Abstract
Organisms use the daily cycles of light and darkness to synchronize their internal circadian clocks with the environment. Because they optimize physiological processes and behavior, properly synchronized circadian clocks are thought to be important for the overall fitness. In Drosophila melanogaster, the circadian clock is synchronized with the natural environment by light-dependent degradation of the clock protein Timeless, mediated by the blue-light photoreceptor Cryptochrome (Cry). Here we report identification of a genetic variant, Veela, which severely disrupts this process, because these genetically altered flies maintain behavioral and molecular rhythmicity under constant-light conditions that usually stop the clock. We show that the Veela strain carries a natural timeless allele (ls-tim), which encodes a less-light-sensitive form of Timeless in combination with a mutant variant of the F-box protein Jetlag. However, neither the ls-tim nor the jetlag genetic variant alone is sufficient to disrupt light input into the central pacemaker. We show a strong interaction between Veela and cryptochrome genetic variants, demonstrating that the Jetlag, Timeless, and Cry proteins function in the same pathway. Veela also reveals a function for the two natural variants of timeless, which differ in their sensitivity to light. In combination with the complex array of retinal and extraretinal photoreceptors known to signal light to the pacemaker, this previously undescribed molecular component of photic sensitivity mediated by the two Timeless proteins reveals that an unexpectedly rich complexity underlies modulation of this process.
Keywords: F-box, polymorphism, photoreception
Most organisms live throughout the year in light/dark (LD) cycles. This natural fluctuation represents a crucial stimulus to adjust the internal circadian clocks to operate in synchrony with the environment (1). Exposure to constant light dramatically affects biological rhythms and molecules comprising the circadian clock in many organisms. In Drosophila, the chronic presence of light (constant light, called LL) usually results in behavioral arrhythmicity and a breakdown of molecular oscillations in the circadian clock (2–4). Although the experimental LL situation is artificial, mutations that abolish this LL effect define essential components of light-signaling pathways that synchronize the central pacemaker to the external world. So far, this has been shown for the Cryptochrome (Cry) mutations cryb and crym (5–7). Crys are related to photolyases, blue-light photoreceptors that use harvested light energy to repair UV-damaged DNA (8). In animals and plants, Cry proteins have been shown to function in the circadian system as photoreceptors, clock factors, or both (8, 9).
Opsin-mediated retinal, extraretinal, and Cry-independent photoreception contributes to light synchronization of the circadian clock in Drosophila (5, 10, 11). However, the main entrainment pathway is believed to involve light-dependent Cry and Timeless (Tim) and perhaps Period (Per) interactions within the behavioral pacemaker neurons of the fly brain (7, 12, 13). Upon light activation, Cry is thought to undergo a conformational change that allows it to bind to Tim in a way that irreversibly targets this clock protein for degradation by the proteasome (7, 14, 15). This light-induced degradation of Tim is crucial for molecular and behavioral clock resetting. If Tim is degraded prematurely by light pulses given at the end of the night, as a consequence the molecular feedback loops comprising the circadian clock and regulating rhythmic locomotor behavior are phase-advanced. Vice versa, the clock reacts with phase delays in case Tim is degraded by light exposure in the early night (reviewed, for example, in ref. 16).
By characterizing the genetic variant Veela, which, like cry mutants, behaves abnormally rhythmically in constant light, we identified a factor involved in the Cry-dependent light-input pathway of Drosophila. Veela genetically interacts with cryb and shows decreased light sensitivity of Tim degradation. We demonstrate that these effects are caused by the simultaneous presence of a natural (less light-sensitive) form of Tim and a mutation in the F-box protein Jetlag (Jet). Importantly, the same jetlag(jet) mutant in combination with another natural and common variant of Tim behaves like wild type (WT). Therefore, previous findings attributing observed light-input defects solely to mutations in the jet gene (17) need to be revised.
Results
Isolation and Initial Mapping of Veela.
During behavioral analysis of potential light-synchronization mutants in D. melanogaster, we identified a strain that exhibits robust rhythmicity in LL (Fig. 1 1and Table 1). The variant mapped to chromosome 2 and, because of its elusive nature (see Supporting Text, which is published as supporting information on the PNAS web site), was named Veela (18). The mutant showed a semidominant effect: ≈30% of Veela/+ flies exhibited weak rhythmicity in LL (Table 1). Behavior in constant darkness was not abnormal (Table 2, which is published as supporting information on the PNAS web site). Mapping by meiotic recombination placed Veela between the aristaless (al) and dumpy (dp) markers, close to the clock gene timeless (refs. 19 and 20; see also Materials and Methods).
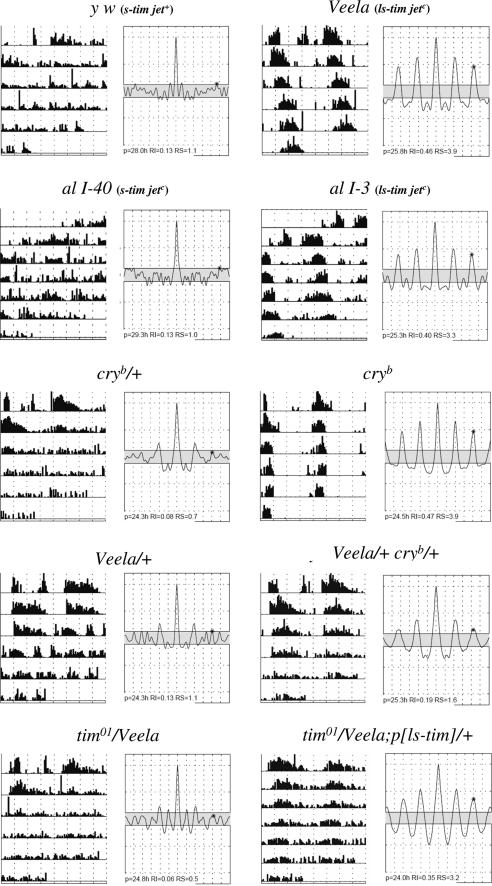
Fig. 1.
Locomotor activity of individual flies during constant light. Genotypes are indicated on top of each actogram accompanied by its corresponding autocorrelogram. Actograms show the raw activity of each fly, in which the height of each bar indicates the amount of locomotion during a 30-min interval. Autocorrelograms show period and rhythmicity index values of each fly as an objective way to determine rhythmicity (see Materials and Methods). The upper four flies also have their polymorphisms in regard to the tim and jet variants indicated (see text). Flies to the left are arrhythmic; individuals to the right are rhythmic. al I-40 and al I-3 each designate recombinants resulting from meiotic crossing over between two second chromosomes in females heterozygous for Veela and the marker combination al dp b pr. Both the I-40 and the I-3 recombinants carry the al marker and the jetc variant but differ with respect to tim. Note that al I-40 flies behaved arrhythmically in LL (like the controls), although they carry jetc.
Table 1.
Locomotor activity rhythms in constant light
| Genotype | NT | nRhy | Period | RI | % Rhy |
|---|---|---|---|---|---|
| y w | 70 | 2 | 22.3 | 0.30 | 3 |
| wild-type Athens | 8 | 0 | — | — | 0 |
| cryb/cryb | 22 | 21 | 24.3 ± 0.1 | 0.33 ± 0.03 | 96 |
| cryb/+ | 23 | 1 | 27.5 | 0.30 | 4 |
| Veela/Veela | 75 | 64 | 25.7 ± 0.1 | 0.33 ± 0.02 | 85 |
| Veela/+ | 35 | 11 | 24.4 ± 0.4 | 0.18 ± 0.01 | 31 |
| Veela/+ cryb/+ | 26 | 20 | 25.9 ± 0.6 | 0.24 ± 0.02 | 77 |
| al I-3/al I-3 (ls-tim jetc) | 10 | 9 | 26.2 ± 0.6 | 0.33 ± 0.04 | 90 |
| dpp 20/dpp 20 (ls-tim jetc) | 8 | 6 | 25.9 ± 0.3 | 0.20 ± 0.04 | 75 |
| dp b pr I-28/dp b pr I-28 (ls-tim jetn) | 13 | 1 | 25.8 | 0.27 | 7 |
| dpp ed 23/dpp ed 23 (s-tim jetc) | 8 | 0 | — | — | 0 |
| al I-40/al I-40 (s-tim jetc) | 12 | 2 | 24.2 | 0.16 | 16 |
| al I-40/al I-40 | 40 | 2 | 23.2 | 0.25 | 5 |
| tim01/+ | 24 | 0 | AR | — | 0 |
| Veela/tim01 | 42 | 13 | 25.9 ± 1.1 | 0.21 ± 0.02 | 22 |
| tim01/tim01; P[ls-tim]/P[ls-tim] | 14 | 1 | 25 | 0.20 | 7 |
| Veela/tim01; P[ls-tim]/+ | 46 | 29 | 24.6 ± 0.2 | 0.25 ± 0.02 | 63 |
NT, total number of flies tested; nRhy, number of individuals behaving rhythmically in LL. Period, free-running cycle durations (hr ± SEM), determined by autocorrelation; RI, Rhythmicity Index, indicating the significance level associated with a given period (see Materials and Methods); % Rhy, percentage of rhythmic flies of a given genotype. Male flies of the indicated genotypes were analyzed. As controls, flies with the X-chromosomal body- and eye-color markers y and w were used (Materials and Methods). The y w strain used here carries the s-tim allele (Fig. 4). The wild-type Athens strain carries the ls-tim allele. Genotypes containing a digit in conjunction with a recessive marker designation (e.g., al I-40) specify recombinants obtained from crossing homozygous Veela flies to multimarker chromosomes (Materials and Methods). The P[ls-tim] transgene is inserted on chromosome 3 and contains the ls-tim version of timeless including tim promoter sequences (25).
Veela Genetically Interacts with cryb and Stabilizes Tim in the Light.
Photic responsiveness of Tim is mediated by its light-induced interaction with Cry, resulting in rapid degradation of Tim, thus representing a crucial mechanism by which Drosophila's clock synchronizes to LD cycles (5, 7, 21). Because a mutation in the cryptochrome gene (cryb) also results in anomalous rhythmicity in LL (ref. 6; see also Fig. 1 and Table 1), we reasoned that Veela may interfere with the Tim–Cry interaction, perhaps, given our mapping results, because of a mutation in tim itself. To examine this possibility, we analyzed the behavior of flies that carried one copy of cryb in combination with one copy of Veela. Strikingly, these double heterozygotes exhibited robust rhythmicity in LL (Fig. 1 and Table 1). Because this phenotype is never observed in heterozygous cryb/+ flies and is much stronger compared with that of Veela/+ flies, our results indicate a strong genetic interaction between Veela and cryb (Fig. 1 and Table 1).
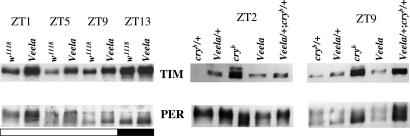
These findings suggested that Veela impairs light inputs to the clock because of interference with the usual Tim–Cry interaction. If true, light-induced degradation of Tim should be affected in Veela flies, and we therefore analyzed Tim abundance during the light portion of the LD cycle [during Zeitgeber times (ZT)0–12; Fig. 2], during which Tim levels are normally very low (5). As expected, Tim abundance in head protein extracts was increased in Veela flies (compared with controls monitored between ZT1 and ZT9), whereas we did not see any significant differences during the night (ZT13; Fig. 2). Next, we analyzed Tim levels in Veela/+ cryb/+ flies, which also showed robust behavioral rhythms in LL (Fig. 1 and Table 1). Here, too, the doubly heterozygous flies showed increased Tim levels at ZT2 and ZT9 compared with both kinds of singly heterozygous controls (Fig. 2). Importantly, Tim signals in the Veela/+ cryb/+ extracts were stronger compared with those of homozygous Veela flies and more similar to those of homozygous cryb flies. Thus, at the molecular level as well as behaviorally, Veela and Cry interact robustly (Figs. 1 and 2). We have no explanation for why Tim signals in the heterozygous Veela/+ flies were stronger compared with those in homozygous Veela flies. Similar effects of Veela and of the combination of Veela with cryb were observed for the Per protein (Fig. 2 Lower), which is thought to be stabilized by heterodimerization with Tim (16).
Fig. 2.
Veela stabilizes Tim during the light portion of LD cycles. Western blots of head extracts from flies collected during LD cycles. Genotypes and time points of collection (ZT) are indicated above the blots. (Upper) Anti-Tim. (Lower) Anti-Per. Below the left blots, white and black bars indicate when the lights were on and off, respectively. Veela and cryb have similar effects on stabilization of Tim as on Per. Heterodimerization of both proteins has been proposed to stabilize Per (16) and is probably the reason for increased levels of Per in Veela, cryb and Veela/+ cryb/+ flies.
Tim and Per Are Rhythmically Expressed in the Pacemaker Neurons and Glia Cells of Veela Flies in Constant Light.
Rhythmic locomotor activity is driven by clock gene expression within certain neurons of the fly brain (22). Based on their location within the lateral and dorsal brain, respectively, these neurons are historically divided into lateral neurons (LNs) and dorsal neurons (DNs): three groups of both LNs [small (s-LNvs), large (l-LNvs), and dorsal LNs] and DNs (cell groups 1–3; ref. 22). Because Veela individuals behave rhythmically in LL, we asked which subset of the clock neurons would drive this behavior or whether all neurons would be affected equally by this genetic variant. To answer this question, we stained whole-mounted brains of Veela adults during the second day in LL with anti-Tim and -Per. We observed robust rhythmic expression of Tim and Per in all clock-neuronal cell types, indicating that Veela disrupts light inputs into all six groups (Fig. 3). In all of these LNs and DNs, peak expression of both Tim and Per were observed at the end of the subjective night (a term for the second half of L in an LL “cycle”) through the beginning of the subjective day, demonstrating synchronized expression of clock genes among these cells. As expected, in the LNs of control flies that are behaviorally arrhythmic after 2 days in LL (Fig. 1), no rhythmic expression or accumulation of the two clock proteins was observed, indicating light-induced Cry- and Veela+-mediated constitutive degradation of Tim (Fig. 3B). A noticeable exception in the controls involved the l-LNv cells, which showed peaks of Tim and Per signals at the end of the subjective day and an additional Per peak early in the subjective day (Fig. 3B). Interestingly, Tim and Per signals in the l-LNs of y w controls flies were always cytoplasmic, perhaps explaining why this coordinated clock protein expression in the l-LNvs is not able to drive rhythmic behavior (see Fig. 3A Inset and B; see also Fig. 1).
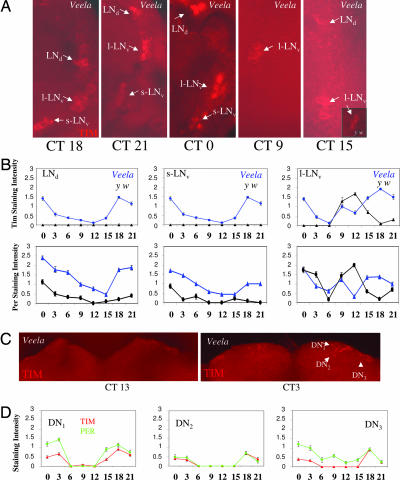
Fig. 3.
Rhythmic Tim and Per expression in constant light in clock neurons of Veela flies. Control and Veela flies were synchronized to 12-h:12-h LD cycles and subsequently released into LL. After 2 days, males were killed at the indicated circadian times (CTs; below the images or x axis), and whole-mounted brains were stained with anti-Tim and -Per. (A) Rhythmic Tim immunoreactivity during LL in all LN of Veela flies and in the cytoplasm of the l-LNvs of y w controls (see Inset, CT15). (B) Quantifications of anti-Tim (A) and anti-PER stainings, including y w control flies. Note that controls show rhythmic Tim and Per accumulation in the l-LNv. Error bars indicate SEM. (C) Rhythmic Tim accumulation in the DNs of Veela flies during the second day of constant light. (D) Quantification of Per and Tim immunoreactivity in DNs of Veela flies during LL. No staining was observed in y w control flies under these conditions. Between 12 and 20 (y w) or 16 and 25 (Veela) brain halves for each time point were analyzed. Error bars indicate SEM.
Rhythmic and prominent Tim and especially Per expression in LL was also observed in glia cells of the medulla optic lobe in Veela flies (Fig. 6, which is published as supporting information on the PNAS web site). cryb mutant flies also behave rhythmically in LL, and consequently rhythmic accumulation of Per and Tim occurred in the LNs of this mutant in LL (Fig. 7, which is published as supporting information on the PNAS web site). We did not observe rhythmic or significant Tim accumulation within glia cells of cryb flies, although Per levels did cycle (Fig. 6).
Veela Flies Express a Less-Light-Sensitive Form of Tim and Carry a Mutation in the jet Gene.
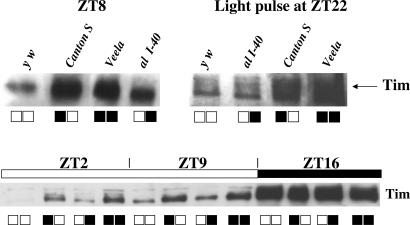
Because Veela mapped to the same genetic interval as tim and genetically interacts with cry (Figs. 1 and 2, Table 1, and Materials and Methods), we sequenced the ORF of tim in the Veela strain but did not find any changes compared with several published WT tim sequences (data not shown). We did notice, though, that the tim form (ls-tim) in the Veela strain encodes both the “long” and “short” forms of Tim, in which the longer form contains 23 additional amino acids at its N terminus by use of alternative translation-start codons (ref. 23; Fig. 4). This ls-tim variant is common to many strains of D. melanogaster, as is exclusive production of the short form (s-tim) for other naturally occurring WTs (ref. 23; F. Sandrelli, E. Tauber, M. Pergorano, G. Mazotta, P. Cisotto, et al., unpublished results). We performed Western blot analysis to see whether there are any differences in regard to Tim expression between the ls-tim and s-tim variants. Tim protein levels in ls-tim flies collected during the day were substantially higher compared with s-tim (Fig. 5). This was also the case when flies with WT eye color were compared (Fig. 5 Lower). Therefore, missing pigments in the white-eyed y w flies are not the cause of increased light exposure and degradation of Tim. The same was true when both genotypes where exposed to a light pulse late at night (Fig. 5). Because Tim levels were similar in both genetic variants during the night portion of flies kept in LD cycles (Fig. 5 Lower), we conclude that the ls-tim allele produces a less-light-sensitive form of Tim. In a recent study, we showed that this difference is caused by a reduced affinity of “long-Tim” to CRY (F. Sandrelli, E. Tauber, M. Pergorano, G. Mazotta, P. Cisotto, et al., unpublished results). However, because flies of the ls-tim type do become arrhythmic in LL (Figs. 1 and 4 and Table 1; ref. 2), there must be an additional light-input defect in Veela that further stabilizes Tim.
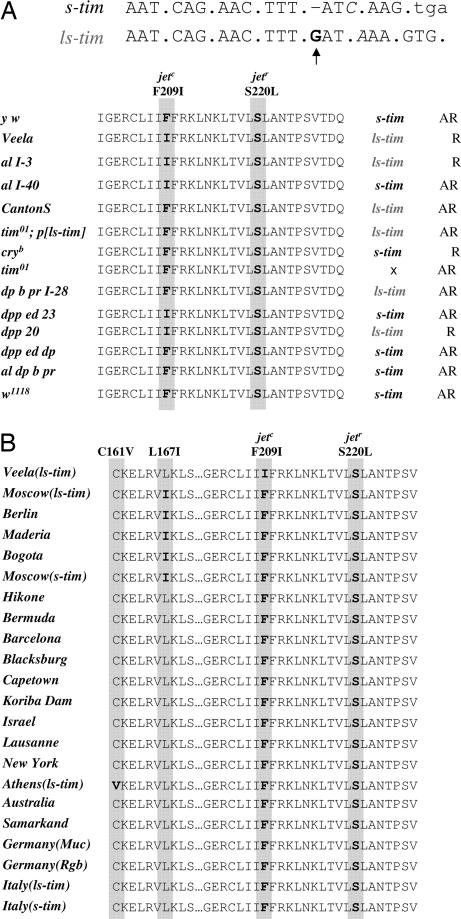
Fig. 4.
tim and jet polymorphisms in Veela and other fly strains. (A) (Upper) Nucleotide sequence of the two tim polymorphisms. A deletion of the G nucleotide at position 294 of the tim cDNA (19) results in the generation of a stop codon immediately 5′ of the translational start of s-tim (23). Note the additional polymorphism three nucleotides downstream of the G deletion (indicated in italics). Fly lines carrying the single base-pair G deletion produce only the short (more light-sensitive) form of Tim. (Lower) Amino acid residues of the fourth and fifth LRR domains of the Jet protein. Highlighted in gray and bold are positions 209 and 220, respectively, which carry the common (F209I) or rare (S220L) polymorphisms (17). Genotypes expressing the respective forms are indicated to the left; their associated tim polymorphism along with the LL-behavioral phenotype is indicated to the right (R, rhythmic; AR, arrhythmic in LL). Meiotic recombinants are listed with their second chromosomal markers and a numerical indicator (e.g., al I-3). (B) Jet polymorphisms in WT fly strains. Displayed are the amino acid residues of the fourth and fifth LRR domains of the Jet protein from different WT fly strains. The fly strains' origin is shown on the left along with the nature of the tim allele, if determined (see Materials and Methods for details). Natural polymorphisms occurring at positions 161 and 167 are highlighted in gray and bold (Cys to Val and Leu to Ile, respectively). None of the WT strains carried the jetc or jetr variant.
Fig. 5.
ls-tim flies express a less-light-sensitive form of Tim, which is independent of jet. Flies of the indicated genotypes (Upper) were raised in 12-h:12-h LD cycles and either collected at the indicated ZT or subjected to a 2-min light pulse at ZT22 and collected at ZT23. Below each lane, the respective allele of tim (left square) and jet (right square) is indicated for each genotype. Open squares indicate s-tim (Left) and jet+ (Right), black squares indicate ls-tim and jetc. (Lower) Four genotypes, from left to right: Italy (s-tim); Canton S; al I-40; b I-27), all flies had WT eye color. Note that the presence of ls-tim leads to a drastic increase in Tim levels, irrespective of jet or eye color.
During our mapping experiments, we learned that a genetic variant with a similar LL phenotype to that of Veela is located close to our mutant but mapped to the right of dp instead (17). Koh et al. (17) mapped the phenotype to a gene (CG8873, now called jetlag) that encodes an F-box protein with leucine-rich repeats (LRR). Although Veela mapped to the other side (leftward) of dp, we decided to sequence the jetlag (jet) gene in Veela mutant flies because of the similar phenotype of the two mutant strains. Koh et al. (17) found four different variants of jet in various fly stocks that exist in unknown frequencies in various fly strains of D. melanogaster. The variants that correlated with the LL-rhythmic behavioral phenotype were reported to involve a phenylalanine-to-isoleucine substitution in one LRR domain or a serine-to-leucine replacement in the neighboring LRR (ref. 17; Fig. 4). Because they found the former substitution in six of seven behaviorally mutant stocks, the Ile variant was named the common (c) mutation, and the Leu isoform was named the rare (r) mutation. The WT variants would apparently encode Phe and Ser at these positions within Jet; however, the strain frequency for these alleles of jet was not determined (17). Our sequence analysis of this region in Veela revealed that its jet variant belongs to the c variant type. No other coding changes were found in the jet gene of Veela flies compared with the published WT sequence, suggesting that the LL-rhythmic phenotype of Veela is solely caused by this particular jetc variant.
jetc Flies Behave Normally in Constant Light When They also Carry the s-tim Allele.
Our meiotic mapping placed Veela to the left of dp (and jet). Crucially, two of our genetic recombinants (generated with two different marker chromosomes) carried the c form of the jet gene but did not show any phenotype in the LL assay; their behavior was indistinguishable from WT controls (Figs. 1 and 4, Table 1, and Materials and Methods). These behavioral-genetic results unequivocally demonstrate that the c variant alone is not sufficient to block light input into the circadian clock. Instead, careful inspection of all our genetic recombinants (n = 26) revealed that the presence of both the ls-tim form and the jetc variant was correlated one to one with abnormally rhythmic behavior in LL (n = 11; Fig. 4 and see Materials and Methods). Because our two mapping stocks carry the s-tim form (Fig. 4 and see Materials and Methods), this explains why we were not able to map Veela to the jet locus per se; the relevant meiotic crossovers would have linked (and did link, in two cases) s-tim with jetc, and this combination is phenotypically normal (Figs. 1 and 4, Table 1, and Materials and Methods).
In theory, it would be possible that another factor instead of ls-tim is responsible for the light-response defect in combination with jetc. To firmly establish that the presence of the less-light-sensitive form of Tim is required, we compared the phenotypes of heterozygous jetc flies, expressing either one copy of ls-tim and one copy of s-tim (Veela/+) or two copies of ls-tim, whereby one is carried on a transgene (Veela/tim01;P[ls-tim]; ref. 24, Fig. 1, and Table 1). Strikingly, the presence of two copies of ls-tim resulted in doubling the proportion of LL-rhythmic individuals (31–63%; Table 1), further demonstrating that both the form of Tim associated with reduced light sensitivity (Fig. 5; F. Sandrelli, E. Tauber, M. Pergorano, G. Mazotta, P. Cisotto, et al., unpublished results) and jetc must be present to block light input into the circadian clock.
jetc and jetr Variants Are Most Likely Not Natural Polymorphisms.
We investigated the possibility that the jet variants described here and in a previous study (17) represent a natural polymorphism as described for the s-tim and ls-tim alleles (ref. 23; F. Sandrelli, E. Tauber, M. Pergorano, G. Mazotta, P. Cisotto, et al., unpublished results). To this end, we sequenced genomic DNA from 15 WT stocks available from the stock centers as well as from flies collected at five locations in central Europe (F. Sandrelli, E. Tauber, M. Pergorano, G. Mazotta, P. Cisotto, et al., unpublished results; Fig. 4). No strains carrying the jetc or jetr variants were identified, but in one case (WT Athens), a base-pair change leading to a single amino change (Cys to Val) at position 161 was identified (Fig. 4). Moreover, several strains harbored a conservative Leu to Ile change at position 167. We analyzed the locomotor behavior of these variants in LL conditions, but their behavior was indistinguishable from other WT controls (Table 1 and data not shown). Although we cannot rule out that the substitutions have subtle consequences for the light sensitivity of the circadian clock, which are not detected in our LL assay, our results indicate they do not grossly alter the function of the Jet protein. Given that we did not identify any WT flies carrying either the jetc or jetr alleles, we conclude that both mutations occurred independently and spontaneously in laboratory stocks or were coinduced along with other mutations by chemical mutagenesis. Nevertheless, because we found two new variants in the region harboring the jetc and jetr polymorphisms, we cannot rule out that, in other natural strains, additional base-pair changes have occurred in different parts of the jet gene, which might more drastically affect the Jet protein and ultimately the light sensitivity of Tim.
Discussion
We identified a genetic variant, Veela, that is abnormally rhythmic in constant light, similarly as is shown for mutations affecting the blue-light photoreceptor Cry (6, 7, 10). Veela's phenotype is due to the simultaneous presence of the ls-tim allele (encoding a less-sensitive form of Tim) and the jetc variant encoding a mutant form of the F-box protein Jet (17). We show that Veela genetically and molecularly interacts with cryb, indicating that Tim, Jet, and Cry function in the same circadian light-synchronization pathway. Our findings show that additional factors are necessary to elicit the phenotypes previously associated with jet variants (17). In particular, we show that only when jetc is linked to the ls-tim allele, which encodes a less-light-sensitive form of Tim, can abnormal behavioral rhythmicity in LL be observed. The importance of the Jet protein per se in the light-entrainment process remains unclear, also when considering certain aspects of the original jet study in conjunction with the findings presented here. All control flies used by Koh et al. (17) came from a y w genetic background (see Supporting Text), which we show here carries the s-tim allele (Fig. 4). Contrarily, all jetc or jetr mutant flies carried the ls-tim allele (necessarily; otherwise, they would have behaved like WT). It follows that behavioral and molecular differences between control and mutant flies reported by Koh et al. (17) in fact reflect the combined effects of ls-tim (vs. s-tim) and jetc (vs. jet+). In conjunction with our Western blot data showing an increased jet-independent stability of the larger Tim form compared with the smaller one (Fig. 5), it seems that the effects on Tim degradation previously attributed to jet variants are mainly a reflection of the different features of the two Tim proteins. This may also explain why Koh et al. (17) saw only very subtle effects of their mutant Jet proteins on Tim degradation in vitro.
Nevertheless, it is clear that jet influences the light-input pathway of the circadian clock; WT flies behave arrhythmically in LL, even though they carry ls-tim. Moreover, Veela strongly interacts with Cry, a crucial protein for circadian light input in flies. Importantly, our findings reveal that, with the current knowledge, an in vivo function for jet's F-box protein can be demonstrated only when the available jet variants are combined with ls-tim. To ultimately resolve the specific function of the Jet protein in the light-input pathway, loss-of-function jet mutants (25) or specific RNAi transgenics need to be generated and analyzed chronobiologically (26).
Characterization of Veela also led to the assignment of a biological function for the two natural tim variants that were identified many years ago (23). We show that Tim encoded by the ls-tim allele is more stable after light exposure, and that this increased stability has behavioral consequences when flies are exposed to constant light; if the ls-tim allele is linked to jetc, these flies behave abnormally rhythmically in LL. If jetc is linked to s-tim, the flies behave like WT and become arrhythmic in LL. Therefore, the less-light-sensitive Tim form encoded by ls-tim is necessary and sufficient to block light input into the circadian clock of jetc flies. In nature, the natural polymorphism at the tim (and perhaps jet) locus might be used to fine-tune the light sensitivity of Drosophila's circadian clock on a purely molecular level (F. Sandrelli, E. Tauber, M. Pergorano, G. Mazotta, P. Cisotto, et al., unpublished results). In conjunction with various anatomical light-input routes that are known to send light to Drosophila's circadian pacemaker (10, 27), our findings reveal a glimpse of the potential complexity of this process. The frequent and random occurrence of tim and jet variants in currently used laboratory strains also speaks to a more cautious strain selection and genotyping in all studies concerning light-input pathways to the circadian clock.
Materials and Methods
Fly Strains.
Stocks of D. melanogaster and chromosomal markers were as described (5, 20, 28). y Df (1)w (y w) flies have yellow body color and white eyes and were initially used as control flies. Because y w carries the s-tim allele, the WT strain Canton S (carrying ls-tim, like Veela flies) was also used as control during this study. The Veela variant was isolated from a stock containing the ninaB360d mutation (29) on chromosome 3 (which was replaced by a WT third chromosome in all Veela flies analyzed here). The ls-tim-encoding transgene contains the full-length tim cDNA (generated by PCR from WT Canton S flies) and 6 kb of genomic 5′-flanking material (24). The various WT strains that were used to identify potential natural polymorphisms in the jet gene were obtained from the Bloomington stock center. The four WT strains from Moscow and two locations in northern Italy are isofemale lines, which were generated from individuals collected in 1997 and 2004, respectively. From each of the three locations, a s-tim and ls-tim line was generated (F. Sandrelli, E. Tauber, M. Pergorano, G. Mazotta, P. Cisotto, et al., unpublished results). The two WT variants from Germany stem from individuals collected in 2006 in Regensburg and near Munich.
Behavioral Analysis.
Locomotor rhythms of individual male flies were recorded as described (5). Flies were kept for at least 3 days in 12 h/12 h LD cycles before being transferred to either constant light (300–400 lux LL) or to constant-dark conditions, in which they remained for at least 5 days. Rhythmicity was determined by using autocorrelation and Matlab software as described (30). Flies with period values in the circadian range and with a rhythmicity index value >0.15 were considered rhythmic (see ref. 30).
Chromosomal Mapping of Veela.
Rhythmic or arrhythmic behavior in LL conditions (indicating the presence or absence of Veela, respectively) was used to map the position of Veela on chromosome 2. Initially, homozygous Veela flies were crossed to the marker stock al (map position 0.4) dp (13.0) b (48.5) pr (54.5), all on the left arm of chromosome 2 (20). A total of 22 recombinants was obtained, and all b pr (n = 7), or single marker b (n = 1) and pr (n = 1) recombinants exhibited the Veela phenotype. All al dp (n = 4) dp b pr (n = 4) al dp b (n = 3) recombinants were WT, suggesting that Veela maps close to dp. The two al recombinants were WT (I-40) and Veela (I-3), respectively (Fig. 1 and Table 1), placing Veela between al and dp, the region that includes the tim locus (8.0; ref. 19). Sequence analysis revealed that Veela carries the ls-tim variant but no other changes in its ORF, which do not occur in other WT or laboratory stocks (Fig. 4 and data not shown). Because other stocks carrying ls-tim do not show the Veela phenotype, we continued our mapping experiments with a dpp (4.0) ed (11.0) dp (13.0) marker stock in an attempt to separate Veela from tim. This was not accomplished; of the four total recombinants obtained from Veela/dpp ed dp females, the one dpp recombinant showed the mutant phenotype (Table 1), so that Veela must map to the right of dpp. All dpp ed (n = 1) and ed dp (n = 2) flies were WT (Table 1 and data not shown), placing Veela between dpp and dp, again the region containing tim.
Sequence analysis of jet in Veela flies and several recombinants revealed a correlation between the jetc variant, ls-tim, and the mutant phenotype in all cases (n = 11; Fig. 4). Because our two mapping stocks express the s-tim variant along with jet+, our recombinants separated ls-tim from jetc resulting in LL-arrhythmic flies (Figs. 1 and 4; Table 1). Correct meiotic mapping of jetc would be possible only with both the marker stock and the jetc strain expressing ls-tim.
Western Blot Analysis.
Flies of the indicated genotypes were first kept in LD cycles for at least 3 days and collected on dry ice during the indicated ZT in LD. For the light-pulse experiment in Fig. 5, flies were raised identically but exposed to a 2-min light pulse (300–400 lux) at ZT22, allowed to recover for 1 h, and collected on dry ice at ZT23. Preparations of head extracts and protein blots were performed by using anti-PER and -TIM as well as dilutions of these antibodies, as described (5), except that for the blots shown in Fig. 5, a different anti-TIM antibody was applied (31).
Immunohistochemistry.
Flies were raised under the same conditions as described above and collected at the indicated circadian times (CTs) during the second day in LL (CTs refer to hours corresponding to ZT in the preceding LD cycles). Whole-mounted brains were prepared and incubated with anti-TIM and -PER, as described (32). Preparations were viewed by using Leica TCS NT (Leica, Deerfield, IL) and Zeiss Meta 510 (Zeiss, Oberkochen, Germany) confocal microscopes. Quantification of stainings was performed (observer blind with regard to genotype) by calculating a staining index, which reflects the number of immunoreactive cells and the staining intensity (on an arbitrary scale from 0 to 4), as described (32).
DNA Sequencing.
The tim gene of Veela flies was sequenced by using genomic DNA and reverse-transcribed RNA fragments, generated by PCR using the methods and oligonucleotides described to sequence the timblind mutant allele (33). To distinguish between s-tim and ls-tim in the various fly stocks, the following oligonucleotides were applied to amplify genomic DNA by PCR: 5′-GTGGTTGCGTAATGCCCTGG-3′ (sense) and 5′-GCACCGTCAGATTGACGA-3′ (antisense). Sequencing of jet genomic DNA was performed by using oligonucleotides 5′-TGGGATAGAAGTCGTTCAAGT-3 (sense) and 5′-TGCCGATGGCTAACAGAT-3′ (antisense) to determine the variants at the common and rare sites within two LRR-encoding domains. The remaining jet DNA sequence was determined by using genomic DNA from Veela flies and standard sequencing protocols.
Supplementary Material
Acknowledgments
We thank C. Helfrich-Förster (University of Regensburg) for help with staining analysis and figure preparations and Michael Rosbash (Brandeis University, Waltham, MA) and Isaac Edery (Rutgers University, Piscataway, NJ) for providing samples of anti-TIM. We thank A. Sehgal for communicating unpublished results and C. P. Kyriacou (University of Leicester, Leicester, U.K.) for WT flies from Moscow and Italy. A. Giesecke supplied comments on the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grant Sta 421/4-2 (to R.S.). N.P. and S.V. were supported by the Deutsche Forschungsgemeinschaft Graduate College 640/1 “Sensory Photoreceptors in Natural and Artificial Systems” grant. Our work is supported by EUCLOCK, an Integrated Project (FP6) funded by the European Commission.
Abbreviations
- LD
light/dark
- LL
constant light
- Cry
Cryptochrome
- Per
Period
- Tim
Timeless
- LN
lateral neurons
- DN
dorsal neurons
- l-LNv
large LN
- LRR
leucine-rich repeat
- ZT
Zeitgeber time.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ. Sunderland, MA: Sinauer; 2004. Chronobiology: Biological Timekeeping. [Google Scholar]
- 2.Konopka RJ, Pittendrigh C, Orr D. J Neurogenet. 1989;6:1–10. doi: 10.3109/01677068909107096. [DOI] [PubMed] [Google Scholar]
- 3.Price JL, Dembinska ME, Young MW, Rosbash M. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marrus SB, Zeng H, Rosbash M. EMBO J. 1996;15:6877–6886. [PMC free article] [PubMed] [Google Scholar]
- 5.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 6.Emery P, Stanewsky R, Hall JC, Rosbash M. Nature. 2000;404:456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- 7.Busza A, Emery-Le M, Rosbash M, Emery P. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- 8.Partch CL, Sancar A. Methods Enzymol. 2005;393:726–745. doi: 10.1016/S0076-6879(05)93038-3. [DOI] [PubMed] [Google Scholar]
- 9.Green CB. Curr Biol. 2004;14:R847–R849. doi: 10.1016/j.cub.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 10.Helfrich-Förster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. Neuron. 2001;30:249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- 11.Emery P, Stanewsky R, Helfrich-Förster C, Emery-Le M, Hall JC, Rosbash M. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 12.Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Kay SA. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 13.Rosato E, Codd V, Mazzotta G, Piccin A, Zordan M, Costa R, Kyriacou CP. Curr Biol. 2001;11:909–917. doi: 10.1016/s0960-9822(01)00259-7. [DOI] [PubMed] [Google Scholar]
- 14.Dissel S, Codd V, Fedic R, Garner KJ, Costa R, Kyriacou CP, Rosato E. Nat Neurosci. 2004;7:834–840. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- 15.Naidoo N, Song W, Hunter-Ensor M, Sehgal A. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- 16.Stanewsky R. J Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- 17.Koh K, Zheng X, Sehgal A. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowling JK. London, UK: Bloomsbury; 2000. Harry Potter and the Goblet of Fire. [Google Scholar]
- 19.Myers MP, Wager-Smith K, Wesley CS, Young MW, Sehgal A. Science. 1995;270:805–808. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- 20.Lindsley DL, Zimm GG. San Diego, CA: Academic; 1992. The Genome of Drosophila melanogaster. [Google Scholar]
- 21.Lin FJ, Song W, Meyer-Bernstein E, Naidoo N, Sehgal A. Mol Cell Biol. 2001;21:7287–7294. doi: 10.1128/MCB.21.21.7287-7294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang DC. Behav Processes. 2006;71:211–225. doi: 10.1016/j.beproc.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Rosato E, Trevisan A, Sandrelli F, Zordan M, Kyriacou CP, Costa R. Nucleic Acids Res. 1997;25:455–458. doi: 10.1093/nar/25.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutila JE, Maltseva O, Rosbash M. J Biol Rhythms. 1998;13:380–392. doi: 10.1177/074873098129000200. [DOI] [PubMed] [Google Scholar]
- 25.Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 26.Manoli DS, Baker BS. Nature. 2004;430:564–569. doi: 10.1038/nature02713. [DOI] [PubMed] [Google Scholar]
- 27.Rieger D, Stanewsky R, Helfrich-Förster C. J Biol Rhythms. 2003;18:377–391. doi: 10.1177/0748730403256997. [DOI] [PubMed] [Google Scholar]
- 28.Sehgal A, Price JL, Man B, Young MW. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 29.von Lintig J, Dreher A, Kiefer C, Wernet MF, Vogt K. Proc Natl Acad Sci USA. 2001;98:1130–1135. doi: 10.1073/pnas.031576398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine JD, Funes P, Dowse HB, Hall JC. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidote D, Majercak J, Parikh V, Edery I. Mol Cell Biol. 1998;18:2004–2013. doi: 10.1128/mcb.18.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veleri S, Brandes C, Helfrich-Förster C, Hall JC, Stanewsky R. Curr Biol. 2003;13:1758–1767. doi: 10.1016/j.cub.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 33.Wülbeck C, Szabo G, Shafer OT, Helfrich-Förster C, Stanewsky R. Genetics. 2005;169:751–766. doi: 10.1534/genetics.104.036244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.