Abstract
Follicle-stimulating hormone (FSH) is essential for human reproduction. The unique functions of this hormone are provided by the FSH receptor-binding beta-subunit encoded by the FSHB gene. Resequencing and genotyping of FSHB in three European, two Asian and one African population, as well as in the great apes (chimpanzee, gorilla, orangutan), revealed low diversity and significant excess of polymorphisms with intermediate frequency alleles. Statistical tests for FSHB showed deviations from neutrality in all populations suggesting a possible effect of balancing selection. Two core haplotypes were identified (carried by 76-96.6% of each population's sample), the sequences of which are clearly separated from each other. As fertility most directly affects an organism's fitness, the carriers of these haplotypes have apparently had more success in human history to contribute to the next generation. There is a preliminary observation suggesting that the second most frequent FSHB haplotype may be associated with rapid conception success in females. Interestingly, the same haplotype is related to an ancestral FSHB variant shared with the ancestor of the great apes. The determination of the functional consequence of the two core FSHB variants may have implications for understanding and regulating human fertility, as well as in assisting infertility treatments.
Keywords: Follicle-stimulating hormone beta gene, haplotype structure, ‘yin-yang’ haplotypes, balancing selection, ancestral gene variant in primates, rapid conceiving in women
Introduction
Follicle stimulating hormone (FSH) is a pituitary-expressed glycoprotein hormone that regulates reproduction in mammals. In females FSH is responsible for the proliferation and survival of follicular somatic cells, and the cyclic recruitment of ovarian follicles into development from early antral stage through maturation to ovulation (McGee & Hsueh, 2000). In males FSH is found to be essential for Sertoli cell proliferation and maintenance of sperm quality in testes (Plant & Marshall, 2001). Transgenic models have accentuated the essential function of FSH in female reproduction. FSH-deficient female mice are infertile and demonstrate small ovaries resulting from a block in folliculogenesis at the preantral stage (Kumar et al. 1997). FSH-overexpressing female mice mimic the features of human ovarian hyperstimulation and polycystic ovary syndromes (Kumar et al. 1999). FSH is widely used for the treatment of human infertility (Baccetti et al. 1997; Howles, 2000; Rose et al. 2000), and the fact that recombinant FSH was developed as the first recombinant product for this condition (Recombinant Human FSH Product Development Group, 1998) underscores its importance in reproductive medicine.
All glycoprotein hormones (FSH, Thyroid Stimulating Hormone, Luteinizing Hormone and Chorionic Gonadotropin) consist of a common alpha subunit, synthesized from the evolutionarily conserved CGA gene at 6q12-21, and a hormone-specific beta subunit. The interactions between the glycoprotein hormones and their corresponding receptors are highly selective, with very few cases of cross activity (Themmen & Huhtaniemi, 2000). Although both FSH subunits contribute to binding to the FSH receptor (FSHR), the β-subunit dictates binding specificity (Fan & Hendrickson, 2005; Fox et al. 2001). In addition to the requirement for a suitable binding surface, it appears that receptor binding and activation by the hormone is accompanied by a concerted conformational change in FSH (Fan & Hendrickson, 2005). The FSHB gene (MIM 136530; 11p13; genomic sequence 4.2 kb) coding for the FSH β-subunit consists of one non-coding exon plus two translated exons that encode the 129-aminoacid preprotein (Fig. 1a). Consistent with its essential function in reproduction only eight subjects (both men and women) with inactivating FSHB mutations have been described, all exhibiting impaired fertility and severely decreased production of mature gametes (Berger et al. 2005; Huhtaniemi, 2003).
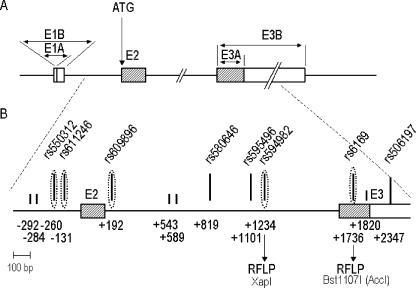
Figure 1.
A,B Schematic representation of the re-sequenced human FSHB region. a Gene structure drawn to an approximate scale. Exons (E1-E3) are indicated by boxes with the coding and non-coding sequences denoted by grey and open areas, respectively. E1 contains an alternate splice donor site resulting in two forms, A and B; E3 contains alternate polyadenylation sites, A and B (Jameson et al. 1988). The intron processing and polyadenylation are regulated independently. b Human SNPs (vertical black bars) identified in Estonians, Mandenkalu and Han are marked as long bars for common SNPs (MAF >10%) and short bars for rare SNPs (<10%). The SNP positions are relative to ATG. Five tag-SNPs (dotted ellipses) were chosen for genotyping the Korean, Czech, CEPH and STP-sample. Two of the tag-SNPs (rs594982 and rs6169) also represent RFLPs.
Despite 30 years of research on the nature and molecular function of human FSH (Moyle & Campbell, 1996; Pierce & Parsons, 1981), including sequencing of the human FSH protein (Shome & Parlow, 1974) and cloning and mapping of the human FSHB gene (Watkins et al. 1987), there are little data on sequence variation of FSHB within human populations. These studies have been restricted to consideration of only single mutations (Liao et al. 1999) or of the protein-coding exons (Lamminen et al. 2005).
We address the detailed haplotype structure of human FSHB by a resequencing study, comparing the human and great ape sequences, and discuss the possible functional consequences of the identified core variants of human FSHB.
Materials and Methods
Samples
The study was approved by the Ethics Committee of Human Research of the University Clinic of Tartu, Estonia (permission no. 117/9, 16.06.03). Estonian samples (n = 47) originated from the DNA bank of the authors' laboratory. Mandenka (n = 24) and Han Chinese (n = 25) samples were obtained from the HGDP-CEPH Human Genome Diversity Cell Line Panel (http://www.cephb.fr/HGDP-CEPH-Panel/). Czech (n = 50), and Korean (n = 45) population samples were shared by Dr. Viktor Kozich (Charles University First Faculty of Medicine, Institute of Metabolic Disease) and Dr. Woo Chul Moon (GoodGene Inc. Seoul, Korea), respectively. We used unrelated individuals from the CEPH/Utah families (n = 30) as a reference. Common chimpanzee (Pan troglodytes) DNA was extracted from sperm material obtained from Tallinn Zoo, Estonia. The sources of orangutan (Pongo pygmaeus) and gorilla (Gorilla gorilla) DNAs were primary cell lines AG12256 and AG05251B, purchased from ECACC. Blood samples for DNA analysis of pregnant women (n = 48, mean age 26.6 ± 6.9), who had conceived during sexual debut or within three months after stopping contraception (STP-sample), were collected at the Tartu University Women's Clinic, after informed consent was obtained from every participant. None of the women or their partners had used fertility treatments and the group consisted of both primi- and multigravidae (n = 22 and n = 26, respectively) with singleton fetus. Multigravidae who had previously experienced unsuccessful gestations were excluded.
PCR, Resequencing and RFLP Analysis
For full resequencing the FSHB gene (2909 bp: 1898 bp of coding region, 456 bp upstream and 555 bp downstream; Fig. 1a) genomic DNA (100 ng) was amplified in four overlapping fragments, using primers designed with the Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and Smart-Taq Hot DNA polymerase (Naxo, Estonia). The conditions for PCR and product purification are described elsewhere (Hallast et al. 2005). PCR and sequencing primers are listed in Supplementary Table S1.
The sequence data was assembled into a contig using phred and phrap software (Ewing & Green, 1998), and the contig was edited in the consed package (Gordon et al. 1998) to ensure that the assembly was accurate (http://www.phrap.org/phredphrapconsed.html). Polymorphisms were identified using the polyphred program, version 4.2 (Nickerson et al. 1997) and confirmed by manual checking. A genetic variant was verified only if it was observed in both the forward and the reverse orientations. Allele frequencies were estimated and conformance to Hardy-Weinberg equilibrium (HWE) was computed by an exact test (α = 0.05) using Genepop 3.1d program (Raymond & Rousset, 1995).
Alternatively, as FSHB markers were in strong linkage disequilibrium (LD) we used a tag-SNP approach (5 SNPs), combining genotyping by re-sequencing (from −456 to 2447 relative to ATG) and RFLP analysis. LD was evaluated by a descriptive statistic r2 estimated for pairs of common SNPs (minor allele frequency, MAF > 0.1) using Arlequin 2.000 (Schneider et al. 2000), and the significance of LD between markers was computed with Genepop 3.1d (Raymond & Rousset, 1995). SNPs rs594982 and rs6169 were typed by RFLP analysis as they result in the formation of recognition sites for restriction enzymes XapI (MBI Fermentas) and Bst1107I (MBI Fermentas), respectively. Allelic status of SNPs rs550312, rs611246, and rs609896 was determined by resequencing.
Data Analysis
Haplotypes were inferred from unphased genotype data using the Bayesian statistical method in program PHASE 2.1 (Stephens et al. 2001; http://www.stat.washington.edu/stephens/). For haplotype reconstruction the model allowing recombination was used. Running parameters were: number of iterations = 1000, thinning interval = 1, burn-in = 100; for increasing the number of iterations of the final run of the algorithm the –X10 parameter, making the final run 10 times longer than other runs, was used. We ran the algorithm 10 times, resulting in identical outputs of the parallel analysis; thus we used the median of the values obtained from one of the runs. Relationships between inferred haplotypes were investigated using the Median-Joining (MJ) network algorithm (Bandelt et al. 1999) within NETWORK 4.0 software.
Sequence diversity parameters were calculated with DnaSP 4.10.3 (Rozas & Rozas, 1999). The direct estimate of per-site heterozygosity (π) was derived from the average pairwise sequence difference, and Watterson's θ represents an estimate of the expected per-site heterozygosity based on the number of segregating sites (S). Tajima's D (DT), Fu and Li's DFL, and Fu and Li's FFL statistics were calculated to determine if the observed patterns of intraspecies diversity are consistent with the standard neutral model. Significant positive DT, DFL and FFL values may indicate an excess of high-frequency SNPs, referring to either balancing selection or population bottlenecks. Conversely, significant negative DT, DFL and FFL values may reflect an excess of rare polymorphisms in a population, indicating either positive selection or an increase in population size.
The relative amount of within-species polymorphism should reflect the amount of between-species fixation under neutrality (Kimura, 1983). The interspecies data was used for the Hudson, Kreitman, and Aguade (HKA) tests (Hudson et al. 1987), to determine whether the ratio of polymorphism to divergence across FSHB coding regions was consistent with that of the noncoding regions (Verrelli & Tishkoff, 2004). Neutrality of FSHB was tested by comparing genetic diversity of the human gene with fixed differences between human and primate sequences.
Alignment of human and great apes FSHB genomic sequences was performed with a web-based implementation of CLUSTALW at the EBI (http://www.ebi.ac.uk/clustalw/).
Exact tests for SNP locus differentiation (both allelic and genotypic) between all pairs of populations were computed with the Genepop 3.1d program.
Results
Statistical Tests for FSHB Reveal Deviations from Neutrality
Resequencing of 2,909 bp of FSHB genomic sequence for 192 human chromosomes originating from three continents supported the conservative nature of the gene, as we identified no non-synonymous mutations. The resulting FSHB genomic sequence collection contained seven common SNPs (MAF > 10%) seen in all populations and only five singleton variants (Fig. 1b; Supplementary Tables S2 & S3; ss49785048 – ss49785060). To test whether patterns of DNA sequence variation in FSHB fit the expectations under the hypothesis of neutrality we analyzed the data with a number of neutrality tests (Table 1). The direction of DT, DFL and FFL statistics is potentially informative about the evolutionary and demographic forces that a population has experienced. The estimated positive DT, DFL and FFL values for the FSHB gene (Table 1) fell into the upper range of the distribution determined in a recent study for 132 different human genes in European- and African-Americans (Akey et al. 2004), where only a few of the analyzed genes (e.g. ABO, ACE2, IL10RB, IL1A) resulted in estimates as high as that determined for FSHB. This indicates an enrichment of intermediate-frequency alleles for FSHB polymorphisms, consistent with either balancing selection or population demography characterized by subdivision or reduction in size (reviewed by Bamshad & Wooding, 2003). For Estonians and Mandenkalu significant positive values in two tests (DT; FFL) rejected the hypothesis of neutrality. Failure to reject the hypothesis of neutrality by Fu and Li's DFL test may be due to weaker power (Simonsen et al. 1995).
Table 1.
FSHB nucleotide diversity parameters and neutrality tests
| Sample size | Estonians 47 | Mandenkalu 24 | Han 25 |
|---|---|---|---|
| Diversity estimates and neutrality tests | |||
| π1 | 0.00123 | 0.00109 | 0.00079 |
| θ1 | 0.00048 | 0.00056 | 0.00055 |
| Tajima's DT | 3.2242 | 2.5233 | 1.130 |
| Fu and Li's DFL | 1.198 | 1.244 | 1.242 |
| Fu and Li's FFL | 2.3974 | 1.9384 | 1.416 |
Estimate of nucleotide diversity per site from average pairwise difference among individuals (π) and number of segregating sites (θ).
p < 0.01
p < 0.05 for Tajima's DT statistics.
p < 0.02 for Fu and Li's DFL and FFL statistics.
FSHB has Two Major Worldwide ‘Yin-Yang’ Haplotypes
Haplotype frequency estimates by PHASE algorithm revealed one prevalent FSHB gene variant (no. 1a, HAP1; Table 2) spread around the world with frequencies ranging from 51.1% in Estonians to 68.0% in Han. Interestingly, the second most frequent gene variant (no. 13a, HAP13; Table 2) is composed of completely mismatching SNP alleles compared to the dominant HAP1. For this haplotype pair nucleotides differ at every SNP, a feature termed yin-yang haplotypes (Zhang et al. 2003). The second gene variant (HAP13) is represented in Estonians with a frequency (38.3%) more than two times greater than that observed in the other re-sequenced population samples (Mandenka 12.5%, Han 14%).
Table 2.
Haplotype structure of FSHB gene
Consistent with low haplotype diversity, FSHB exhibited strong intergenic linkage disequilibrium for both applied statistics: p-values from Fisher's exact test and correlation coefficient r2. Allelic associations were significant and strong throughout the gene for Estonians (p < 0.0001, 0.795 < r2 < 1), Mandenkalu (p < 0.001, 0.62 < r2 < 1) and Han (p < 0.001, 0.53 < r2 < 1). Thus, we could choose five tag-SNPs that were sufficient to represent all major FSHB gene variants (Fig. 1b; Table 2).
The five FSHB htSNPs were typed for additional populations from Europe (Czech, unrelated CEPH/Utah individuals) and Asia (Koreans) (Supplementary Tables S2 & S3). The data confirmed the presence of the two principal gene variants (HAP1, HAP13), composed of completely mismatching SNP alleles in all populations, together covering 96.6% of CEPH, 96% of Czech, 92.6% of Estonian, 86% of Han, 79.2% of Mandenka, and 76% of Korean 5-SNP haplotypes (Supplementary Table S4). HAP1 was the dominant gene variant for all populations except among CEPH individuals. HAP13 appeared to be enriched in populations of European-origin, from 39.4% (Estonians) to 68.3% (CEPH), compared to non-Europeans for which frequencies ranged from 14% (Han) to 21% (Koreans). In the Median-Joining network of all FSHB haplotypes the two groups of haplotypes are clearly separated, with HAP1 approximately equally distributed among Europeans and non-Europeans, and HAP13 enriched in populations of European origin (Fig. 2).
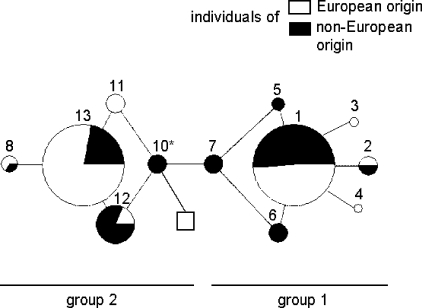
Figure 2.
Median-Joining (MJ) network for predicted FSHB haplotypes. Haplotypes have been constructed based on five FSHB tag-SNPs (Fig. 1b; Supplementary Table S4) typed for three European (Estonian, Czech, unrelated CEPH/Utah), two Asian (Han, Korean) and one African population (Mandenka). The size of each node is proportional to the haplotype frequency in the total dataset. Branch lengths represent one nucleotide substitution. The relative distribution of each haplotype among individuals of non-European and European origin is indicated by black and white, respectively. Haplotype nomenclature is identical to Table 2 and Supplementary Tables S4. Haplotypes form two groups, clustering around the major variants HAP1 and HAP13 differing at every SNP. *Human HAP10 is shared between chimpanzee and gorilla FSHB sequence. Orangutan haplotype is denoted by a square.
Comparison of Human FSHB with Great Apes' Gene Sequences
In order to uncover the ancestral FSHB variant among primates we sequenced the chimpanzee (C), gorilla (G) and orangutan (O) gene (Supplementary Fig. S1; Pan troglodytes FSHB Genbank Acc. No. DQ302103; Gorilla gorilla FSHB DQ304480; Pongo pygmaeus FSHB DQ304481). Divergence of primate FSHB from the human (H) sequence falls into the range of previous estimates: for H/C 1.28%, for H/G 1.92% and for H/O 3.38% compared to a report for 53 intergenic regions 1.24 ± 0.07%, 1.62 ± 0.08% and 3.08 ± 0.11%, respectively (Chen & Li, 2001). We identified three amino acid differences in great apes relative to human FSHB: a change in the signal peptide (amino acid no. 4) from Leu to Val between human and orangutan, and changes in the mature protein from Tyr to His (exon 2, amino acid no. 49) between human and chimpanzee, and Lys to Asn (exon 3, amino acid no. 64) between human and all of the other studied species (Supplementary Fig. S1). None of the differences are located within the region of the hormone adjacent to the receptor in the recently described FSH-FSHR co-crystal structure (Fan & Hendrickson, 2005).
Primate haplotypes formed from the positions of common human SNPs were identical among chimpanzee, gorilla and orangutan, except for a single basepair deletion in the orangutan gene for SNP rs611246 (Table 2). When compared to human FSHB the conserved great apes' haplotype is seen to be most similar to human HAP13, as opposed to HAP1 (Fig. 2). Only 2 changes are required from the conserved great ape haplotype to human HAP13, whereas HAP1 differs in five positions from other primates (Table 2).
The three primate (chimpanzee, gorilla and orangutan) sequences were used as references for the HKA test (Hudson et al. 1987), which resulted in failure to reject the null hypothesis of neutrality in all three re-sequenced human populations (H/C χ2 = 0.825-0.869, 0.35 < p < 0.37; H/G χ2 = 0.063-0.085, 0.77 < p < 0.81; H/O χ2 = 0.415-0.470, 0.49 < p < 0.52). The results of HKA tests could either indicate lack of positive selection or lack of statistical power to detect selection by this test.
HAP13 May Favor Rapid Conceiving in Women
FSH deficiency or overexpression has been shown to influence female fertility. In order to explore the hypothesis that the identified FSHB core haplotypes might have functional consequences for human reproduction, we analyzed Estonian women (n = 48) who had conceived within three months after stopping contraception (STP = short time to pregnancy) and compared them with an unselected population sample of Estonian women (n = 47) collected in the same University Clinic. In different circumstances short intervals between births, greatly regulated by the female's ability to conceive, may be either advantageous or disadvantageous and thus subjected to balancing selection. We are aware that comparison with a population sample might weaken the power of the analysis, as it could also include potential STP-women. However, in this case the usage of a traditional matched control group of women with a longer conceiving period is inappropriate, and may also be a source of bias as the time to pregnancy is influenced by several factors: hormonal status and gamete quality of both partners, anatomy of genital tract, infections, sexual behaviour, etc.
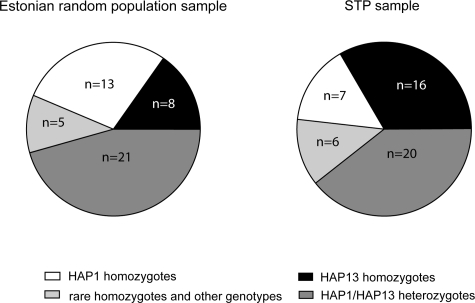
The frequency of the worldwide variant (HAP1) was 38.5% in STP-women and 53.2% in the random population sample, whereas the prevalence of HAP13 was 52.1% in the STP group and 39.4% in the random sample (χ2 = 3.982; p < 0.05). Notably, the difference in distributions of the homozygotes for these haplotypes was even more pronounced (χ2 = 10.471, p < 0.002; Fisher's exact test p < 0.001; Fig. 3). In single locus comparisons two of the five tag-SNPs (rs611246, rs594982) differed significantly in allelic (Fisher's exact test, p < 0.05) and genotypic (Fisher's exact test, p < 0.005) distributions among the studied groups. The latter remained significant (p < 0.01) after correcting for multiple comparisons.
Figure 3.
The distribution of homo- and heterozygous individuals for FSHB haplotypes in a random Estonian sample, and in Estonian women who conceived within three months after stopping contraception (STP = short time to pregnancy).
Interestingly, the HAP13 carriers were not only enriched among Estonian STP-women, but also in unrelated CEPH individuals. In the CEPH sample the frequency of HAP13 was 68.3% and HAP13 homozygotes 37%, whereas no homozygotes for HAP1 were detected. It may be noteworthy that CEPH individuals stem from the Utah pedigrees originally selected for study on the basis of having a high number (minimum 8 sibs) of children (Dausset et al. 1990).
Based on these observations we postulate a hypothesis that in females the FSHB core haplotypes may have been subjected to balancing selection due to an effect on intervals between births. However, this preliminary, although significant, finding based on a modest number of individuals needs to be confirmed with an expanded sample set involving several populations.
Discussion
Is the FSHB Gene Subject to Balancing Selection?
Despite its essential and non-redundant role in human reproduction, the genetic variation across FSHB and its possible effect on gene function has not been previously studied. Possible selection on the maintenance of FSHB gene function is revealed by its evolutionarily conserved sequence in mammals and teleost fish (Li & Ford, 1998) as well as by the very small number of non-synonymous mutations (n = 4, cases = 8, all leading to infertility) identified in humans (Berger et al. 2005). Consistent with this evidence for conservation our re-sequencing study of 96 individuals, representing populations from 3 continents, did not uncover any non-synonymous changes. The majority of the FSHB SNPs were represented as common polymorphisms with worldwide occurrence. Two of the variants overlapped with previous studies: (i) a common synonymous change Tyr-Tyr (rs6169) in exon 3 present in Asia (Han Chinese, Malays Indians, Koreans), Europe (Finns, Danes, Estonians, Czech, CEPH/Utah) and Africa (Mandenka) (Lamminen et al. 2005; Liao et al. 1999); and (ii) rs609896 also found in Asia (Han Chinese, Japanese, Koreans), Europe (Estonians, Czechs, CEPH/Utah, Finns, Danes) and Africa (Mandenkalu, Yoruba) (The International HapMap Consortium, 2005; Lamminen et al. 2005).
We identified two worldwide FSHB core haplotypes (HAP1, HAP13 carried by 76 to 96.6% of each population's individuals), the sequences of which are clearly separated from each other (Fig. 2). Although the ‘yin-yang’ haplotypes with mismatching alleles at every SNP sites can also be explained by neutral evolution (Zhang et al. 2003), we have provided evidence that this is probably not the case for FSHB. Statistical tests suggest significant deviations for FSHB from neutrality. Three neutrality tests (DT, DFL and FFL) indicated an enrichment of SNPs with intermediate minor allele frequencies. The significantly positive Tajima's DT values for FSHB stand out when compared to the distribution of DT values for 313 other human genes, 281 of which showed a negative DT value interpreted as a strong evidence for a recent expansion of the human population (Stephens et al. 2001). The results are consistent with the scenario of balancing selection acting on the FSHB gene (reviewed by Bamshad & Wooding, 2003). Although similar results can be caused by population subdivision, in which haplotypes are restricted to specific subpopulations or reduction in population size, this scenario is less supported by the FSHB data as all the studied populations share the two major haplotypes, as well as having similar results for the neutrality tests. However, in Europe the possible effect of past continent-specific demographic events contributing to the observed FSHB haplotype distributions cannot be ruled out. The observed enrichment of the HAP13 in Europe compared to the other continents may have roots in its demographic history, characterized by founder effect(s) and population expansion(s) through climate changes, demic movements (migration and expansion), wars, crusades and epidemics (Ammerman & Sforza, 1984; Gamble et al. 2004; reviewed by Barbujani & Goldstein, 2004). Interestingly the same haplotype, not the dominant HAP1, is the human gene variant most closely related to the ancestral FSHB inferred from the sequences present in all the extant great apes.
As fertility most directly affects an organism's fitness, the carriers of FSHB HAP1 and HAP13 have apparently been successful in human history at contributing to the next generation. Balancing selection has favoured the maintenance of the two human FSHB haplotypes potentially having a different effect on the mechanism of dominant follicle development, thus guaranteeing successful reproductive ability for a population in different climatic, social and economic situations. However, the functional consequences of these alternative haplotypes are still to be discovered. Based on an observation from a pilot study, we have postulated a hypothesis that FSHB gene variants may have an effect on female conception efficiency.
What Might be the Functional Consequence of the Selected FSHB Haplotypes?
None of the polymorphisms defining the two major FSHB haplotypes, apparently enriched in human evolution due to selective advantage in reproduction, are non-synonymous mutations that would directly affect the structure of the FSHB protein. However, the analyzed SNPs may represent regulatory polymorphisms, or alternatively may be associated with polymorphisms up- or downstream of the studied region, potentially influencing transcription levels, transcript stability or alternative splicing of the mRNA. Several recent studies have highlighted the functional importance of regulatory polymorphisms, such as those that influence gene expression (Boffelli et al. 2004; Morley et al. 2004; Pastinen & Hudson, 2004) or alternative splicing (Pagani & Baralle, 2004; Sorek et al. 2004).
The rate of transcription of FSHB is a limiting factor in the production and eventual secretion of pituitary FSH. The transcription and subsequent changes in serum FSH levels are largely controlled by the production of gonadotropin releasing hormone (GnRH) by neurons of the hypothalamus (Culler & Negro-Vilar, 1987). Although the human FSHB promoter has not been characterized in detail, GnRH induction of FSH production is partially dependent on AP-1–like enhancers, known to exist in sheep and rat FSHB promoters (−4741 to +759 relative to mRNA transcription site) that are also conserved among other mammals (Miller et al. 2002; Strahl et al. 1997). Due to strong linkage disequilibrium the identified FSHB core haplotypes could have been driven to high frequency by hitchhiking with one or more unstudied upstream regulatory variants affecting gene transcription. Alternatively, the FSHB variants may be associated with alternative splicing. At least four distinct species of mRNA transcripts, all encoding identical peptides, are processed from the FSHB gene (Jameson et al. 1988). Although all the FSHB mRNAs share the same transcriptional initiation site from a single promoter, alternative splicing of non-coding exon 1 provides two variants of the 5'UTR differing by 30 bp. Two different polyadenylation signals are also used: one coinciding with the stop codon and the other (used in >80% of transcripts) located about 1kb downstream. The functional significance of these regulatory regions and alternative mRNA products of the human FSHB gene remains a subject for future studies.
In conclusion, we have identified the presence of two core haplotypes for the human FSHB gene, and shown that the spread of these gene variants is consistent with balancing selection. Interestingly, a pilot study suggests that the second most frequent human haplotype may be associated with rapid conception. The determination of the functional consequence of the FSHB variants may have implications for understanding and regulating human fertility, as well as in assisting infertility treatments.
Acknowledgments
We thank Dr. Margus Punab for fruitful discussions on human male and female fertility issues; Dr. Robert K. Campbell for comments, encouragement and editing of the English language; and Dr. Francesc Calafell for notes on human demography. Drs. Howard Cann, Viktor Kozich, Woo Chul Moon are thanked for providing Mandenka and Chinese Han, Czech and Korean DNA samples, respectively. We are grateful to Imbi Taniel (Tallinn Zoo, Estonia) for collecting chimpanzee sperm sample. Tõnu Margus is thanked for assistance in sequence assembly and Viljo Soo for technical help. The study was supported by a Wellcome Trust International Senior Research Fellowship (grant no. 070191/Z/03/Z) in Biomedical Science in Central Europe and Estonian Science Foundation grant no 5796 (to M.L.). K.R. was supported by the Estonian Ministry of Education and Science Core grant no. 0182641s04 and a scholarship from the World Federation of Scientists.
Supplementary Material
DNA sequence alignments of human, chimpanzee, gorilla and orangutan FSHB gene. Location of exons, identified SNPs and fixed differences among species causing nonsynonymous changes are indicated.
Primers for FSHB PCR amplification and sequencing reactions.
SNP genotypes of FSHB gene in Estonian (A), Mandenka (B), Han (C), Korea (D), Czech (E), CEPH (F) individuals, as well as for Estonian women who have conceived within 3 months after stopping contraception (STP-women, G). For Estonians, Mandenkalu and Han the data was obtained from full resequencing, and for the other samples from a combination of resequencing and RFLP approaches (see Materials and Methods).Genotypes are given on the sense strand (with direction from centromere to telomere, according to accession AL358944). Empty cells denote markers which have not been genotyped in the respective population.
FSHB gene SNP minor allele frequencies.
FSHB haplotypes de-fined by five ‘tag’ SNPs.
References
- Akey JM. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman AJ. The Neolithic Transition and the Genetics in Europe. Princetown, NJ: Princeton University Press; 1984. [Google Scholar]
- Baccetti B. The effect of follicle stimulating hormone therapy on human sperm structure (Notulae seminologicae 11) Hum Reprod. 1997;12:1955–1968. doi: 10.1093/humrep/12.9.1955. [DOI] [PubMed] [Google Scholar]
- Bamshad M. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barbujani G. Africans and Asians abroad: genetic diversity in Europe. Annu Rev Genomics Hum Genet. 2004;5:119–150. doi: 10.1146/annurev.genom.5.061903.180021. [DOI] [PubMed] [Google Scholar]
- Berger K. Clinical and hormonal features of selective follicle-stimulating hormone (FSH) deficiency due to FSH beta-subunit gene mutations in both sexes. Fertil Steril. 2005;83:466–470. doi: 10.1016/j.fertnstert.2004.06.069. [DOI] [PubMed] [Google Scholar]
- Boffelli D. Comparative genomics at the vertebrate extremes. Nat Rev Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- Chen FC. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culler MD. Pulsatile follicle-stimulating hormone secretion is independent of luteinizing hormone-releasing hormone (LHRH): pulsatile replacement of LHRH bioactivity in LHRH-immunoneutralized rats. Endocrinology. 1987;120:2011–2021. doi: 10.1210/endo-120-5-2011. [DOI] [PubMed] [Google Scholar]
- Dausset J. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- Ewens WJ. The sampling theory of selectively neutral alleles. Theor Popul Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- Ewing B. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Fan QR. Structure of human follicle-stimulating hormone in complex with its receptor. Nature. 2005;433:269–277. doi: 10.1038/nature03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox KM. Three-dimensional structure of human follicle-stimulating hormone. Mol Endocrinol. 2001;15:378–389. doi: 10.1210/mend.15.3.0603. [DOI] [PubMed] [Google Scholar]
- Gamble C. Climate change and evolving human diversity in Europe during the last glacial. Philos Trans R Soc Lond B Biol Sci. 2004;359:243–253. doi: 10.1098/rstb.2003.1396. discussion 253–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Hallast P. Segmental Duplications and Gene Conversion: Human Luteinizing Hormone/Chorionic Gonadotropin Beta Gene Cluster. Gen Res. 2005;15:1535–1546. doi: 10.1101/gr.4270505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howles CM. Role of LH and FSH in ovarian function. Mol Cell Endocrinol. 2000;161:25–30. doi: 10.1016/s0303-7207(99)00219-1. [DOI] [PubMed] [Google Scholar]
- Hudson RR. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhtaniemi I. Mutations affecting gonadotropin secretion and action. Horm Res. 2003;60(S3):21–30. doi: 10.1159/000074496. [DOI] [PubMed] [Google Scholar]
- Jameson JL. Human follicle-stimulating hormone beta-subunit gene encodes multiple messenger ribonucleic acids. Mol Endocrinol. 1988;2:806–815. doi: 10.1210/mend-2-9-806. [DOI] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, MA: Cambridge University Press; 1983. [Google Scholar]
- Kumar TR. Transgenic models to study gonadotropin function: the role of follicle-stimulating hormone in gonadal growth and tumorigenesis. Mol Endocrinol. 1999;13:851–865. doi: 10.1210/mend.13.6.0297. [DOI] [PubMed] [Google Scholar]
- Kumar TR. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- Lamminen T. Human FSH{beta} subunit gene is highly conserved. Mol Hum Reprod. 2005;11:601–605. doi: 10.1093/molehr/gah198. [DOI] [PubMed] [Google Scholar]
- Li MD. A comprehensive evolutionary analysis based on nucleotide and amino acid sequences of the alpha- and beta-subunits of glycoprotein hormone gene family. J Endocrinol. 1998;156:529–542. doi: 10.1677/joe.0.1560529. [DOI] [PubMed] [Google Scholar]
- Liao WX. New AccI polymorphism in the follicle-stimulating hormone beta-subunit gene and its prevalence in three Southeast Asian populations. Hum Hered. 1999;49:181–182. doi: 10.1159/000022869. [DOI] [PubMed] [Google Scholar]
- McGee EA. Initial and cyclic recruitment of ovarian follicles. Endocr Rev. 2000;21:200–214. doi: 10.1210/edrv.21.2.0394. [DOI] [PubMed] [Google Scholar]
- Miller WL. The nature of FSH induction by GnRH. Trends Endocrinol Metab. 2002;13:257–263. doi: 10.1016/s1043-2760(02)00614-8. [DOI] [PubMed] [Google Scholar]
- Morley M. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle WR. Gonadotropins. In: Adashi EY, editor. Reproductive Endocrinology, Surgery and Technology. Philadelphia: Lippincott-Raven Publishers; 1996. pp. 684–724. [Google Scholar]
- Nickerson DA. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- Pastinen T. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- Pierce JG. Glycoprotein hormones: structure and function. Annu Rev Biochem. 1981;50:465–495. doi: 10.1146/annurev.bi.50.070181.002341. [DOI] [PubMed] [Google Scholar]
- Plant TM. The functional significance of FSH in spermatogenesis and the control of its secretion in male primates. Endocr Rev. 2001;22:764–786. doi: 10.1210/edrv.22.6.0446. [DOI] [PubMed] [Google Scholar]
- Raymond M. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J Heredity. 1995;86:248–249. [Google Scholar]
- Recombinant Human FSH Product Development Group. Recombinant follicle stimulating hormone: development of the first biotechnology product for the treatment of infertility. Hum Reprod Update. 1998;4:862–881. doi: 10.1093/humupd/4.6.862. [DOI] [PubMed] [Google Scholar]
- Rose MP. Definition and measurement of follicle stimulating hormone. Endocr Rev. 2000;21:5–22. doi: 10.1210/edrv.21.1.0388. [DOI] [PubMed] [Google Scholar]
- Rozas J. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Schneider S. Genetics and Biometry Lab, Dept. of Anthropology, University of Geneva; 2000. Arlequin: A software for population genetics data analysis. Ver 2.000. [Google Scholar]
- Shome B. Human follicle stimulating hormone: first proposal for the amino acid sequence of the hormone-specific, beta subunit (hFSHb) J Clin Endocrinol Metab. 1974;39:203–205. doi: 10.1210/jcem-39-1-203. [DOI] [PubMed] [Google Scholar]
- Simonsen KL. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics. 1995;141:413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Stephens JC. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–493. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- Stephens M. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD. Two proximal activating protein-1-binding sites are sufficient to stimulate transcription of the ovine follicle-stimulating hormone-beta gene. Endocrinology. 1997;138:2621–2631. doi: 10.1210/endo.138.6.5205. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortuim. A haplotype map of the genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Themmen APN. Mutations of gonadotropins and gonadotropin receptors: elucidating the physiology and pathophysiology of pituitary-gonadal function. Endocr Rev. 2000;21:551–583. doi: 10.1210/edrv.21.5.0409. [DOI] [PubMed] [Google Scholar]
- Verrelli BC. Signatures of selection and gene conversion associated with human color vision variation. Am J Hum Genet. 2004;75:363–375. doi: 10.1086/423287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux Y. Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proc Natl Acad Sci U S A. 2002;99:9650–9655. doi: 10.1073/pnas.112324299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PC. DNA sequence and regional assignment of the human follicle-stimulating hormone beta-subunit gene to the short arm of human chromosome 11. Dna. 1987;6:205–212. doi: 10.1089/dna.1987.6.205. [DOI] [PubMed] [Google Scholar]
- Watterson G. The homozygosity test of neutrality. Genetics. 1978;88:405–417. doi: 10.1093/genetics/88.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Genomewide distribution of high-frequency, completely mismatching SNP haplotype pairs observed to be common across human populations. Am J Hum Genet. 2003;73:1073–1081. doi: 10.1086/379154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequence alignments of human, chimpanzee, gorilla and orangutan FSHB gene. Location of exons, identified SNPs and fixed differences among species causing nonsynonymous changes are indicated.
Primers for FSHB PCR amplification and sequencing reactions.
SNP genotypes of FSHB gene in Estonian (A), Mandenka (B), Han (C), Korea (D), Czech (E), CEPH (F) individuals, as well as for Estonian women who have conceived within 3 months after stopping contraception (STP-women, G). For Estonians, Mandenkalu and Han the data was obtained from full resequencing, and for the other samples from a combination of resequencing and RFLP approaches (see Materials and Methods).Genotypes are given on the sense strand (with direction from centromere to telomere, according to accession AL358944). Empty cells denote markers which have not been genotyped in the respective population.
FSHB gene SNP minor allele frequencies.
FSHB haplotypes de-fined by five ‘tag’ SNPs.