Abstract
Genome-scale models of Escherichia coli K-12 MG1655 metabolism have been able to predict growth phenotypes in most, but not all, defined growth environments. Here we introduce the use of an optimization-based algorithm that predicts the missing reactions that are required to reconcile computation and experiment when they disagree. The computer-generated hypotheses for missing reactions were verified experimentally in five cases, leading to the functional assignment of eight ORFs (yjjLMN, yeaTU, dctA, idnT, and putP) with two new enzymatic activities and four transport functions. This study thus demonstrates the use of systems analysis to discover metabolic and transport functions and their genetic basis by a combination of experimental and computational approaches.
Keywords: constraint-based, flux balance analysis, functional genomics, metabolic reconstruction, systems biology
Current genome annotations include a substantial fraction of ORFs with unknown function (1, 2); methods are needed to provide insight into the possible function of these genes, without the need for screening individual gene products across a multitude of possible activities. Metabolic and regulatory networks are reconstructed from genome annotations and scientific literature to integrate and represent our current knowledge of network components and interactions (3). Dual-perturbation methods have been developed to study regulatory networks (4), which can be used to reconcile model predictions and experimental data, thus leading to possible iterative model refinements and experimentally testable hypotheses (4, 5). Iterative model-building can be systematized through the use of computational algorithms (6). Such an approach is presented here, and it consists of four steps. First, computational analysis identified discrepancies between model predictions and growth phenotyping data by using a reconstructed genome-scale Escherichia coli metabolic network. Second, an algorithm then identified enzymatic and transport reactions that likely were missing from the current metabolic reconstruction that could reconcile model predictions and experimental observations. Third, ORFs that might be responsible for these missing activities then were identified by using literature searches, sequence-homology searches (7), context-based homology methods (8, 9), and in some cases unpublished microarray data. Fourth, experimental verification of the algorithm's predictions then were carried out by evaluating growth phenotypes of single-deletion strains available in the Keio collection (10) and gene-expression measurements. Here we present a comprehensive combined computational and experimental approach to analyze phenotypic data and genome annotation information in a global manner to uncover individual ORF function.
Results
Growth phenotyping data (11), available from Biolog (Hayward CA; www.biolog.com), were used to identify missing reactions from the reconstructed genome-scale metabolic network of E. coli MG1655 (iJR904) (12). Using a flux balance model of E. coli, we identified 54 minimal medium conditions with different carbon or nitrogen sources that support growth experimentally in the Biolog data but for which the reconstructed metabolic network could not explain a growth state. Four of these discrepancies (d-allose, glucose 1-phosphate, fructose 1-phosphate, and l-cysteine) could be reconciled with available literature for E. coli metabolism. For the remaining discrepancies, we then used an algorithm (step two in the procedure described above; for details see Materials and Methods) to identify the minimal number of reactions [from a universal database of known metabolic transformations (13, 14)] that needed to be added to the reconstructed network so that cellular growth could be computed by the model (Fig. 1). Additional solutions also were found, which could have as many or more missing reactions as prior solutions, by repeating the algorithm and excluding previous solutions.
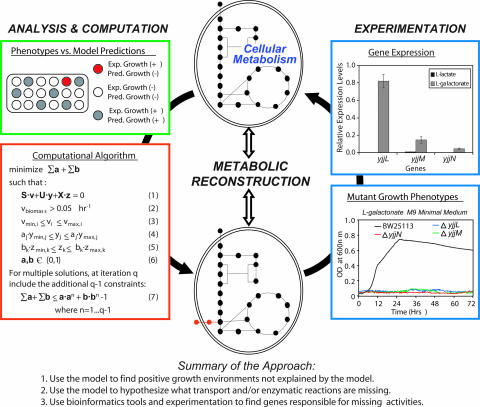
Fig. 1.
Systematic procedure for identifying missing reactions and ORF assignments. With a metabolic network reconstruction, growth predictions can be made and compared with experimental growth phenotypes (green box). For medium conditions where the microorganism grows and the model does not predict growth (red wells), reactions from a universal database can be identified by an optimization algorithm (red box) that, if added to the current reconstruction, allow for growth predictions by the model (see Materials and Methods and Supporting Text for algorithm details). Experimental testing of mutant strains can be used to identify genes responsible for the added enzymatic activities, and subsequent studies of gene-expression and biochemical characterizations can be used to further support conclusions (blue boxes). Data shown in the blue boxes is for the genes identified as being involved in l-galactonate catabolism: yjjL (putative transporter), yjjM (putative regulator), and yjjN (putative oxidoreductase). Reported gene-expression levels are relative to the internal control gene, acpP.
The algorithm identified metabolic and/or transport activities whose addition would allow the reconstructed network to reproduce growth in 26 of the 50 identified environments (see Table 2, which is published as supporting information on the PNAS web site, for details). The algorithm found 10 or more solutions for 14 of these 26 cases and a single unique solution for 11 of these 26 cases (for d-malate, there were two possible solutions; see text below). Eight of the 26 cases only require a transporter for the metabolite of interest, indicating that the enzymatic transformations needed are already present in the reconstruction.
A subset of the 26 minimal medium conditions was chosen for further experimentation to verify the existence of the predicted missing transformations and to identify the genes responsible for them. Fourteen minimal medium conditions were initially screened, but we were unable to confirm growth in five of the minimal medium conditions (xanthine and xanthosine as nitrogen sources and lyxose, α-ketobutyrate, and α-hydroxybutyrate as carbon sources). E. coli grew slowly on agmatine and citrulline as nitrogen sources and showed a 3-day lag period when grown on glyoxylate as a carbon source (data not shown). We describe the remaining six cases in more detail, and the results are summarized in Table 1.
Table 1.
Summary of experimental results for growth on various carbon sources
| Carbon source | Missing functions | Genes | Mutant phenotypes | RT-PCR* |
|
|---|---|---|---|---|---|
| Control | Carbon source | ||||
| l-galactonate | Transporter | yjjL | Lethal | 8.9 × 10−5 | 0.82 |
| Oxidoreductase | yjjN | Lag,† μ | 1.1 × 10−5 | 4.4 × 10−2 | |
| Regulator | yjjM | Lethal | 9.4 × 10−3 | 0.14 | |
| d-malate | Transporter | dctA | Lag, μ | 0.12 | 0.62 |
| Dehydrogenase | yeaU | Lethal | 1.9 × 10−3 | 1.1 | |
| Regulator | yeaT | Lag,† μ | 6.5 × 10−3 | 1.5 | |
| Propionate | Transporter | putP | μ | 0.13 | 0.32 |
| 5-keto-d-gluconate | Transporter | idnT | μ | 3.0 × 10−2 | 7.51 |
| Thymidine | Transporter | tolC | Yx/s, μ | 3.6 × 10−2 | 5.0 × 10−2 |
Lag, no growth was observed for a period of 2 days; lethal, no growth was observed for a period of 3 days; Yx/s, overall biomass yields per substrate were substantially reduced; μ, growth rates were substantially reduced (see text for reported values).
*RT-PCR levels are the mean relative expression levels to the internal control gene acpP for the control condition (see Materials and Methods for details) and the respective carbon source condition. See Table 4 for the means and standard deviations.
†For ΔyjjN with l-galactonate and ΔyeaT with d-malate as a carbon source, no growth was observed in the Bioscreen C analysis, but growth was observed after 2 days in Erlenmeyer flasks, which is likely attributable to aeration differences within the different culture conditions.
Transport Activities.
In silico growth of E. coli on propionate and 5-keto-d-gluconate requires the addition of a transporter for these metabolites to the reconstructed metabolic network because there are currently no existing annotated transporters for these compounds in E. coli. Eight genes with predicted or known transport activities were investigated by evaluating growth phenotypes of single-gene deletion strains (see Table 3, which is published as supporting information on the PNAS web site, for list of mutants screened). Of eight single-gene deletion mutants screened, only the following showed reduced growth rates: ΔputP on propionate and ΔidnT on 5-keto-d-gluconate (see Fig. 3, which is published as supporting information on the PNAS web site). Additionally, both putP and idnT show increased expression levels when grown on propionate and 5-keto-d-gluconate minimal medium, respectively (Table 1). Although IdnT is a known d-gluconate and l-idonate transporter, a prior study has shown that 5-keto-d-gluconate inhibits d-gluconate transport (15). PutP is a proline transporter that belongs to the same family of transporters as a propionate transporter (MctP) from Rhizobium leguminosarum (16), and the deletion of putP reduces the growth rate (μ) on propionate (μ = 0.02 ± 0.002 h−1 for ΔputP as compared with 0.06 ± 0.002 h−1 for the parental strain). This combined evidence suggests that putP and idnT are responsible for the transport of propionate and 5-keto-d-gluconate, respectively.
Growth on Thymidine.
The Biolog experimental data indicates that E. coli also can grow on thymidine as a sole carbon source but not as a nitrogen source. The iJR904 metabolic network includes a reaction cleaving thymidine into deoxy-ribose-1-phosphate and thymine. Although the sugar can be converted into central metabolic intermediates, the network has no way of dissipating thymine, resulting in a no growth prediction. Accordingly, the simplest solution identified by the algorithm calls for the secretion of thymine (some of the other solutions use the thymine reductive pathway coupled with secretion of pathway intermediates). HPLC analysis of the batch culture medium was in agreement with this hypothesis, because thymidine consumption was accompanied by corresponding thymine secretion into the growth medium (see Fig. 4, which is published as supporting information on the PNAS web site). A recent study has shown that E. coli strains with ntrB(Con) mutations can use thymine as a nitrogen source at room temperature (but not at 37°C and not in the parental strain) (17). The HPLC results are in agreement with this study because we only observed secretion of thymine in cells grown at 37°C, which lack the ntrB(Con) mutation. Only 1 mutant (ΔtolC), of the 29 screened mutants, exhibited a substantially reduced growth rate and biomass yield, but the mutant still secreted thymine [an independent screen of the ΔtolC mutant found it had defective growth on 19 of 38 evaluated carbon sources (18)]. It remains unclear which transporter is responsible for transport of thymine across the inner membrane and if tolC plays a role, but our data indicate that a secretion mechanism must exist for thymine.
d-Malate Catabolism.
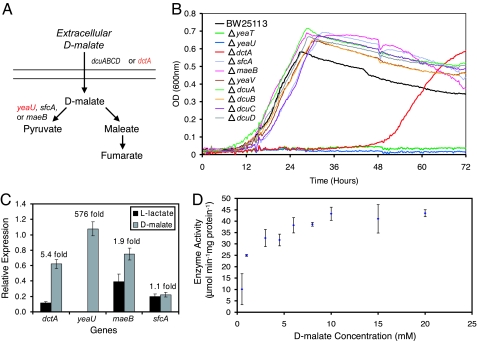
Transport and enzymatic reaction(s) are needed to reconcile d-malate catabolism; the computational algorithm developed here identified the decarboxylation of d-malate into pyruvate as the simplest explanation (Fig. 2A). Three mutants (ΔdctA, ΔyeaT, and ΔyeaU) showed altered growth on d-malate. The ΔdctA mutant was only able to grow after a 2-day lag period (μ = 0.23 ± 0.006 h−1 for ΔdctA as compared with 0.35 ± 0.006 h−1 for the parental strain), whereas the ΔyeaU and ΔyeaT mutants showed no growth over 3 days (Fig. 2B) in the Bioscreen C (a 200-well growth-monitoring system). The ΔyeaT mutant was able to grow after 2 days in Erlenmeyer flasks but at a much slower growth rate (μ = 0.01 ± 0.001 h−1, based on two replicates). The eventual growth of the ΔdctA mutant indicates that another transporter eventually compensates for the dctA deletion (Fig. 2); in fact, dcuA and dcuB show increased expression levels in the ΔdctA strain grown on d-malate (see Table 4, which is published as supporting information on the PNAS web site). In addition to mutant phenotyping data, both dctA and yeaU show increased expression levels during growth on d-malate as compared with l-lactate (Fig. 2C), providing further evidence for their involvement in d-malate catabolism. DctA is one of four dicarboxylic acid transporters that are known to transport l-malate. Based on the combined evidence provided by mutant growth phenotypes and gene-expression data, dctA appears to be responsible for the transport of d-malate as well. YeaU has significant homology with tartrate dehydrogenase from Pseudomonas putida, which has been shown to oxidize d-malate into pyruvate (19). Biochemical assays of the partially purified E. coli YeaU protein showed that the enzyme does oxidize d-malate by using NAD as an electron acceptor (Fig. 2D). YeaT is annotated as a putative regulator and, based on its chromosomal location and lethal phenotype, it appears to positively regulate yeaU. This conclusion was further supported by RT-PCR measurements of DNA isolated by chromatin immunoprecipitation (ChIP) from a yeaT-8x myc-tagged strain grown on d-malate, which shows enrichment for the upstream region of yeaU (see Table 5, which is published as supporting information on the PNAS web site). Our experimental data thus shows that d-malate catabolism involves the dctA, yeaT, and yeaU gene products.
Fig. 2.
Utilization of d-malate as a sole carbon source. (A) The two pathways that are predicted by the algorithm and the corresponding genes whose mutants were screened for altered phenotypes (red genes indicate that corresponding mutant strains showed altered growth). (B) Growth phenotype screens for the parental strain (BW25113), three mutants with altered phenotypes (ΔyeaU, ΔdctA, and ΔyeaT), and other screened mutants for growth on d-malate. (C) Expression levels were determined for BW25113 grown on l-lactate and d-malate by using RT-PCR. The values reported are the expression levels relative to the internal control gene acpP and the fold expression changes (d-malate to l-lactate) across the two conditions. (D) Results from kinetic assays of overexpressed YeaU protein show enzymatic activity levels as a function of increasing concentrations of d-malate.
Galactonate γ-Lactonate and Galactonate Catabolism.
The Biolog data indicates that both d- and l-galactonate γ-lactone can support growth as sole carbon sources; however, the hydration of galactonate γ-lactone into galactonate occurs nonenzymatically (at pH 7; data not shown), making it difficult to assess growth solely on galactonate γ-lactone. d-Galactonate catabolism has been well characterized in the literature (20) and already is represented in the metabolic reconstruction. Currently, l-galactonate is not present in the KEGG database (14), so the algorithm in its current implementation cannot resolve the observed phenotype; however, the catabolism of l-galactonate has been reported in E. coli, and the pathway has been identified (21). The genes responsible for the transport and oxidation of l-galactonate have not been found and subsequently were targeted in this study.
From Affymetrix gene-expression data of MG1655 grown on l-galactonate (data not shown), we were able to identify two genes (yjjL and yjjN) that were strongly up-regulated compared with growth on other carbon sources. Knockout mutants of these two genes and the putative regulator (yjjM) located between these loci were unable to grow on l-galactonate in a Bioscreen C analysis. In batch growth experiments conducted in flasks, the ΔyjjL and ΔyjjM strains still were unable to grow; however, the ΔyjjN strain did grow after a 2-day lag phase but at a slower growth rate than the parental strain (μ = 0.36 ± 0.02 h−1 for BW25113 and 0.17 ± 0.01 h−1 for ΔyjjN). Subsequent analysis of expression levels for all three genes by using RT-PCR showed that their expression is strongly up-regulated on l-galactonate as compared with l-lactate (data shown in Fig. 1 and Table 1). From these results, we conclude that the putative transporter (yjjL) and putative oxidoreductase (yjjN) are likely to be responsible for the missing transport and enzymatic (l-galactonate oxidoreductase) activities and that yjjM regulates their gene expression.
Discussion
The results from this study demonstrate that the synergy between experimentation and in silico modeling allows for the discovery of the roles individual genes play in an organism's behavior. In this study, a metabolic model was used to evaluate growth phenotypes and when discrepancies were found the model was used to generate testable hypotheses concerning the presence of metabolic and transport functions. The algorithm presented here can be used to identify missing metabolic and transport reactions based on growth phenotyping data, which, as shown here, can subsequently lead to the identification of gene functions. This algorithmic method can be used to direct the search for enzymes and transporters within a genome, and a variety of experimental methods can be used to find and confirm the roles of the responsible genes (7–9, 22). Phenotypic screens of single-deletion mutants have been carried out (18, 23), and these will be important for narrowing down likely gene candidates as well as pointing out alternate metabolic routes [requiring the identification of synthetic lethal gene interactions (24)]. Because predictions could be made for only 26 of the 50 cases in this study, a limitation to the computational approach in its current implementation is that the missing reactions must belong to the universal database. As such, undiscovered metabolic reactions will not be identified; however, the inclusion of computational methods that calculate novel and thermodynamically feasible metabolic transformations (25) will expand the scope of this study.
We have shown that the presented approach (combining data analysis, hypothesis generation, and experimental testing) can be used to systematically discover missing gene functions in microbial organisms. Although these efforts have centered on E. coli, the approach is likely to be more useful for less characterized organisms where new growth phenotypes are observed and many more genes with unknown functions are present.
Materials and Methods
Computational Methods.
For each minimal medium growth condition (supplemented with a carbon or nitrogen source) the algorithm calculates the minimum number of reactions that need to be added from U (matrix of known metabolic reactions) and X (a matrix of exchange reactions) to allow for growth. The optimization objective and constraints are detailed in Fig. 1 (and discussed in greater depth in Supporting Text, which is published as supporting information on the PNAS web site). The computational algorithm is a hybrid of those developed for calculating the minimum number of reactions required for cellular growth and for designing strains for metabolic engineering purposes (13, 26). The matrices S, U, and X, respectively, contain the stoichiometric coefficients for the reactions in the current metabolic reconstruction (iJR904) (12), the universal database (13) [derived from KEGG (14)], and the exchange reactions allowing intracellular metabolites to be added to or depleted from the system. The two binary vectors, a and b, will have only nonzero entries if the corresponding reaction(s) need to be added to the network to enable growth from U and X, respectively. The vectors v, y, and z contain the fluxes through the iJR904, universal database, and exchange reactions, respectively. The algorithm was used iteratively, with each iteration excluding previous solutions, to identify up to 15 possible solutions (13, 26). Mutants were identified for phenotypic screens based on literature searches, BLAST sequence-homology searches, context-based homology searches [via the Prolinks database (9)], available Biolog results for E. coli mutants [ASAP database (23)], and unpublished gene-expression microarray data (for l-galactonate).
Strains and Culture Conditions.
The Keio collection of in-frame single-gene deletion strains (10) was the source for nearly all of the mutant strains used in this study. The individual strains from the collection were obtained from Andrei Osterman at The Burnham Institute (La Jolla, CA). The parental strain for all of the mutants was BW25113 (27) and was obtained from the E. coli Genetic Stock Collection (New Haven, CT). A ΔyjjM mutant was generated and used in this study because the Keio collection mutant lacks the predicted promoter for yjjN (28). To generate the ΔyjjM mutant, the same method used to make the knockout collection (27) was used but with the following primers: 5′-TTAATGAGCATAACGCGTGTTCTCATTAATGGATCGAATCATTGATTGTTTAGCTGAATTATTCCGGGGATCCGTCGACC-3′ and 5′-ATGTACAACATCAGCCGCACCACAGTGCGTCATATTCTCAGCCACTTACGCGAATGCGCGGTGTAGGCTGGAGCTGCTTC-3′. All mutants for genes listed in Table 1 were confirmed with PCR. A yeaT-8x myc-tagged strain used for ChIP experiments was generated from the BW25113 parental strain by using pBOP508 as described previously (29).
All strains were screened for growth by using the Bioscreen C platereader system (Growth Curves USA, Piscataway, NJ) by monitoring optical density at 600 nm over 72 h. For strains that showed altered phenotypes relative to the parental strain (such as lethal phenotypes, prolonged lag periods, reduced biomass yields, or reduced growth rates), additional batch experiments were conducted in triplicate in 250-ml Erlenmeyer flasks because aeration rates can be limiting in the Bioscreen C. For most nonlethal strains with reduced growth rates (ΔyjjN, ΔtolC, ΔdctA, ΔidnT, and ΔputP), Erlenmeyer flask experiments were conducted after the removal of the antibiotic resistance gene (for methods see ref. 27). Growth rates are reported for flask experiments as mean ± standard deviation for three replicates, with the exception of duplicates for ΔyeaT. Strains were precultured overnight in 2 g/liter glucose-supplemented M9 minimal medium (strains tested for their ability to use propionate as a carbon source were precultured in LB medium), resuspended in M9 minimal medium with no carbon source, and transferred to a new medium with a starting OD (at 600 nm) equal to 0.05. All carbon sources were tested in M9 minimal medium (6.8 g of Na2HPO4, 3 g of KH2PO4, 0.5 g of NaCl, 1 g of NH4Cl, 2 ml of 1M MgSO4, and 100 μl of 1 M CaCl2 per liter) with a concentration of 2 g/liter. Nitrogen sources (2 g/liter) were screened in W-salts minimal medium (30) containing succinate (2 g/liter).
HPLC Analysis of Medium.
HPLC analysis of the growth medium was conducted for parental and mutant strains growing on thymidine as a sole carbon source. Cells were filtered from the medium by using a 0.22-μm filter. An aminex HPX-87H anion-exchange column (Bio-Rad Laboratories, Hercules, CA) then was used to separate medium components on a Waters HPLC system. A 5 M degassed solution of sulfuric acid was used as the mobile phase with a flow rate of 0.5 ml/min. Each sample was run for 35 min, and standards for thymine and thymidine showed that they eluted off the column at 25 and 30 min, respectively. UV absorbances were measured at 260 nm and 210 nm.
Enzymatic Assays.
An expression vector containing the yeaU gene was generated with the pCRT7 TOPO TA expression kit (Invitrogen, Carlsbad, CA) by using the manufacturer's recommended protocols. Transcription of the gene was induced by the addition of 75 mM isopropyl β-d-thiogalactoside (IPTG). Cells were lysed, and the overexpressed protein in crude extract was partially purified by using an Amicon μm 30,000-molecular-weight cut-off centrifugal filter (Millipore, Billerica, MA). Kinetic assays were carried out at room temperature as previously reported (at pH 8 instead of 8.4) (31) with d-malate concentrations between 0.5 and 20 mM. All tests were additionally performed on control lysates from cells harboring a reverse-insert vector, where the yeaU gene was inserted in the backward orientation.
RT-PCR Measurements of Expression Levels.
BW25113 precultures were grown in 250-ml Erlenmeyer flasks until reaching mid-log phase and then transferred in triplicate into fresh media. Cells then were harvested during mid-log phase (OD at 600 nm between 0.2 and 0.5). Samples were RNA-stabilized by using RNAProtect Bacterial Reagent, and a RNeasy mini kit was used to isolate total RNA (both from Qiagen, Valencia, CA). cDNA was prepared from the total RNA, and QIAquick PCR Purification Kits were used to clean up the cDNA synthesis product. The resulting cDNA then was quantified and used in subsequent RT-PCR assays. Nine replicate RT-PCR measurements (three replicates for each of the triplicate cDNA samples) were made for each gene (including the reference gene, acpP) under the specified growth conditions. cDNA from cells grown on l-lactate was used as a control. A standard curve (generated by using different amounts of genomic DNA instead of cDNA with fixed primer concentrations) was used to determine the primer efficiencies. The relative gene-expression levels were determined by normalizing the amount of a cDNA product to the quantity of acpP cDNA from the same cDNA sample.
ChIP.
BW25113 and YeaT-8x myc-tagged cells were grown aerobically in M9 minimal medium with d-malate (2 g/liter) at 37°C until an OD of 0.6 at 600 nm was reached. Triplicate experiments were conducted for each strain. Cells then were treated with 1% formaldehyde at room temperature for 30 min to allow cross-linking to occur. YeaT-8x myc bound DNA was isolated for all six samples by using previously described immunoprecipitation methods (29). Quantitative PCR then was carried out on these samples by using primers for the −155 to −2 region upstream of yeaU and the promoter region of acpP. Triplicate quantitative PCR measurements were made for each of the six samples.
Supplementary Material
Acknowledgments
We wish to thank Tomoya Baba and Hirotada Mori for generously providing us access to the Keio E. coli gene deletion strain collection and Byung-Kwan Cho for help with the ChIP experiments. Use of this collection greatly facilitated the detailed results generated in this work and was a valuable resource. These studies were supported by National Institutes of Health Grant GM57089. J.L.R. also received financial support as a Faculty Fellow at the University of California at San Diego.
Abbreviation
- μ
growth rate.
Footnotes
Conflict of interest statement: B.O.P. and the University of California at San Diego have a financial interest in Genomatica, Inc. Although this grant has been identified for conflict of interest management based on the overall scope of the project and its potential to benefit Genomatica, Inc., the research findings included in this publication may not necessarily relate directly to the interests of Genomatica, Inc.
This article is a PNAS direct submission.
References
- 1.Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, et al. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 3.Reed JL, Famili I, Thiele I, Palsson BO. Nat Rev Genet. 2006;7:130–141. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 4.Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 5.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 6.King RD, Whelan KE, Jones FM, Reiser PG, Bryant CH, Muggleton SH, Kell DB, Oliver SG. Nature. 2004;427:247–252. doi: 10.1038/nature02236. [DOI] [PubMed] [Google Scholar]
- 7.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 9.Bowers PM, Pellegrini M, Thompson MJ, Fierro J, Yeates TO, Eisenberg D. Genome Biol. 2004;5:R35. doi: 10.1186/gb-2004-5-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Mol Syst Biol. 2006;2:2006–0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochner BR, Gadzinski P, Panomitros E. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed JL, Vo TD, Schilling CH, Palsson BO. Genome Biol. 2003;4:R54. doi: 10.1186/gb-2003-4-9-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pharkya P, Burgard AP, Maranas CD. Genome Res. 2004;14:2367–2376. doi: 10.1101/gr.2872004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bausch C, Peekhaus N, Utz C, Blais T, Murray E, Lowary T, Conway T. J Bacteriol. 1998;180:3704–3710. doi: 10.1128/jb.180.14.3704-3710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosie AH, Allaway D, Poole PS. J Bacteriol. 2002;184:5436–5448. doi: 10.1128/JB.184.19.5436-5448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh KD, Gyaneshwar P, Markenscoff Papadimitriou E, Fong R, Kim KS, Parales R, Zhou Z, Inwood W, Kustu S. Proc Natl Acad Sci USA. 2006;103:5114–5119. doi: 10.1073/pnas.0600521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito M, Baba T, Mori H. Metab Eng. 2005;7:318–327. doi: 10.1016/j.ymben.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Tipton PA, Peisach J. Biochemistry. 1990;29:1749–1756. doi: 10.1021/bi00459a013. [DOI] [PubMed] [Google Scholar]
- 20.Cooper RA. Arch Microbiol. 1978;118:199–206. doi: 10.1007/BF00415730. [DOI] [PubMed] [Google Scholar]
- 21.Cooper RA. FEBS Lett. 1979;103:216–220. doi: 10.1016/0014-5793(79)81330-7. [DOI] [PubMed] [Google Scholar]
- 22.Kharchenko P, Vitkup D, Church GM. Bioinformatics. 2004;20(Suppl 1):I178–I185. doi: 10.1093/bioinformatics/bth930. [DOI] [PubMed] [Google Scholar]
- 23.Glasner JD, Liss P, Plunkett G III, Darling A, Prasad T, Rusch M, Byrnes A, Gilson M, Biehl B, Blattner FR, Perna NT. Nucleic Acids Res. 2003;31:147–151. doi: 10.1093/nar/gkg125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Measday V, Baetz K, Guzzo J, Yuen K, Kwok T, Sheikh B, Ding H, Ueta R, Hoac T, Cheng B, et al. Proc Natl Acad Sci USA. 2005;102:13956–13961. doi: 10.1073/pnas.0503504102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatzimanikatis V, Li C, Ionita JA, Henry CS, Jankowski MD, Broadbelt LJ. Bioinformatics. 2005;21:1603–1609. doi: 10.1093/bioinformatics/bti213. [DOI] [PubMed] [Google Scholar]
- 26.Burgard AP, Vaidyaraman S, Maranas CD. Biotechnol Prog. 2001;17:791–797. doi: 10.1021/bp0100880. [DOI] [PubMed] [Google Scholar]
- 27.Datsenko KA, Wanner BL. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salgado H, Gama-Castro S, Martinez-Antonio A, Diaz-Peredo E, Sanchez-Solano F, Peralta-Gil M, Garcia-Alonso D, Jimenez-Jacinto V, Santos-Zavaleta A, Bonavides-Martinez C, Collado-Vides J. Nucleic Acids Res. 2004;32:D303–D306. doi: 10.1093/nar/gkh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho BK, Knight EM, Palsson BO. Biotechniques. 2006;40:67–72. doi: 10.2144/000112039. [DOI] [PubMed] [Google Scholar]
- 30.Smith GR, Halpern YS, Magasanik B. J Biol Chem. 1971;246:3320–3329. [PubMed] [Google Scholar]
- 31.Kohn LD, Jakoby WB. Methods Enzymol. 1966;9:236–240. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.