Abstract
zfh-1 is a zinc finger/homeodomain transcriptional repressor in Drosophila that regulates differentiation of muscle and gonadal cells and is also expressed in the central nervous system (CNS). Binding sites for zfh-1 overlap with those for snail, and like snail, it recruits the corepressor CtBP-1. The protein ZEB-1 appears to be a vertebrate homologue of zfh-1 and is expressed in several tissues including muscle, CNS, and T lymphocytes, and during skeletal differentiation. Mutation of the ZEB-1 gene led to a severe T cell phenotype and skeletal defects but, interestingly, no defects were evident in other ZEB-1-expressing tissues. These results suggested that another ZEB-1-related factor may compensate for the loss of ZEB-1 in other tissues. Here, we characterize such a ZEB-1-related protein, which we have termed as ZEB-2. The overall organization of ZEB-2 is similar to ZEB-1 and zfh-1 and it has similar biochemical properties: it binds E boxes and interacts with CtBP-1 to repress transcription. However, there are also differences between ZEB-1 and ZEB-2, both in activity and tissue distribution. Whereas ZEB-1 and ZEB-2 overlap in skeletal muscle and CNS (providing an explanation for why mutation of ZEB-1 alone has little effect in these tissues), they show a different pattern of expression in lymphoid cells. ZEB-1, but not ZEB-2, is expressed in T cells from the thymus ZEB-2 appears to be expressed on splenic B cells. Additionally, ZEB-2 inhibits a wider spectrum of transcription factors than ZEB-1.
A wide number of biological processes involve regulation of gene expression by transcriptional repression (1). Whereas some factors repress transcription by displacement of activators (passive repression), most have an intrinsic transcriptional repressor activity. A number of transcriptional repressors contain zinc finger motifs as the DNA binding domain. Among these are snail, ZEB-1 (and its Drosophila homologue, zfh-1), Hairy, BKLF, and FOG. Similarity in these zinc fingers allows some of these proteins to bind the same DNA sequence. For example, snail, ZEB-1, and zfh-1 recognize a similar subset of E box and E box-like sequences in their target genes (2).
Interestingly, for all of the above zinc finger proteins, repression is partly mediated by binding to the corepressor CtBP-1 through a specific sequence [the CtBP interacting domain (CID)] (3–7, 29). However, binding and repression via CtBP-1 does not fully explain the activity of factors such as ZEB-1, zfh-1, or Hairy (4, 6, 8). For example, Hairy has been shown also to interact with the corepressor Groucho to mediate a separate pathway of transcriptional repression (4, 6), and ZEB-1 also contains repressor regions other than the CID (8). One repressor domain of ZEB-1 (region 1), which inhibits hematopoietic genes, is located in a region close to the N-terminal zinc fingers, whereas a second repressor domain (region 3), near the C-terminal zinc fingers, represses the myogenic transcription factor MEF2C and blocks muscle differentiation (8).
ZEB-1 is expressed in tissues such as the central nervous system (CNS), heart, skeletal muscle, and hematopoietic cells (9–13). Despite this wide pattern of expression, ZEB-1−/− mice survive until birth, and the only phenotype observed was skeletal deformities and severe T cell deficiency (no neural, heart or skeletal muscle phenotype was evident) (12, 13). This finding suggested that a ZEB-1-related gene(s) may compensate for the loss of ZEB-1 in the CNS and muscle.
We found that a recently cloned human cDNA denoted KIAA0569 (14) shows an identical overall gene structure and significant sequence similarity to zfh-1 and ZEB-1. KIAA0569 was identified along with ≈800 other genes in a screen for cDNAs encoding large proteins expressed in the CNS (14, 15). However, no more information about KIAA0569 was available.
Here, we present evidence that KIAA0569 is ZEB-2. Like other zinc finger repressors, we demonstrate that ZEB-2 binds a subset of E box and E box-like sequences and it actively represses transcription, at least in part, through its association with the corepressor CtBP-1. However, there are differences between ZEB-1 and ZEB-2 in organization of the repressor domain and in the set of transcription factors that they repress. Although ZEB-1 and ZEB-2 overlap in CNS and muscle, the pattern in lymphocytes is quite distinct: ZEB-1 is expressed in the thymocytes whereas ZEB-2 is not. Thus, ZEB-2 could compensate for ZEB-1 in muscle and nervous system, but not in T lymphocytes where a dramatic phenotype is observed in ZEB-1−/− mice (13). In contrast, ZEB-2 appears to be highly expressed in the B cells of the spleen.
Materials and Methods
Cell Culture.
C33a cervical carcinoma cells were obtained from the American Type Culture Collection (ATCC) depository (Manassas, VA) and were maintained in DMEM (Life Technologies) containing 5% FBS and 5% calf serum (Life Technologies). 293T cells (obtained from J. S. Korsmeyer, Dana–Farber Cancer Institute, Boston) were maintained in the same medium.
Plasmid Construction.
The clone pBluescript-KIAA0569 (GenBank accession number AB011141) was kindly provided by T. Nagase (Kazusa DNA Research, Chiba, Japan) (14). To construct the LexA expression vector (CS2-LexA), LexA protein was cloned into ClaI/EcoRI sites of the cytomegalovirus-driven CS2 vector (obtained from R. Kopan, Washington University School of Medicine, St. Louis, MO). A nuclear localization signal from the simian virus 40 (SV40) T antigen plus a stop codon (CCT AAG AAG AAG AGG AAG GTT TAA) was cloned in the XbaI/SnaBI sites of CS2. Then, the fragments of ZEB-2 corresponding to the central repressor domain (RD) (aa 337–996), domain 1 (aa 337–595), domain 2 (aa 594–752), domain CID (aa 751–871), and domain 3 (aa 870–996) were cloned into the corresponding EcoRI/XbaI site of CS2-LexA. Expression of all fragments was assessed by Western blot analysis (see below) to ensure equal levels of expression (data not shown).
To construct Gal4–ZEB-2 fusion proteins, ZEB-2 cDNA regions were amplified by PCR and fused in-frame with the DNA binding domain of Gal4 protein by cloning into the EcoRI/XbaI sites of the PM1 vector (16). Gal4–ZEB-1, Gal4-zfh-1, and Gal4-snail constructs were previously described (2, 7, 16).
To construct the FLAG-tagged region CID of ZEB-2, an oligonucleotide containing a Kozak sequence, an ATG codon plus a FLAG sequence (GCC ACC ATG GAC TAC AAG GAC GAC GAT GAC AAG) was cloned into the XhoI/MluI sites of pCI-neo (Promega). Then, a PCR fragment corresponding to the region between aa 979 and 1109 (C-terminal zinc fingers of ZEB-2) was cloned into the MluI/XbaI sites of pCI-neo, and a PCR fragment encoding aa 751–871 (CID) of ZEB-2 was then cloned into the XbaI/NotI site. In the reverse primer for amplification of the CID of ZEB-2, the nuclear localization signal from the SV40 T antigen and a stop codon were included (see above). The constructs for the FLAG-tagged snail, FLAG-tagged CID of ZEB-1, FLAG-tagged C-terminal zinc fingers of ZEB-1, and FLAG-tagged CID of zfh-1 were previously described (7).
pGxSV-CAT [containing four Gal4 binding sites upstream of the SV40 enhancer driving the chloramphenicol acetyltransferase (CAT) gene] and pLG (containing six LexA sites 30 bp upstream of five Gal4 sites and the E1B TATA box driving the CAT gene) were previously described (16, 17). Most Gal4 activators were either previously described (8, 16, 17) [Gal4–CTF; Gal4–VP16; Gal4–MEF2C(1–465); Gal4—NF-κB–p65; Gal4–PU.1; Gal4–ITF-1 (aa 1–427); Gal4–TFE-3 (aa 2–216)] or in the case of Gal4-myoD (amino acids 1–318) obtained from G. Tomaselli (Johns Hopkins University, Baltimore).
Vectors encoding fusion proteins for glutathione S-transferase (GST) and the N-terminal zinc fingers (aa 203–344) of ZEB-2 were obtained by PCR amplification and cloning of the resulting product into the BamHI/NotI site of PGEX–4T-1 (Pharmacia). The GST–C-terminal zinc fingers (aa 979-1109) of ZEB-2 were obtained by PCR amplification and cloning into the BamHI/NotI site of PGEX–2T (Pharmacia). A prokaryotic expression vector for the N-terminal zinc fingers of ZEB-1 was obtained from T. Kadesch (University of Pennsylvania, Philadelphia).
A myc-tagged expression vector for CtBP-1 (CS2—MT–CtBP-1) was previously described (7).
Production of Recombinant Proteins.
Bacteria transformed with PGEX constructs encoding the N- and C-terminal zinc finger domains of ZEB-2 and the N-terminal domains of ZEB-1 were induced to produce recombinant proteins by incubation with 0.1 mM isopropyl β-d-thiogalactoside (IPTG) following the manufacturer's instructions (Pharmacia). After incubation with IPTG, bacteria were lysed in NETN [20 mm Tris-HCl (pH 8.0), 100 mM NaCl, 0.5% Nonidet P-40, and 1 mM EDTA], sonicated, and the lysate tested and quantified for the expression of the different GST fusion proteins by Western blot for GST by using an anti-GST–horseradish peroxidase-conjugated Ab (Santa Cruz Biotechnologies, Santa Cruz, CA).
Gel Shift Experiments.
Probes in the gel shift experiments were created by annealing oligonucleotides containing two copies of a ZEB site (E361/E399) from the α4 integrin promoter (16, 17) and end-labeling with [γ-32P]ATP by using T4 polynucleotide kinase. Bacterial lysates containing recombinant proteins for the zinc fingers of ZEB-1 and ZEB-2 were incubated with 1 μg of BSA and 0.5 μg of poly(dI·dC) in 25 μl of reaction mix containing 10 mM Tris·HCl (pH 7.9), 50 mM NaCl, 1 mM EDTA, and 10% glycerol for 10 min on ice in the presence or absence of 50 times excess of unlabeled probe. After 10 additional min at room temperature, 6 fmol of labeled probe was added and the mixture was incubated for 10 additional min at room temperature. Then, samples were subjected to electrophoresis as described (16, 17).
Northern Blot Analysis.
Blots containing poly(A)+ RNA from different tissues were purchased from Clontech. Blots were hybridized with 1.5 × 106 cpm/ml of a probe specific for either human ZEB-1 (cDNA fragment from nt 961 to 1218) (10) or ZEB-2 (a cDNA fragment from nt 215 to 489) (GenBank accession number AB011141). Probes were labeled by random priming (Boehringer Mannheim). Hybridization was performed by incubation for 2 h at 70°C in Express Hyb solution (Clontech). After hybridization, blots were washed in 0.05× SSC and 0.1% SDS at 70°C.
Transient Transfections and CAT Assays.
Cells were transfected by the calcium phosphate method (16). After 48 h, lysates were collected and CAT assays were performed as described (16). Transfection efficiency was corrected by the cotransfection of a thymidine kinase-driven luciferase reporter vector as described (16).
Western Blot Assays.
Forty-eight hours after transfection, cells were lysed in ELB buffer [150 mM NaCl, 50 mM Hepes (pH 7.0), 5 mM EDTA, and 0.1% Nonidet P-40] and sonicated briefly. The precleared lysates were boiled in sample buffer with 5% 2-mercaptoethanol and then loaded onto a 4–15% polyacrylamide gradient gel (Bio-Rad). In the case of the coimmunoprecipitation for CtBP-1, lysates were immunoprecipitated with anti-FLAG M2 Ab (IBI, Kodak) for 2 h at room temperature. Gels were then transferred to a poly(vinylidene difluoride) membrane (Immobilon-P, Millipore) by using a 10 mM CAPS (3-cyclohexylamino-1-propanesulfonic acid) (pH 11.0) transfer buffer with 10% methanol. After transferring for at least 6 h, the membrane was incubated with the corresponding Abs: anti-Gal4 polyclonal Ab (Santa Cruz Biotechnology), anti-LexA polyclonal Ab (Upstate Biotechnology, Lake Placid, NY), anti-Flag polyclonal Ab (Santa Cruz Biotechnology), or anti-myc 9E10 mAb (Santa Cruz Biotechnology). After incubation with the corresponding anti-mouse- or anti-rabbit-horseradish peroxidase-conjugated secondary Abs (Jackson ImmunoResearch), the Western blots were developed by using the chemoluminescence technique (NEN Life Sciences) according to the manufacturer's instructions.
Results
ZEB-2 Is a Second Human Member of the zfh-1 Family.
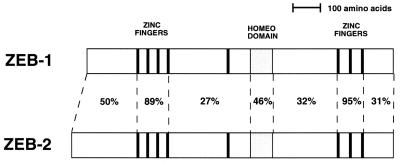
Members of the zfh-1 family have been shown to repress muscle differentiation both in mammalian and in Drosophila systems (refs. 2, 11, and 16 and references therein). In addition, ZEB-1 has been shown to play an important role in T cell differentiation (8, 10, 12, 17). ZEB-1/zfh-1 are expressed in other tissues, including heart and CNS, and, in the case of ZEB-1, during skeletal differentiation (9, 13). Mice carrying a targeted deletion of ZEB-1 show skeletal deformities and severe T cell deficiency, but no muscle or CNS phenotype was observed (12, 13). This result suggested that a ZEB-1-related gene may compensate for the loss of ZEB-1 in these other tissues. In our search for such ZEB-1-related proteins, we found that a gene cloned from a screening for large cDNAs in the brain (KIAA056; GenBank accession number AB011141) (14), showed similarity both in overall gene structure and in sequence to ZEB-1 and zfh-1, suggesting that it may be related to ZEB-1. Thus, we have designated this gene as ZEB-2. ZEB-2 shows a high level of sequence similarity to ZEB-1 in the zinc finger domains (that serve as DNA binding domains) and the homeodomain (whose function is still unclear), but little similarity was evident in the rest of the molecule, including the central RD (in between both zinc finger regions) (Fig. 1).
Figure 1.
ZEB-2 is a member of the zfh-1 family of vertebrate zinc finger/homeodomain proteins. Shown is the scheme of the structure of human ZEB-1 and ZEB-2 (KIAA0569) genes. Percentage indicates identity at the amino acid level (GenBank accession numbers U12170 and AB011141, respectively).
We have previously suggested that zfh-1 is a Drosophila homologue of vertebrate ZEB-1 (2). Like ZEB-1 and ZEB-2, zfh-1 and ZEB-1 show also sequence similarity in the zinc fingers and homeodomain but little similarity is evident elsewhere, including the RD (2, 10, 18). However, despite the low level of sequence similarity in the RD, zfh-1 can replace ZEB-1 in regulating muscle differentiation in mammalian cells, and the pattern of transcription factors repressed by the two proteins is similar (2). The degree of sequence similarity between zfh-1 and ZEB-1 (2) and zfh-1 and ZEB-2 is equivalent, although ZEB-1 is closer to ZEB-2 than either is to zfh-1 (data not shown). Therefore, it is likely that both ZEB-1 and ZEB-2 originated late in evolution from a single zfh-1 gene. This is quite common in other families of genes in mammalians that evolve from single genes in Drosophila [e.g., muscle regulatory family (MRF) proteins, MEF2, integrins, etc.)] (19).
ZEB-1 and ZEB-2 Show an Overlapping but Distinct Pattern of Expression.
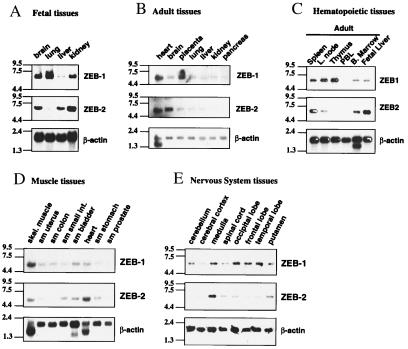
ZEB-1 expression has been examined by immunohistochemistry in different species by using Abs against segments of the protein that also recognize ZEB-2 (9, 12, 20). Therefore, the expression pattern so far ascribed to ZEB-1 is actually the combination of ZEB-1 plus ZEB-2. We then examined the pattern of ZEB-1 and ZEB-2 mRNA for both ZEB-1 and -2 distribution by Northern blot analysis by using specific probes.
In fetal tissues, expression of mRNA for both ZEB-1 and -2 was similar in most tissues including the brain, but ZEB-2 was very low in the lung (Fig. 2A). In adult tissues, ZEB-1 and ZEB-2 show also a similar and overlapping pattern including a set of muscle and CNS tissues (Fig. 2 D and E), explaining the lack of phenotype in these tissues in the ZEB-1−/− (13). However, a number of differences were also evident (Fig. 2B). ZEB-1 was expressed at high levels in the placenta where ZEB-2 expression was only marginal. Expression of ZEB-2 in the brain and heart was higher than ZEB-1.
Figure 2.
ZEB-1 and ZEB-2 show a reciprocal pattern of expression in lymphocyte tissues. In other tissues they have an overlapping, but not identical, pattern of expression. Northern blots of different sets of human tissues were hybridized with cDNA probes specific for ZEB-1 and ZEB-2 as described in Materials and Methods. A probe for human β-actin was used as internal control for mRNA loading. sm: smooth muscle.
In view of the role of ZEB-1 in T cell differentiation (2, 10, 11, 17), we performed a more detailed analysis of ZEB-1 and ZEB-2 mRNAs in different hematopoietic tissues. Both ZEB-1 and ZEB-2 were expressed in the fetal liver and bone marrow (Fig. 2C). Whereas ZEB-1 was highly expressed in the thymus, ZEB-2 was not evident, indicating that T cells express specifically ZEB-1 but not ZEB-2. In contrast, ZEB-2 appears to be expressed in the B cells and it was highly expressed in the spleen when compared with ZEB-1 (data not shown and Fig. 2C). Expression of both mRNAs was low in the peripheral blood leukocytes. The lack of ZEB-2 expression on thymocytes may explain why there is a severe atrophy in the thymus and, consequently, a severe deficit in the total number of T lymphocytes in ZEB-1−/− mice (12) (e.g., ZEB-2 is not present to compensate for the loss of ZEB-1).
This analysis provides evidence of the individual patterns for ZEB-1 and ZEB-2. Our results demonstrate that ZEB-1 and ZEB-2 have a different pattern of expression in lymphoid tissues.
ZEB-2 Binds to E Box and E Box-Like Sequences.
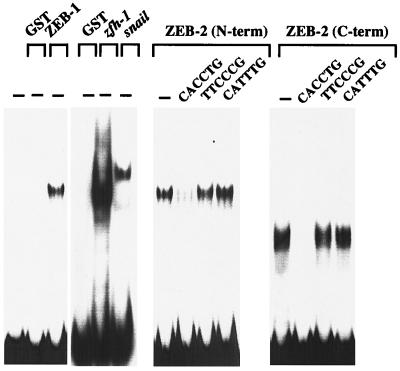
Zinc fingers are common DNA binding motifs among transcription factors. We and others have demonstrated that zinc fingers of repressors such as snail, zfh-1, and ZEB-1 serve as DNA binding domains with the highest affinity for a subset of E box and E box-like sequences, specially the CACCTG sequence (refs. 2, 10, and 11 and references therein). To determine whether ZEB-2 might bind a similar DNA sequence, we tested the binding of the N- and C-terminal zinc finger domains of ZEB-2. As shown in Fig. 3, both zinc finger regions of ZEB-2 strongly bind to the CACCTG sequence. This binding was abolished by competition with an excess of an unlabeled CACCTG sequence but not by a mutated sequence. Moreover, ZEB-2 does not bind to all E box sequences and, as previously reported for ZEB-1 (10), ZEB-2 does bind to the CATTTG E box sequence (Fig. 3).
Figure 3.
ZEB-2 binds to a subset of E box sequences. Gel retardation assays by using a probe containing ZEB sites (E361/E399) from the α4 integrin promoter (16, 17). Recombinant proteins encoding the C- and N-terminal zinc fingers of ZEB-2, the N-terminal zinc fingers of ZEB-1, C-terminal zinc fingers of zfh-1, and full-length snail were obtained by expression in bacteria as described in Materials and Methods. ZEB-1 and ZEB-2 binding was competed with a 50-fold excess of unlabeled probe, but not with the mutant non-E box probe (TTCCCC) or an unrelated E box sequence (CATTTG).
These results indicate that ZEB-1, ZEB-2, zfh-1, and snail all bind to a similar DNA sequence, suggesting that these proteins may target the same genes when they are coexpressed or they may regulate similar genes in a different temporal pattern as suggested for snail and zfh-1 (2).
ZEB-2 Is a Transcriptional Repressor That Interacts with the Corepressor CtBP-1.
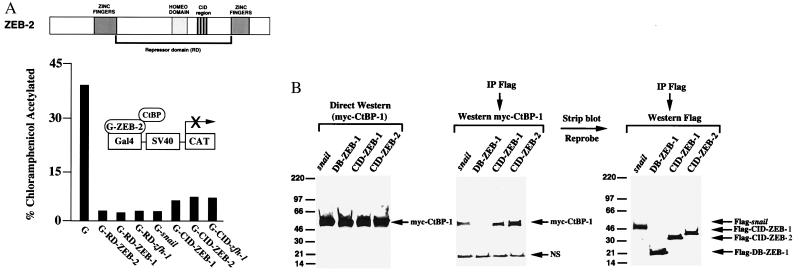
Active transcriptional repressors are defined by the existence of a repressor domain that is able to function independently of its DNA binding domain (1). We investigated whether ZEB-2 also contains intrinsic repressor activity. When the corresponding region of ZEB-2 was fused to the DNA binding domain of the yeast protein Gal4 (to construct G-RD-ZEB-2), we found that ZEB-2 repressed the SV40 promoter/enhancer (Fig. 4A). Repression by other zinc finger repressors (snail, ZEB-1, and zfh-1) was included as a control (Fig. 4A). Expression of Gal4 alone did not have any effect on the activity of the SV40 enhancer (Fig. 4A).
Figure 4.
ZEB-2 is an active transcriptional repressor. (A) The central RD and the CID mediate transcriptional repression independently of the DNA binding domain. The RD (aa 337–996) and CID (aa 751–871) regions of ZEB-2 were fused to the DNA binding domain of the yeast Gal4 protein and tested for their ability to repress the SV40 enhancer/promoter. The activity of the corresponding regions of ZEB-1 [RD–ZEB-1 (aa 302–903) and CID–ZEB-1 (aa 700–776)] are shown for comparison. A total of 1.3 μg of the Gal4 vector (G), 2 μg of wild-type G-RD–ZEB-1, G-RD–ZEB-2, G-RD-zfh-1, or G-snail, and 1.3 μg of G-CID–ZEB-1, G-CID–ZEB-2, or G-RD-zfh-1 were cotransfected with 0.6 μg of a reporter containing the SV40 enhancer/promoter (16). Transfection and assessment of CAT activity was performed as described in Materials and Methods. CAT results are an average of duplicate assays and representative of at least five separate experiments. Standard deviations were below 15%. (B) Flag-tagged constructs for the CID regions of ZEB-1 and ZEB-2, the DNA binding domain (C-terminal zinc fingers) of ZEB-1 (DB–ZEB-1), and full length snail were cotransfected in C33a cells with myc-tagged CtBP-1. After 48 h, cells were lysed as described in Materials and Methods. Twelve percent of the lysate was loaded onto the gel without immunoprecipitation as input control (direct Western blot in the figure), whereas the remaining lysate was immunoprecipitated with anti-Flag mAb. Binding to CtBP-1 was detected by Western blot by using 9E10 anti-myc mAb. Blots were then stripped of the Abs and incubated with anti-Flag Ab to detect levels of DB–ZEB-1, CID–ZEB-1, CID–ZEB-2, and snail, NS, nonspecific band.
A number of zinc finger repressors from Drosophila to mammals repress transcription through interaction with the recently described corepressor, CtBP-1 (3–7, 21, 29). CtBP-1 binds to a consensus sequence P-L-X-L-S/T (with the higher affinity for PLDLS) [the CID (21)] in these repressors. We found that as with the above proteins, ZEB-2 also contains CtBP-1 binding sites. With the exception of snail (which contains two CID sites) and ZEB-1 (which contains three CID sites), most CtBP binding proteins so far described, contain only one CtBP binding site. In the case of ZEB-2, there are four sites (757, PLRLT; 785, PLNLS; 815, PLDLS; 859, PLNLT). Accordingly, we found that ZEB-2 bound CtBP-1 even more efficiently than ZEB-1 (Fig. 4B). When fused to the Gal4 DNA binding domain, the CID region of ZEB-2 (aa 751–871) was a strong repressor of the SV40 transcriptional activity (Fig. 4A).
ZEB-2 Targets a Wider Subset of Transcription Factors Than Other Members of the ZEB Family.
Some repressors contain more than one repressor domain. This feature allows versatility in their mechanism of repression and, therefore, in their targets of repression. Thus, Drosophila Hairy has been shown to be able to interact with CtBP-1 as well as with the corepressor Groucho (4, 6). We have also shown that ZEB-1 contains multiple RDs and that interaction with CtBP-1 does not account for all of its repressor activity (8).
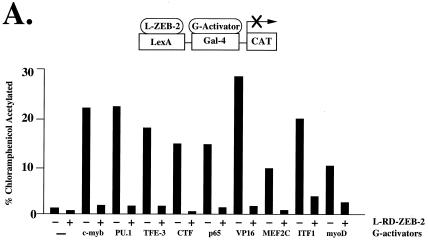
The overall sequence similarity between ZEB-1 and ZEB-2 in the RD is quite limited outside the CID region (≈30%) (Fig. 1A). However, the same is also true for ZEB-1 and zfh-1 despite the fact that their RDs appear similar in activity (around 12% sequence similarity in their RD) (2). We therefore investigated whether ZEB-2 may share the same pattern of repressor activity as ZEB-1 and zfh-1. For that purpose, we fused RD–ZEB-2 to the DNA binding domain of the bacterial protein LexA and checked its ability to repress a number of transcriptional activators. In Fig. 5A, we show that ZEB-2, as we previously found for ZEB-1 (8), represses the activity of c-myb, the ets protein PU.1, NF-κB p65, MEF2C, CTF, and VP16. However, factors that were refractory to ZEB-1, such as the B cell transcription factor ITF-1 and muscle transcription factor myoD, were also repressed by ZEB-2. This finding suggests that ZEB-1 and ZEB-2 may use distinct mechanisms of repression and ZEB-1 may only target a subset of genes targeted by ZEB-2.
Figure 5.
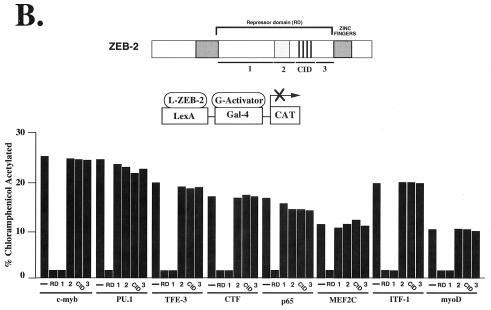
ZEB-1 and ZEB-2 have different transcriptional repressor specificity. (A) RD–ZEB-2 was fused to the DNA binding domain of the bacterial protein LexA (L-ZEB-2) and tested for its activity to repress various transcriptional activators fused to the DNA binding domain of Gal4 (G-activators). A total of 0.8 μg of the reporter pGL construct (16) containing LexA sites 30 bp upstream of Gal4 sites was cotransfected into C33a cells (293T cells in the case of G-myoD) with 2 μg of L-RD–ZEB-2 and 0.1–0.3 μg of different G-activators. After 36–48 h, cells were harvested and the CAT activity was determined as described in Materials and Methods. Equal molar amounts of the control LexA expression vector did not affect the activity of any of the different G-activators (data not shown). (B) Region 1 of ZEB-2 repressed transcription of several factors whereas others (e.g., MEF2C, PU.1) required the entire RD. Regions 1, 2, CID, and 3 of RD–ZEB-2 (indicated by numbers 1, 2, CID, and 3) were fused to the DNA binding domain of LexA and tested for their ability to repress transcriptional activators fused to the DNA binding domain of Gal4. A total of 0.8 μg of the pGL was cotransfected in C33a cells (293T cells in the case of G-myoD) with 2 μg of L-ZEB-2 constructs and 0.1–0.5 μg of different G-activators. After 36–48 h, cells were harvested and the CAT activity was determined. Equal molar amounts of the control LexA expression vector did not affect the activity of any of the different Gal4-activators (data not shown). Expression of all ZEB-2 regions was assessed by Western blot analysis to ensure equal levels of expression (data not shown). CAT results in this figure are an average of duplicate assays and are all representative of at least five separate experiments for each G-activator tested. In every case standard deviations were below 15%.
These results also indicate more functional similarity between zfh-1 and ZEB-1 than to ZEB-2, suggesting that although sequence-wise both ZEB-1 and ZEB-2 are equally similar to zfh-1, somehow during evolution the pattern of transcriptional repressor specificity segregate more to ZEB-1 than to ZEB-2 and that ZEB-2 is the more divergent family member.
ZEB-1 and ZEB-2 Have Distinct Organization in Their Repressor Domains.
ZEB-1 repressor activity is segregated to different regions of the protein (8). While most factors, including hematopoietic factors, are repressed by region 1 (between the N-terminal zinc fingers and the homeodomain), region 3 (close to the C-terminal zinc fingers) represses exclusively the myogenic factor MEF2C and inhibits muscle differentiation (8). We investigated whether a similar functional organization is present in ZEB-2 (see Materials and Methods for details of the sequence of the different regions). We designed constructs encoding the corresponding regions of ZEB-1 and ZEB-2, fused them to LexA, and checked their ability to repress different transcriptional activators. Some factors, such as c-myb, TFE-3, CTF, or NF-κB p65 were repressed by the same region of ZEB-2 and ZEB-1 (region 1) (Fig. 5B). However, other factors were repressed by different regions of the proteins. For example, the ets factor PU.1 and the myogenic factor MEF2C were repressed by regions 1 and 3 of ZEB-1, respectively; however, repression of these factors by ZEB-2 seemed to require the entire RD (Fig. 5A and data not shown).
ZEB-1/zfh-1 regulates myogenic differentiation in both mammalian cells and Drosophila (2, 11, 16, 22). In mammals, muscle differentiation is regulated by two families of positive factors: (i) MRF proteins that induce muscle differentiation by binding E box sequences in the promoter regions of muscle genes and activating their transcription (23) and (ii) the MEF2 proteins, which synergize with MRF proteins to regulate muscle differentiation and activate transcription either by binding to specific DNA sequences or through interaction with the MRF proteins (24). Another difference between ZEB-1 and ZEB-2 is that ZEB-2 was able to repress myoD whereas ZEB-1 cannot (Fig. 5 A and B). Repression of the MRF member myoD by ZEB-2 is the result of the activity of region 1 (Fig. 5B). Although ZEB-1 can block myogenic differentiation, this appear to be due to its ability to repress the activity of MEF2C (2, 8, 11, 16). ZEB-2 also blocked MEF2C transcriptional activity (Fig. 5 A and B).
Another specific target of ZEB-2 repression is the B cell factor ITF-1. ITF-1, along with TFE-3, has a key role in the regulation of the heavy chain Ig enhancer (25). Both ITF-1 and TFE-3 are repressed by region 1 of ZEB-2 (Fig. 5B). This may have significance because ZEB-1 is expressed in T cells, whereas ZEB-2 appears to be restricted to B cells (Fig. 2).
Discussion
Here, we demonstrate that ZEB-2 is an active transcriptional repressor that binds to similar DNA sites as ZEB-1, zhf-1, and snail. The pattern of expression of ZEB-1 and ZEB-2 is largely overlapping, suggesting significant redundancy in function. However, one exception is T lymphocytes where ZEB-1 is expressed with little ZEB-2 evident. Accordingly, there is a T cell phenotype in the knock out mice (12, 13). At this point, it is not known what the phenotype of the ZEB-2 knock-out mice will be, but based on the pattern of expression and activity of ZEB-2, one may predict that the lack of ZEB-2 may be important in B cell function in view of its differential expression pattern in thymus versus spleen. In addition, ZEB-2 represses the activity of the B cell transcription factor, ITF-1, whereas ZEB-1 cannot. ITF-1 has been shown to be a critical factor in the regulation of the Ig heavy chain (IgH) enhancer (25).
Several transcription factors have been shown previously to be important in regulating T versus B cell lymphocyte fate (reviewed in ref. 26). TCF-1, LEF-1, and GATA-3 are involved in T cell development, whereas E2A, EBF, and Pax-5 are involved in B cell regulation. It is unclear at what level ZEB-1 and ZEB-2 function in lymphoid differentiation. However, it is interesting to note that ZEB-1 has been shown to regulate GATA-3 in T cells (27), suggesting that the zfh-1 family members could be controlling the pattern of transcription factors in T and B cell progenitor cells.
It is of note that a number of factors repressed by both ZEB-1 and ZEB-2 are actually repressed by distinct domains in these two proteins. These results suggest that some factors may be repressed through different mechanisms by the two proteins. Thus, although the activities of ZEB-1 and ZEB-2 may be overlapping in some tissues, the mechanisms of repression by the two family members may be distinct.
All three proteins, ZEB-1, ZEB-2 and zfh-1, belong to a wider zfh family characterized by the presence of zinc finger clusters and homeodomains, although the number and distribution of these domains vary from member to member. The human homologue of Drosophila zfh-2 appears to be ATBF-1, which has been shown to repress the α-fetoprotein gene (although the mechanism of repression is not understood) (28). Therefore, it is likely that the zfh family will eventually be found to comprise a number of transcriptional repressors involved in differentiation of various tissues.
Acknowledgments
We thank Dr. Nagase for providing the cDNA for KIAA0569 shortly after his discovery. We also thank Drs. Chinnaduray, T. Genetta, and R. Kopan for providing us with cell lines and plasmids. This work was supported by grants from the National Institutes of Health to D.C.D.
Abbreviations
- CID
CtBP interacting domain
- CNS
central nervous system
- SV40
simian virus 40
- CAT
chloramphenicol acetyltransferase
- GST
glutathione S-transferase
- RD
repressor domain
- MRF
muscle regulatory family
References
- 1.Ogbourne S, Antalis T M. Biochem J. 1998;331:1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Postigo A A, Ward E, Skeath J, Dean D C. Mol Cell Biol. 1999;19:7255–7963. doi: 10.1128/mcb.19.10.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nibu Y, Zhang H, Levine M. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 4.Poortinga G, Watanabe M, Parkhurst S M. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner J, Crossley M. EMBO J. 1998;17:5229–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Levine M. Proc Natl Acad Sci USA. 1999;96:535–540. doi: 10.1073/pnas.96.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Postigo A A, Dean D C. Proc Natl Acad Sci USA. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Postigo A A, Dean D C. Mol Cell Biol. 1999;19:7961–7971. doi: 10.1128/mcb.19.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funahashi J, Seikido R, Murai K, Kamachi Y, Kondoh H. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 10.Genetta T, Ruezinsky D, Kadesch T. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa-Sehara A, Nabeshima K, Kondoh H. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H. J Exp Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagi T, Moribe H, Kondoh H, Higashi Y. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 14.Nagase T, Ishikawa K I, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. DNA Res. 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- 15.Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. DNA Res. 1999;6:63–70. doi: 10.1093/dnares/6.1.63. [DOI] [PubMed] [Google Scholar]
- 16.Postigo A A, Dean D C. EMBO J. 1997;16:3935–3943. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postigo A A, Sheppard A M, Mucenski M L, Dean D C. EMBO J. 1997;16:3924–3934. doi: 10.1093/emboj/16.13.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortini M E, Lai Z C, Rubin G M. Mech Dev. 1991;34:113–122. doi: 10.1016/0925-4773(91)90048-b. [DOI] [PubMed] [Google Scholar]
- 19.Cossu G, Tajbakhsh S, Buckingham M. Trends Genet. 1996;12:218–223. doi: 10.1016/0168-9525(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 20.Franklin A J, Jetton T L, Shelton K D, Magnuson M A. Mol Cell Biol. 1994;14:6773–6788. doi: 10.1128/mcb.14.10.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaeper U, Boyd J M, Verman S, Uhlmann E, Subramanian T, Chinnadurai G. Proc Natl Acad Sci USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai Z, Rushton E, Bate M, Rubin G M. Proc Natl Acad Sci USA. 1993;90:4122–4126. doi: 10.1073/pnas.90.9.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thayer M J, Weintraub H. Cell. 1990;63:23–32. doi: 10.1016/0092-8674(90)90285-m. [DOI] [PubMed] [Google Scholar]
- 24.Molkentin J D, Black B L, Martin J F, Olson N E. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 25.Ruezinsky D, Beckmann H, Kadesch T. Genes Dev. 1991;5:29–37. doi: 10.1101/gad.5.1.29. [DOI] [PubMed] [Google Scholar]
- 26.Rothenberg E V, Telfer J C, Anderson MK. BioEssays. 1999;21:726–742. doi: 10.1002/(SICI)1521-1878(199909)21:9<726::AID-BIES4>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 27.Gregoire J M, Romeo P H. J Biol Chem. 1999;274:6567–6578. doi: 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- 28.Yasuda H, Mizuno A, Tamaoki T, Morinaga T. Mol Cell Biol. 1994;14:1395–1401. doi: 10.1128/mcb.14.2.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner J, Crossley M. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]