protein and MgATP, because NifEN′ proteins prepared in the absence of one or more of these factors, such as NifENminus Mo/homocitrate, NifENminus homocitrate, NifENminus Mo, NifENminus Fe protein and NifENminus MgATP, are unable to activate the ΔnifB MoFe protein (Table 2). Fe protein-mediated MgATP hydrolysis is also implicated in this process, based on the observations that (i) NifENMgADP, NifENATPγS, and NifENAMPPNP, prepared by replacing MgATP with MgADP or nonhydrolysable ATP analogs (Table 3, which is published as supporting information on the PNAS web site), cannot reconstitute the ΔnifB MoFe protein (Table 2) and (ii) NifENA157S Fe protein, NifENM156C Fe protein and NifENA157G Fe protein, prepared by substituting wild-type Fe protein for variants defective in MgATP hydrolysis (Table 3), show greatly diminished or no capacity to activate the ΔnifB MoFe protein (Table 2). In addition, the requirement of electron transfer for cluster conversion is suggested by the inability of NifENapo Fe protein, prepared by replacing wild-type Fe protein with cluster-deficient apo Fe protein (Table 3), to reconstitute the ΔnifB MoFe protein (Table 2). The metal contents of the NifEN′ proteins indicate considerably less or no Mo (and homocitrate; ref. 24) incorporation into the precursors in cases where one or more of the MoFe protein maturation components are absent or disrupted (Table 1) and correlate well with their respective capacities to activate the ΔnifB MoFe protein (Table 2).
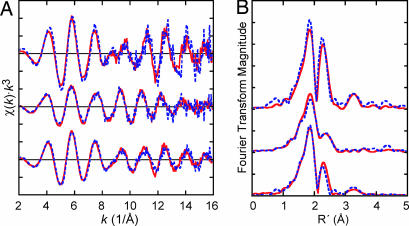
EXAFS analysis provides a structural probe of the processed precursor on NifENcomplete. The Fe K-edge EXAFS of NifENcomplete are slightly shifted in frequency relative to those of NifEN in the high-k region (Fig. 4); whereas the EXAFS of ΔnifB NifENcomplete are largely unchanged from those of ΔnifB NifEN (Fig. 4), indicating that it is the precursor, not the permanent clusters, that is altered. Using an established method (18), the EXAFS for the processed precursor were obtained by subtraction of ΔnifB NifENcomplete from NifENcomplete in an 8/7:15/7 ratio. The Fe EXAFS features of the NifENcomplete precursor are only subtly changed from those of the NifEN precursor (Fig. 4), consistent with a lack of major structural differences between these two species. Importantly, the signature feature of a FeMoco-like cluster, the intense peak at ≈3.5 Å in the Fourier transform (18) is still present in the NifENcomplete precursor (Fig. 4). However, EXAFS fitting results indicate that there is a change in the components that comprises this peak from two Fe–Fe scatterers, at 3.68 and 3.80 Å, in the unprocessed precursor to one at 3.70 Å after processing (Table 4, which is published as supporting information on the PNAS web site). MoFe protein-bound FeMoco is similarly fit by only one peak (Table 4) (18). Compared to FeMoco, the NifENcomplete precursor has slightly longer scattering distances on average, whereas it has slightly shorter differences than those found in the unprocessed NifEN precursor (Table 4). These results confirm that NifENcomplete still contains a FeMoco-like cluster, and furthermore, suggest that the Fe/S core of the processed precursor in NifENcomplete is tightened relative to that of NifEN, although it is still not as compact as FeMoco in its dithionite-reduced state. Given the differences in the spin states of these clusters, it is expected that they would have slightly different bond lengths. Similar differences are observed for the different states of FeMoco in MoFe protein (26). Because the contribution from Mo scattering is small relative to the other components in the average Fe environment, Fe EXAFS fits cannot provide proof of Mo incorporation into the cluster. This proof comes from direct analysis of the Mo environment in NifENcomplete through Mo K-edge XAS. The Mo edge spectrum of NifENcomplete is highly similar to that of MoFe protein (Fig. 5) and consistent with Mo in an octahedral environment of mixed O and S ligation like that found in FeMoco (27). The small, ≈0.6 eV, difference between the edge positions of these proteins (Fig. 5) could be due to subtle differences in the Mo ligand environment, which are expected given the absence of the FeMoco His ligand on NifE (23), or to differences in the cluster oxidation state. Of particular significance is the lack of any preedge features in NifENcomplete (Fig. 5), which provides evidence that there is no free molybdate associated with this protein after repurification (molybdate has a characteristic sharp preedge transition at ≈20,008 eV, Fig. 5) (28). The Mo K-edge EXAFS of NifENcomplete clearly indicate that the majority of Mo in this protein is in an Fe/S cluster. The strong scattering contributions at high-k and the ratio of Fourier transform peaks (Fig. 5) are consistent with Mo in a FeMoco (29, 30) or FeMoco-like environment (31). Accordingly, the NifENcomplete EXAFS are fit with Mo-S and Mo-Fe scattering components at similar distances to those in FeMoco (Table 5, which is published as supporting information on the PNAS web site). The total magnitude of the NifENcomplete Fourier transform is greatly diminished relative to that of the MoFe protein and the long-range features are absent, indicating a significant amount of disorder in the Mo environment of NifENcomplete (Fig. 5). This disorder could be due to asymmetric coordination as compared to that in FeMoco, or weak Mo binding in the Fe-S cluster leading to a mixture of partially incorporated states, contamination from Mo bound elsewhere in the protein, or a combination of these effects. EXAFS fitting results indicate that NifENcomplete is best fit with a reduced number of Mo-S and Mo-Fe scatterers compared to the MoFe protein and with either 3 statically disordered Mo-O/N scatterers at an average distance of 2.13 Å, or two types of Mo-O/N scatterers at 2.00 Å and 2.17 Å (Table 5). Note that the Mo-Fe scattering observed for NifENcomplete must be associated with the Fe in the precursor and not the permanent clusters, as there is no change to the spectroscopic features of ΔnifB NifEN, which represents the permanent clusters upon Mo incubation. In combination, the Fe and Mo K-edge XAS and EXAFS results indicate that the NifENcomplete precursor consists of a FeMoco-like structure with slightly elongated bond lengths and an asymmetrically or loosely bound terminal Mo atom that is also coordinated by a mixture of O/N-containing ligands.
Fig. 4.
Fe K-edge EXAFS (A) and Fourier transforms (B) of (top to bottom), the NifENcomplete precursor (blue) with the NifEN precursor (red); ΔnifB NifENcomplete (blue) with ΔnifB NifEN (red); and NifENcomplete (blue) with NifEN (red). EXAFS data for the NifENcomplete precursor were obtained by subtraction of ΔnifB NifENcomplete EXAFS from NifENcomplete EXAFS in an 8/7:15/7 ratio. NifEN precursor data were obtained by a 1:2 subtraction of ΔnifB NifEN EXAFS from NifEN EXAFS (18).
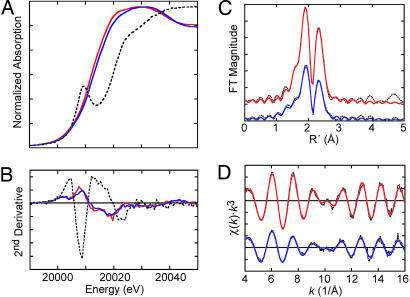
Fig. 5.
Mo K-edge XAS and EXAFS. (A and B) Mo K-edge x-ray absorption spectra (A) and smoothed second derivatives (B) of NifENcomplete (blue), MoFe protein (red), and molybdate in 33% glycerol (dashed black). (C and D) Mo K-edge EXAFS (C) and Fourier transforms (D) of data (dotted black) and fits for NifENcomplete (blue) and MoFe protein (red). A four-component fit is shown for NifENcomplete comprising Mo-O at 2.00 and 2.17 Å, Mo-S at 2.37 Å, and Mo-Fe at 2.70 Å. The MoFe protein fit comprises Mo-O at 2.22 Å, Mo-S at 2.37 Å, and Mo-Fe at 2.69 Å. Complete fit results are given in Table 5.
Discussion
FeMoco biosynthesis is one of the most complicated processes in metalloprotein biochemistry, requiring a multitude of component proteins and encompassing a variety of synthetic strategies. By employing an experimental approach of analyzing only purified proteins, the roles of key components and the timing of key events in this process are unambiguously determined. Furthermore, the combination of spectroscopic characterization with biochemical analyses enables a more detailed picture of the specific FeMoco assembly mechanism. Previously, we identified a Mo-free, NifEN-bound FeMoco precursor with a core structure similar to FeMoco as an early intermediate in this process (18). In this work, we show that Mo and homocitrate are incorporated into the precursor while it is bound to NifEN, resulting in the formation of a Mo/homocitrate-containing, FeMoco-like cluster. We also show that this cluster is transformed into a fully matured FeMoco and inserted into its target location in MoFe protein without the aid of a carrier or chaperone.
The results of the FeMoco maturation assay clearly indicate that Mo, homocitrate, [4Fe-4S]-containing Fe protein and ATP hydrolysis are all required for FeMoco maturation on NifEN. The impact of homocitrate on Mo binding to NifEN (Tables 1 and 2) indicates that homocitrate and Mo are possibly preassociated and concurrently incorporated into the precursor. It has been speculated that homocitrate, the organic moiety of FeMoco responsible for the overall negative charge of the cluster, may assist FeMoco insertion by “steering” it down a positively charged “FeMoco insertion funnel” into its final location in the MoFe protein (32). It is interesting that some of the NifD residues (α subunit of MoFe protein) providing the “FeMoco insertion funnel” are also positively charged in the corresponding NifE residues (α subunit of NifEN), indicating the possible presence of an analogous, positively charged “Mo/homocitrate insertion funnel” in the NifEN complex (11). Mobilization of Mo (and, apparently, homocitrate) for FeMoco biosynthesis has been proposed to be carried out by proteins that can sequester Mo, place Mo in the correct oxidation state for FeMoco formation, and deliver Mo to a FeMoco assembly site through protein-protein interactions with NifEN (11, 12). In a related study we establish that the Fe protein functions as a specific Mo/homocitrate insertase in FeMoco biosynthesis (24).
The combined Mo and Fe EXAFS results indicate that the fully complemented cluster species, formed upon incorporation of Mo and homocitrate into the Mo-free precursor, is structurally very similar to mature FeMoco. The bond distances in this processed form of the precursor are consistent with a compaction of the Fe/S core of the precursor upon processing and are similar to, although not the same as, those of FeMoco. EXAFS studies clearly indicate that Mo is associated with the Fe/S core, but also indicate that Mo binding in this cluster is likely asymmetrical or relatively weak compared to that in FeMoco. These EXAFS results are consistent with EPR results showing that the processed cluster is in a different spin state than both FeMoco and the unprocessed precursor. The final transformation of this cluster probably involves another change in cluster oxidation state, leading to further tightening of the core Fe/S structure and stronger Mo binding. Additionally, the cluster will be changed, especially at the Mo site, upon release from NifEN and ligation to the MoFe protein. This argument is based on the fact that residues that either provide a covalent ligand or tightly pack FeMoco within the MoFe protein are not duplicated in the corresponding region within the NifEN complex. For example, α-His442, which coordinates the Mo atom and anchors FeMoco to MoFe protein, is substituted by Asn at the corresponding position in NifEN.
It is likely that the differences in protein environments not only account for some of the minor differences between the clusters on NifEN and MoFe protein, but also create an “affinity gradient” between NifEN and MoFe protein for cluster binding that enable the negatively charged cluster to “escape” from NifEN (cluster donor), travel down the positively charged “FeMoco insertion funnel” of the MoFe protein and eventually “lock” into the binding site within the MoFe protein (cluster receptor). This “locking” mechanism is likely directed by the change in Mo coordination to a stronger protein ligand upon MoFe protein binding, and potentially, also encompasses a change in cluster oxidation state. Our results indicate that no specific carrier proteins or factors are absolutely required to “escort” the cluster from NifEN to MoFe protein in vitro. Therefore, it is possible that the respective cluster binding sites within NifEN and the MoFe proteins are brought into close vicinity by interactions between the two proteins thereby facilitating the “diffusion” of the cluster between the two binding sites without additional assistance. The proposed diffusion reaction in which MoFe protein interacts with its FeMoco donor may have important implications for the structure of MoFe protein in different states and its mechanism for protecting its air-sensitive clusters. Furthermore, the observed plasticity of Mo in a FeMoco-like structure, and the changes observed with different spin states may provide insights into the structural changes that take place in FeMoco during catalysis. Thus, the results presented herein not only significantly clarify the process of FeMoco maturation; they may also prove useful for understanding other facets of nitrogenase chemistry.
The required components for FeMoco maturation have been identified, but other components are likely required to facilitate this process. For example, if the FeMoco assembly proceeds through an [8Fe-9S] cluster, as has been proposed (18), then there will be a need for a component to remove and sequester toxic Fe upon Mo incorporation into the cluster. Further research is needed to continue to fine-tune the understanding of the final step of FeMoco biosynthesis.
Materials and Methods
Unless otherwise noted, all chemicals and reagents were obtained from Fisher, Aldrich, or Sigma. Cell growth, protein purification, preparation of apo Fe protein, FeMoco maturation assay, metal analysis, visible region absorption, EPR and x-ray spectroscopies were performed as described (13, 17, 18, 24, 33, 34). See Supporting Text, which is published as supporting information on the PNAS web site, for more information on these procedures.
FeMoco Maturation on NifEN.
To monitor FeMoco maturation while it is bound to NifEN, FeMoco precursor-bound NifEN was subjected to FeMoco maturation assays (17) with modified conditions, repurified, and subsequently examined in MoFe protein reconstitution assays (below). Such repurified NifEN proteins are categorically designated NifEN′, with different superscripts indicating different assay compositions. The following NifEN′ proteins were prepared. (i) NifENcomplete. This assay contained, in a 50 ml total volume, 25 mM Tris·HCl (pH 8.0), 2 mM Na2S2O4, 100 mg of FeMoco precursor containing NifEN, 120 mg of Fe protein, 0.4 mM homocitrate, 0.4 mM Na2MO4, 2.4 mM ATP, 4.8 mM MgCl2, 30 mM creatine phosphate, and 24 units/ml of creatine phosphokinase. This mixture was stirred for 1 h at 30°C and then NifEN was repurified as described (17). Note that the ΔnifB MoFe protein was omitted from the assay to allow for accumulation of processed FeMoco on NifENcomplete. (ii) ΔnifB NifENcomplete. This was a control assay with the same conditions as i except that NifEN was replaced by precursor-free ΔnifB NifEN (17). (iii) NifENminus Mo/homocitrate, NifENminus homocitrate, NifENminus Mo, NifENminus MgATP, or NifENminus Fe protein. These were control assays with the same assay conditions as i except that one or two of the components required for FeMoco maturation were omitted. (iv) NifENapo Fe protein, NifENA157 Fe protein, NifENM156C Fe protein, or NifENA157G Fe protein. These were control assays with the same conditions as i except that wild-type Fe protein was replaced by 120 mg of apo, A157S, M156C, or A157G Fe proteins, respectively. (v) NifENMgADP, NifENATPγS or NifENAMPPNP. These were control assays with the same conditions as i except that ATP was replaced by 2.4 mM ADP, adenosine 5′-O-(3-thiotriphosphate) (ATPγS) or 5′-adenylylimidodiphosphate (AMPPNP). Creatine phosphate and creatine phosphokinase were omitted when the function of ADP was evaluated. Table 3 summarizes the designations of repurified NifEN′ and the assay conditions used to obtain these proteins.
MoFe Protein Reconstitution with NifEN′.
To test whether the NifEN′ proteins (above) contain mature FeMoco, the following assay was designed to reconstitute the ΔnifB MoFe protein. Such an assay contained, in a 0.8 ml total volume, 25 mM Tris·HCl (pH 8.0), 20 mM Na2S2O4, 0.5 mg of purified ΔnifB MoFe protein from A. vinelandii strain DJ1143 (32). FeMoco insertion was initiated with the addition of 2 mg of isolated NifEN′ to the mixture above. The reaction mixture was incubated and stopped as described for the FeMoco maturation assay (17). The enzymatic activities were subsequently determined as described (35).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM-67626 (to M.W.R.) and RR-01209 (to K.O.H.). The Stanford Synchrotron Radiation Laboratory (SSRL) is supported by the Department of Energy (DOE) Office of Basic Energy Sciences, and the SSRL X-Ray Absorption facilities are supported by DOE Office of Biological and Environmental Research and National Institutes of Health National Center for Research Resources Biotechnology Training Program.
Abbreviations
- FeMoco
FeMo cofactor
- EXAFS
extended x-ray absorption fine structure
- IDS
indigo disulfonate
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Burgess BK, Lowe DJ. Chem Rev. 1996;96:2983–3011. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Howard JB, Rees DC. Chem Rev. 1996;96:2965–2982. doi: 10.1021/cr9500545. [DOI] [PubMed] [Google Scholar]
- 3.Smith BE. Adv Inorg Chem. 1999;47:159–218. [Google Scholar]
- 4.Rees DC, Tezcan FA, Haynes CA, Walton MY, Andrade S, Einsle O, Howard JB. Philos Trans R Soc A. 2005;363:971–984. doi: 10.1098/rsta.2004.1539. [DOI] [PubMed] [Google Scholar]
- 5.Christiansen J, Dean DR, Seefeldt LC. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:269–295. doi: 10.1146/annurev.arplant.52.1.269. [DOI] [PubMed] [Google Scholar]
- 6.Igarashi RY, Seefeldt LC. Crit Rev Biochem Mol Biol. 2003;38:351–384. doi: 10.1080/10409230391036766. [DOI] [PubMed] [Google Scholar]
- 7.Seefeldt LC, Dance IG, Dean DR. Biochemistry. 2004;43:1401–1409. doi: 10.1021/bi036038g. [DOI] [PubMed] [Google Scholar]
- 8.Peters JW, Szilagyi RK. Curr Opin Chem Biol. 2006;10:1–8. doi: 10.1016/j.cbpa.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Peters JW, Stowell MHB, Soltis SM, Finnegan MG, Johnson MK, Rees DC. Biochemistry. 1997;36:1181–1187. doi: 10.1021/bi9626665. [DOI] [PubMed] [Google Scholar]
- 10.Einsle O, Tezcan FA, Andrade SLA, Schmid B, Yoshida M, Howard JB, Rees DC. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 11.Dos Santos PC, Dean DR, Hu Y, Ribbe MW. Chem Rev. 2004;104:1159–1173. doi: 10.1021/cr020608l. [DOI] [PubMed] [Google Scholar]
- 12.Rubio LM, Ludden PW. J Bacteriol. 2005;187:405–414. doi: 10.1128/JB.187.2.405-414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribbe MW, Hu Y, Guo M, Schmid B, Burgess BK. J Biol Chem. 2002;277:23469–23476. doi: 10.1074/jbc.M202061200. [DOI] [PubMed] [Google Scholar]
- 14.Corbett MC, Hu Y, Naderi F, Ribbe MW, Hedman B, Hodgson KO. J Biol Chem. 2004;279:28276–28282. doi: 10.1074/jbc.M403156200. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y, Corbett MC, Fay AW, Webber JA, Hedman B, Hodgson KO, Ribbe MW. Proc Natl Acad Sci USA. 2005;102:13825–13830. doi: 10.1073/pnas.0506967102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SC, Holm RH. Chem Rev. 2004;104:1135–1158. doi: 10.1021/cr0206216. [DOI] [PubMed] [Google Scholar]
- 17.Hu Y, Fay AW, Ribbe MW. Proc Natl Acad Sci USA. 2005;102:3236–3241. doi: 10.1073/pnas.0409201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbett MC, Hu Y, Fay AW, Ribbe MW, Hedman B, Hodgson KO. Proc Natl Acad Sci USA. 2006;103:1238–1243. doi: 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson AC, Burgess BK, Dean DR. J Bacteriol. 1986;166:180–186. doi: 10.1128/jb.166.1.180-186.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen RM, Chatterjee R, Ludden PW, Shah VK. J Biol Chem. 1995;270:26890–26896. doi: 10.1074/jbc.270.45.26890. [DOI] [PubMed] [Google Scholar]
- 21.Goodwin PJ, Agar JN, Roll JT, Roberts GP, Johnson MK, Dean DR. Biochemistry. 1998;37:10420–10428. doi: 10.1021/bi980435n. [DOI] [PubMed] [Google Scholar]
- 22.Roberts GP, Brill WJ. J Bacteriol. 1980;144:210–216. doi: 10.1128/jb.144.1.210-216.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brigle KE, Weiss MC, Newton WE, Dean DR. J Bacteriol. 1987;169:1547–1553. doi: 10.1128/jb.169.4.1547-1553.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Corbett MC, Fay AW, Webber JA, Hodgson KO, Hedman B, Ribbe MW. Proc Natl Acad Sci USA. 2006:17125–17130. doi: 10.1073/pnas.0602651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madden MS, Kindon ND, Ludden PW, Shah VK. Proc Natl Acad Sci USA. 1990;87:6517–6521. doi: 10.1073/pnas.87.17.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christiansen J, Tittsworth RC, Hales BJ, Cramer SP. J Am Chem Soc. 1995;117:10017–10024. [Google Scholar]
- 27.Conradson SD, Burgess BK, Newton WE, Hodgson KO, McDonald JW, Rubinson JF, Gheller SF, Mortenson LE, Adams MWW, et al. J Am Chem Soc. 1985;107:7935–7940. [Google Scholar]
- 28.Kutzler FW, Scott RA, Berg JM, Hodgson KO, Doniach S, Cramer SP, Chang CH. J Am Chem Soc. 1981;103:6083–6088. [Google Scholar]
- 29.Conradson SD, Burgess BK, Newton WE, Mortenson LE, Hodgson KO. J Am Chem Soc. 1987;109:7507–7515. [Google Scholar]
- 30.Liu HI, Filipponi A, Gavini N, Burgess BK, Hedman B, Di Cicco A, Natoli CR, Hodgson KO. J Am Chem Soc. 1994;116:2418–2423. [Google Scholar]
- 31.Nordlander E, Lee SC, Cen W, Wu ZY, Natoli CR, Di Cicco A, Filipponi A, Hedman B, Hodgson KO, Holm RH. J Am Chem Soc. 1993;115:5549–5558. [Google Scholar]
- 32.Schmid B, Ribbe MW, Einsle O, Yoshida M, Thomas LM, Dean DR, Rees DC, Burgess BK. Science. 2002;296:352–356. doi: 10.1126/science.1070010. [DOI] [PubMed] [Google Scholar]
- 33.Clark LJ, Axley JH. Anal Biochem. 1955;27:2000–2003. [Google Scholar]
- 34.Van de Bogart M, Beinert H. Anal Biochem. 1967;20:325–334. doi: 10.1016/0003-2697(67)90038-3. [DOI] [PubMed] [Google Scholar]
- 35.Ribbe MW, Burgess BK. Proc Natl Acad Sci USA. 2001;98:5521–5525. doi: 10.1073/pnas.101119498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.