Abstract
Arabidopsis COP1 acts to repress photomorphogenesis in the absence of light. It was shown that in the dark, COP1 directly interacts with the bZIP transcription factor HY5, a positive regulator of photomorphogenesis, and promotes its proteasome-mediated degradation. Here we identify a novel bZIP protein HYH, as a new target of COP1. We identify a physical and genetic interaction between HYH and COP1 and show that this interaction results in dark-specific degradation of HYH. Genetic analysis indicates that HYH is predominantly involved in blue-light regulation of development and gene expression, and that the function of HYH in part overlaps with that of HY5. The accumulation of HYH protein, not the mRNA, is dependent on the presence of HY5. Our data suggest that HYH and HY5 can, respectively, act as heterodimers and homodimers, thus mediating light-regulated expression of overlapping as well as distinct target genes. We propose that COP1 mediates light control of gene expression through targeted degradation of multiple photomorphogenesis-promoting transcription factors in the nucleus.
Keywords: Photomorphogenesis, transcription, COP1, bZIP, Arabidopsis, protein degradation
Plants continuously monitor the intensity, spectral composition, duration, and direction of the environmental light to optimize their growth and development. Recently, significant progress has been made with respect to the identity and structure of the light-sensing photoreceptors. However, the signaling pathways emanating from these receptors are only beginning to be unraveled (for review, see Neff et al. 2000; Quail 2000). Light is perceived by wavelength-specific photoreceptors, blue/UV-A light by cryptochromes (CRY1 and CRY2) and phototropins, and far-red/red light by phytochromes (PHYA, PHYB, PHYC, PHYD, and PHYE). Cryptochromes are constitutively nuclear proteins, whereas light activation results in nuclear import of the cytosolic phytochromes, suggesting that a significant portion of photoreceptor-mediated signaling occurs within the nucleus. Furthermore, it was recently found that the phytochrome protein itself contains a ser/thr kinase activity (Yeh and Lagarias 1998).
A dramatic example of light signaling can be seen during Arabidopsis seedling development, when dark-grown etiolated seedlings exposed to light undergo a dramatic morphological change. This developmental process is called photomorphogenesis or de-etiolation, and it involves a regulated change in the expression of an estimated one-third of the genes in Arabidopsis (Ma et al. 2001). Recent results indicate that the massive change in gene expression is the result of a transcriptional cascade (Tepperman et al. 2001). By analyzing the temporal changes of genomic expression profiles during phyA signaling, Tepperman et al. (2000) found that a large percentage of the early response genes (induced in <1 h) are transcription factors, whereas it takes several hours before many of the genes associated with photomorphogenic development are highly expressed. These results suggest that photoreceptors transduce the signal to a set of key transcription factors that, in turn, rapidly induce expression of a new set of transcriptional regulators. This results in an extensive branching of the signal and the promotion of photomorphogenic development.
So far, the transcription factors involved in light-regulated gene expression have been identified by looking for mutants with reduced ability to respond to light signals. Mutations in the bZIP protein HY5 result in an elongated hypocotyl in all light conditions, whereas mutations in the bHLH protein HFR1/REP1/RSF1 have a far-red-light-specific phenotype (Fairchild et al. 2000; Soh et al. 2000; Spiegelman et al. 2000). Three MYB proteins affecting light signaling were also identified: LHY and CCA are involved in circadian rhythm, and LAF1 is specifically impaired in far-red-light signaling (Schaffer et al. 1998; Wang and Tobin 1998; Ballesteros et al. 2001). A second approach identified the bHLH protein PIF3 through direct interaction with a photoreceptor (Ni et al. 1998; Halliday et al. 1999). PIF3 was shown to mediate phytochrome-regulated transcription, and, interestingly, phyB was shown to interact with DNA-bound PIF3 in a light-dependent manner (Martinez-Garcia et al. 2000).
Seedlings carrying mutations in each of 11 different negative regulators of photomorphogenesis, the COP/DET/FUS genes, phenocopy light-grown seedlings when grown in darkness. The recessive nature of the cop/det/fus mutations suggests that photomorphogenesis is the default developmental pathway and that dark-grown seedlings repress this development. The dramatic phenotype of the cop/det/fus mutations indicates that the COP/DET/FUS proteins act as master regulators of this developmental switch, and it has been shown that the loss of these genes results in deregulation of genes determining photomorphogenic development. The recent characterization of the evolutionarily conserved COP/DET/FUS proteins have provided clues to the mechanism by which these proteins function. The COP9 signalosome, a multiprotein complex homologous to the substrate-recognizing lid subcomplex of the proteasome, was recently shown to interact with an E3 ubiquitin ligase (Schwechheimer et al. 2001). Furthermore, the COP9 signalosome, COP10, DET1, and COP1, were recently found to mediate the degradation of HY5 (Osterlund et al. 2000). COP1, a protein containing domains commonly found in a subclass of E3 ubiquitin ligases, is localized to the nucleus in the dark, and the nuclear abundance of COP1 decreases in the presence of light (von Arnim and Deng 1994). All these results lend support to a hypothesis that many, if not all, of the COP/DET/FUS proteins act in a putative pathway aimed at degrading key transcriptional regulators, such as HY5, in the dark, thus inhibiting light-dependent transcription in darkness. The failure of COP/DET/FUS proteins to degrade their target proteins would then allow these proteins to drive photomorphogenic development in the absence of light. The target proteins of the COP/DET/FUS proteins are therefore likely to be key regulators of light-dependent development. Although COP1, the putative substrate-recognizing component of the pathway, has been found to interact with several proteins (Yamamoto et al. 1998, 2001; Torii et al. 1999; Holm et al. 2001; Wang et al. 2001), HY5 is so far the only known target of COP1-mediated degradation.
Here we describe the identification of HYH, a new regulator of light-dependent gene expression. The HYH protein is specifically degraded in the dark in a manner dependent on the activity of the COP/DET/FUS proteins. We identify a physical and genetic interaction between COP1 and HYH. The interaction results in the COP1-specific degradation of HYH in the dark, thus validating the concept of a COP/DET/FUS-mediated pathway for protein degradation. HYH is a G-box-binding member of the bZIP family of transcription factors, and we characterize functional interactions between HYH and HY5 in an effort to understand how they are regulated by COP1.
Results
Identification of HYH, a COP1-interacting protein in Arabidopsis
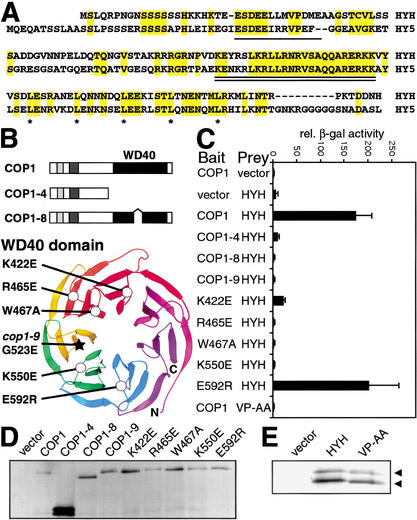
In an effort to obtain additional support for a role of the COP/DET/FUS proteins in protein degradation and to identify novel COP/DET/FUS-regulated protein targets, full-length COP1 protein was used as bait in a yeast two-hybrid screen. Out of ∼170,000 transformants, 2 independent cDNAs encoding a novel protein were recovered. The longest cDNA was 592 nucleotides (accession no. AF453477) and was identical to portions of the predicted gene At3g17610 (Arabidopsis Genome Initiative 2000; see Materials and Methods). The cDNA encodes a 149-amino-acid protein with a predicted molecular mass of 16.9 kD, containing a basic zipper (bZIP) transcription factor domain in the C-terminal half. The bZIP protein, denoted HYH for HY5 homolog, is the closest homolog of HY5 in the Arabidopsis genome. Figure 1A shows an alignment between HYH and HY5. HYH shows 49% (73/149) overall amino acid identity with HY5. The highest level of identity is found in the DNA-binding basic domain, where 21 of the 24 amino acids are identical. Although the N-terminal half of the proteins are less conserved, a sequence motif previously identified in HY5 as a COP1-interacting motif and a caseine kinase 2 (CK2) phosphorylation site (Hardtke et al. 2000; Holm et al. 2001) are conserved in HYH (Fig. 1A).
Figure 1.
HYH interacts with the WD40 domain of COP1 through a COP1-interaction motif. (A) Alignment of HYH and HY5; identical amino acids are shaded in yellow. The COP1-interaction motifs (Holm et al. 2001) are single underlined and the conserved DNA-binding basic domains are double underlined; leucines in the dimerizing leucine zipper are marked by asterisks. (B) Domain organization and mutations in COP1. The genetically identified mutations cop1-4, cop1-8, and cop1-9, as well as the amino-acid-substituted COP1 proteins have been previously described (Holm et al. 2001). The ribbon diagram of the Gβ β-propeller was used to align the COP1 WD40 repeats. The carbon α backbone (Cα) positions of the substituted COP1 residues are marked by circles. The star marks the position of the cop1-9 mutation. (C) Yeast two-hybrid interactions between wild-type or mutated COP1 proteins and HYH proteins. VP-AA indicates alanine substitution of the conserved VP pair in the COP1-interacting motif of HYH. Error bar represent standard deviation, n = 6. (D,E) Immunoblots of yeast expressing indicated Gal4 DNA-binding domain fused COP1 and activation-domain fused HYH proteins, respectively.
HYH interacts with the COP1 WD40 domain through a COP1 interaction motif
To quantify and further characterize the interaction between HYH and COP1, we assayed the interactions between wild-type and mutated COP1 and HYH proteins in liquid β-galactosidase (β-gal) assays (Fig. 1C). The interaction between HYH and COP1 results in 25-fold higher β-gal activity than that observed for the vector control. None of three genetically identified mutations of COP1 was able to interact with HYH. COP1-4 lacks the WD40 domain, whereas COP1-8 and COP1-9 contain a deletion and an amino acid substitution in the WD40 domain, respectively (Fig. 1B), suggesting that the COP1 WD40 domain is required for HYH interaction. To determine if the putative COP1-interacting motif in HYH is required for the COP1 interaction, we introduced alanine substitutions of the conserved VP pair at the core of the motif in HYH. The VP to AA substitution completely abolishes HYH's interaction with COP1, indicating that the motif is, indeed, required for the HYH–COP1 interaction (Fig. 1C).
To further characterize the interaction between HYH and the WD40 domain of COP1, we used a set of amino-acid-substituted COP1 proteins previously described (Fig. 1B; Holm et al. 2001). These single amino acid substitutions were selected because they are likely to interfere with protein–protein interactions of the COP1 WD40 domain. They are unlikely, in contrast to the COP1-9 mutation, to affect the overall structure of the domain. Figure 1C shows that one of the five substitutions, E592R, has no effect on the HYH interaction. On the other hand, the K422E, R465E, W467A, and K550E substitutions dramatically reduce interaction with HYH. The mutated COP1 and HYH proteins were examined in immunoblots, and their expression was found to be at similar levels as the respective wild-type proteins (Fig. 1D,E). In all, we have identified a novel COP1-interacting bZIP protein that interacts with the WD40 domain of COP1 through a COP1-interacting motif.
HYH is a nuclear protein and colocalizes with COP1 in living plant cells
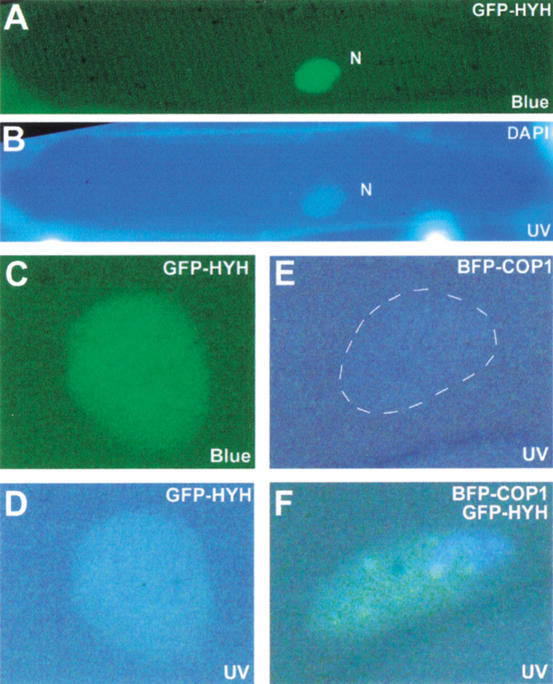
To examine the intracellular localization of HYH, we expressed a green fluorescent protein (GFP) fusion of HYH in onion epidermal cells. As shown in Figure 2A, the GFP–HYH protein displays a diffused and uniform nuclear fluorescence, suggesting that HYH is a nuclear protein. It has previously been shown that COP1 protein expressed from a transgene localizes to distinct foci or speckles in the nuclei of onion epidermal cells as well as in Arabidopsis hypocotyl cells (Ang et al. 1998; Osterlund et al. 1999; Stacey et al. 1999). Furthermore, the HY5 protein has been found to colocalize with COP1 in these nuclear speckles (Ang et al. 1998). Recently, the COP1-dependent localization of HY5 to nuclear speckles was further analyzed through fluorescent resonance energy transfer (FRET) techniques, suggesting a physical interaction between HY5 and COP1 in the speckles (Osterlund et al. 1999). To examine if HYH also colocalizes with COP1 in the nuclear speckles, we expressed GFP–HYH together with BFP–COP1 in onion cells. Figure 2F shows a representative nucleus expressing both proteins excited by UV light in which green speckles are clearly visible. These green speckles represent foci where BFP–COP1 and GFP–HYH are in close proximity, allowing FRET from BFP–COP1 to GFP–HYH to occur. The speckled GFP–HYH fluorescence requires the presence of BFP–COP1 because it was never observed with GFP–HYH alone (Fig. 2A–D). The emission from BFP–COP1 alone is very weak and difficult to detect (Fig. 2E). This result provides an in vivo support for a physical interaction between COP1 and HYH.
Figure 2.
HYH is nuclear and colocalizes with COP1 in onion cells. (A) Blue-light excitation of a DAPI-stained onion epidermal cell expressing GFP–HYH. The diffuse GFP fluorescence is restricted to the nucleus (N). (B) The same cell as in A excited by UV light to show DAPI stain to indicate position of the nucleus. (C) A nucleus expressing GFP–HYH excited by blue light. (D) The nucleus in C excited by UV light. (E) A nucleus expressing BFP–COP1 excited by UV light. (F) A representative onion cell nucleus that expresses both GFP–HYH and BFP–COP1 showing FRET-induced green speckles upon UV light excitation. FRET can occur when BFP and GFP proteins are in close proximity; blue photons emitted by UV-excited BFP can then excite GFP to allow emission of green light.
The HYH–COP1 interaction results in degradation of HYH in the dark
So far the only known target of COP1-mediated protein degradation is the HY5 transcription factor. Because HYH, like HY5, interacts with the WD40 domain of COP1, we set out to examine whether HYH might be a target of COP1-mediated protein degradation in darkness. To this end we grew wild-type seedlings and seedlings overexpressing HYH (HYH-OE) in white light for 3 d and then transferred them to darkness for 24, 48, and 72 h. Protein extracts were prepared 3 days after germination (DAG) and after indicated time in darkness. The protein concentration was normalized, and the levels of HYH protein were assayed by Western blot using HYH-specific polyclonal antibodies.
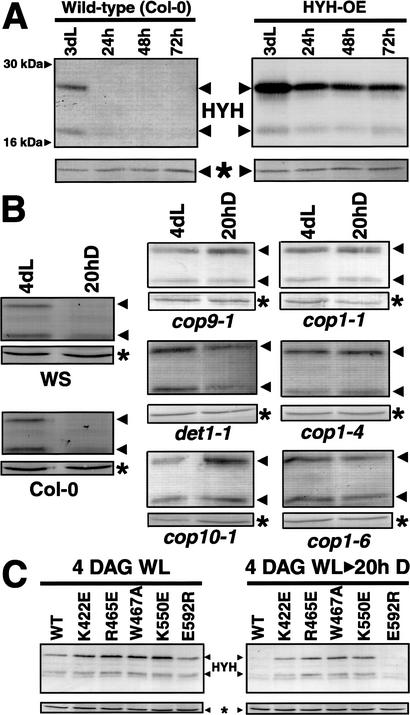
As seen in Figure 3A, the endogenous HYH protein is detected in 3-DAG seedlings. The polyclonal HYH antisera consistently detect two protein bands of ∼27 kD and 21 kD, respectively. Both bands are lost in the hyh mutant (Fig. 4), suggesting that they are encoded by the HYH gene. However, the relative level of the 21-kD protein differed between experiments, suggesting that this protein could be a degradation product. The apparent molecular mass of HYH is higher than the predicted 16.9 kD. We do not know the reason for the higher apparent molecular weight, but it is worth noting that the related HY5 protein, predicted to be ∼18 kD, migrated as a 30-kD band in SDS-PAGE.
Figure 3.
The COP1 interaction results in degradation of HYH in the dark. (A) Both endogenous and overexpressed HYH protein is degraded upon transfer to darkness. The wild-type (Col-0) and HYH-OE seedlings were grown in white light for 3 d and then transferred to darkness. Protein extracts were prepared at day 3 and after 24, 48, and 72 h in darkness, and the protein concentration was normalized before loading. Neither of the 27- and 21-kD bands were detected using preimmune sera (data not shown). The HYH proteins migrate distinct from the 30-kD HY5 protein, and no cross-reactivity to endogenous HY5 was observed in immunoblots. (B) Wild-type (Col-0 and WS ecotypes) and cop9-1, det1-1, cop10-1, cop1-1, cop1-4, and cop1-6 seedlings were grown in white light for 4 d and for an additional 20 h in darkness, respectively. Protein extracts were prepared at 4 DAG and at 4 DAG + 20 h, and the protein concentration was normalized before loading. (C) Seedlings overexpressing wild-type or indicated amino-acid-substituted COP1 protein in a cop1-5 null mutant background were grown in white light for 4 d and then transferred to darkness for 20 h. Protein extracts were prepared at day 4 (4 DAG), and after 20 h in darkness (20h D), the protein concentration was normalized and 15 μg of total extract was loaded in each lane. The amount of HYH protein was assayed by immunoblotting; the asterisk marks a cross-reacting band that serves as an internal loading control. At least two independent lines were used for each transgenic construct, and similar results were observed.
Figure 4.
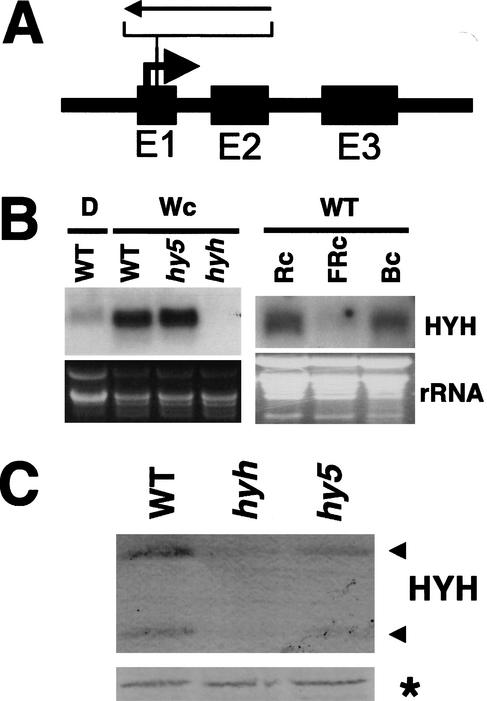
Identification of a knockout mutation in the HYH gene. (A) Schematic illustration of the HYH gene organization with the position of the T-DNA insertion indicated. (B) Northern blot of 20 μg of total RNA prepared from 6-DAG dark-grown wild-type (WS) seedlings (D), and 6-DAG wild-type (WS), hy5-ks50, and hyh seedlings grown in white light, respectively. Right panel shows Northern blot of 20 μg of total RNA prepared from 6-DAG wild-type (Col-0) seedlings grown in continuous red (108 μmole/sec per m2), far-red (160 μmole/sec per m2), and blue (16 μmole/sec per m2) light, respectively. The HYH RNA is detected as a single band at ∼600 bp. (Bottom panels) The ethidium bromide-stained gel serving as a loading control. (C) Immunoblot of 10 μg of total protein prepared from 3-DAG wild-type (WS), hyh, and hy5-ks50 seedlings grown in white light. The two proteins detected with the HYH antibodies are indicated with arrows to the right. The asterisk marks a cross-reacting protein serving as a loading control.
Interestingly, when the 3-DAG light-grown seedlings were transferred to darkness for 24 h, the level of HYH protein was reduced to below the detection level. This indicates a dark-specific reduction of HYH protein levels. When the HYH protein was overexpressed from the constitutive 35S promoter, the level was also dramatically reduced upon transfer to darkness. This suggests that the reduction is regulated at a posttranscriptional level (Fig. 3A).
The reduction of HYH protein levels in the dark requires the COP/DET/FUS proteins and direct interaction with COP1
To examine the role of the COP/DET/FUS proteins in the dark-dependent reduction of HYH protein, we grew wild-type and cop9, det1, cop10 as well as cop1-1, cop1-4, and cop1-6 seedlings in white light for 4 d and then transferred them to darkness for 20 h. Protein extracts were prepared at day 4 and after the additional 20 h in the dark. The protein concentration was normalized, and HYH abundance was examined by Western blot. We found that although the level of HYH protein rapidly decreases in wild-type Col-0 and WS seedlings upon transfer to the dark, this decrease is abolished in all of the tested cop/det/fus mutants (Fig. 3B). This finding suggests that the activities of each of the COP/DET/FUS proteins are required for the reduction of HYH protein levels.
These results are consistent with the hypothesis that HYH, like HY5, is specifically degraded in the dark. To further test this hypothesis, we examined whether the direct protein–protein interaction between COP1 and HYH is involved in the dark-dependent degradation of HYH. To this end, we used cop1-5 null mutant seedlings that overexpress wild type, K422E-, R465E-, W467A-, K550E-, or E592R-substituted COP1 proteins. The K422E, R465E, W467A, K550E, and E592R transgenic lines have been previously described, and overexpression of either of these COP1 proteins is able to rescue the lethality of the cop1-5 mutation (Holm et al. 2001). Seedlings were grown in constant white light for 4 d and then transferred to darkness. Protein extracts were prepared at 4 DAG and after 20 h in the dark. After normalization of protein concentration, HYH protein levels were detected by immunoblot. As shown in Figure 3C, the HYH protein is present at high levels in all 4-DAG seedlings. However, among the seedlings that have been grown in darkness for an additional 20 h, only the seedlings expressing wild type and the COP1-E592R protein were able to degrade the HYH protein. This corresponds well with the yeast two-hybrid results (Fig. 1C), where HYH is able to interact with wild-type and E592R-substituted COP1 proteins but is unable to interact with the K422E-, R465E-, W467A-, and K550E-substituted proteins. This direct correlation between yeast two-hybrid interactions and the degradation of HYH protein in seedlings provides strong evidence that the COP1 interaction is directly mediating the dark-dependent degradation of HYH.
Isolation and characterization of a null mutation in HYH
To examine the role of HYH in light-regulated seedling development, we screened the Arabidopsis knockout collection at Madison, WI, for T-DNA insertions in the HYH gene (Sussman et al. 2000). The HYH gene is located on the top arm of chromosome 3, and we identified a T-DNA insertion within the gene. The T-DNA is inserted in the first exon of the HYH gene at nucleotide position 20 from the translational start site. The T-DNA line was back-crossed into wild type (WS) and crossed into hy5-ks50, cop1-1, cop1-4, and cop1-6 alleles. Analyses of these crosses revealed a single T-DNA locus cosegregating with the hyh phenotype. The insertion results in no detectable levels of both HYH mRNA (Fig. 4B) and protein (Fig. 4C), suggesting that hyh-1 is a null mutation. Transcription of the HYH gene is dramatically higher in light-grown seedlings compared with seedlings grown in darkness (Fig. 4B). The light-dependent activation of HYH transcription is higher than the approximately two- to threefold activation observed for HY5 (Osterlund et al. 2000). To further characterize the light regulation of HYH expression, we examined the relative levels of HYH mRNA in 6-DAG seedlings grown in monochromatic far-red, red, and blue light. We found that the HYH mRNA was present at high levels in red and blue light but that HYH mRNA accumulation was significantly reduced in seedlings grown in far-red light (Fig. 4B). Although the level of HYH RNA in hy5 seedlings is comparable to wild-type levels (Fig. 4B), the HYH protein level is dramatically reduced in an hy5 mutant (Fig. 4C). This suggests that the presence of the HY5 protein is important for the normal accumulation of HYH protein.
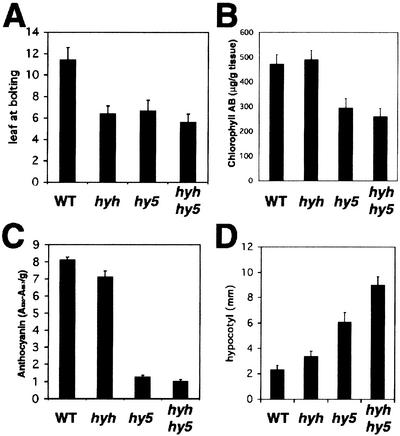
When propagating the hyh plants we found that the hyh mutation causes early flowering. Long-day-grown plants with the hyh mutation flower after producing ∼6 rosette leaves compared to ∼12 in wild type. The hy5 mutation also results in early flowering after ∼6 rosette leaves. No additive effect is seen in the double-mutant plants (Fig. 5A).
Figure 5.
Characterization of the hyh mutant. (A) Number of rosette leaves at bolting for wild-type, hyh, hy5-ks50, and hyh/hy5-ks50 plants grown in long-day conditions (16 h light, 8 h dark). (B) Chlorophyll a and b content in 4-DAG wild-type, hyh, hy5-ks50, and hyh/hy5-ks50 seedlings grown in white light. Error bars represent standard deviation, n = 5. (C) Anthocyanin content in 3-DAG wild-type, hyh, hy5-ks50, and hyh/hy5-ks50 seedlings grown in white light. Error bars represent standard deviation, n = 4. (D) Hypocotyl length of 6-DAG wild-type, hyh, hy5-ks50, and hyh/hy5-ks50 seedlings grown in blue light (∼16 μmole/sec per m2). Error bar represents standard deviation, n = 20.
hyh shows a predominantly blue-light-specific defect in seedling development
The seedling phenotype of hyh is relatively subtle compared with that of hy5. White-light-grown hyh seedlings are slightly paler than wild-type seedlings. No significant difference in chlorophyll content was observed (Fig. 5B), but hyh seedlings accumulate slightly reduced levels of anthocyanin (Fig. 5C). The hy5 mutation results in an ∼6.5-fold reduction in anthocyanin accumulation, and hy5/hyh double-mutant seedlings contains ∼80% anthocyanin accumulation compared with that of the hy5 mutation alone. These results indicate that although the absence of HY5 protein has a dramatic effect on anthocyanin accumulation, the effect is enhanced if the HYH protein is also missing, suggesting an overlapping yet not redundant function between the HY5 and HYH proteins in white light. The hyh mutation has no significant effect on hypocotyl length or morphology of seedlings grown at any fluence of white, red, or far-red light or in the dark (data not shown). However, we found that blue-light-grown hyh seedlings have significantly longer hypocotyls than do wild-type seedlings (Fig. 5D). Furthermore, blue-light-grown hyh/hy5 double-mutant seedlings have a longer hypocotyl than seedlings carrying the hyh or hy5 mutation alone (Fig. 5D), suggesting that the hyh mutation augments the hypocotyl phenotype of hy5, specifically in blue light. These results suggest that HYH and HY5 have partially overlapping functions in light-dependent development and that the role of HYH is more pronounced under the blue-light condition.
Overexpression of HYH can compensate for loss of HY5
The absence of hyh phenotypes in white, red, and far-red light stands in contrast to the hy5 mutant seedlings, which show elongated hypocotyls in all light conditions. It is not clear why mutations in these two related transcription factors give rise to so different phenotypes.
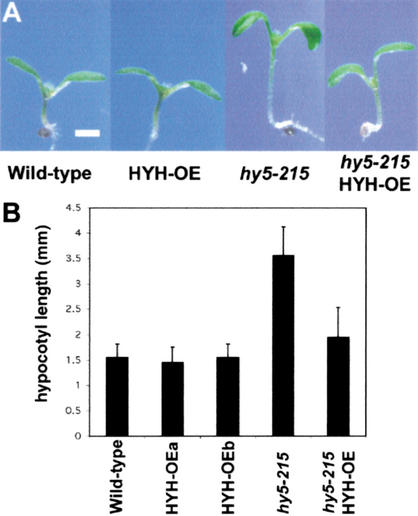
As a first step to address the functional relationship between these two structurally related proteins, we crossed the HYH overexpression transgene (HYH-OE) into the hy5-215 mutant in an effort to determine whether overexpression of HYH can compensate for the loss of HY5. Figure 6A shows 6-DAG seedlings grown in white light. Overexpression of HYH has no significant effect on the hypocotyl length: wild-type and two independent lines overexpressing HYH all have ∼1.5-mm-long hypocotyls. Interestingly, although the average length of hypocotyls in hy5-215 seedlings is ∼3.5 mm under our high-intensity white light condition, the hypocotyl length of hy5-215 seedlings overexpressing HYH is only ∼2 mm, suggesting that increased levels of HYH, indeed, can largely suppress the hy5 phenotype. Furthermore, hy5 seedlings are paler than wild-type seedlings because of a lower anthocyanin level and a reduced ability to accumulate chlorophyll (Fig. 5B,C), and the HYH overexpression is also able to partially suppress both of these phenotypes (data not shown). Taken together, these results show that although the phenotype of hyh is subtle, overexpression of HYH can largely suppress the more dramatic phenotypes of hy5, suggesting a functional overlap also in white light.
Figure 6.
Overexpression of HYH is able to suppress the phenotype of hy5-215. (A) 6-DAG white-light-grown wild-type, HYH-OE, hy5-215, and hy5-215/HYH-OE seedlings. Bar, 1 mm. (B) Graphical representation of the hypocotyl length of the seedlings in A. The columns represent wild-type (WT), two independent transgenic lines overexpressing HYH (HYH-OEa and b), hy5-215 seedlings, and hy5-215 seedlings overexpressing HYH, all in the Col-0 ecotype. Error bars represent standard deviation, n = >30.
The hyh mutation can suppress the block-of-greening phenotype of cop1
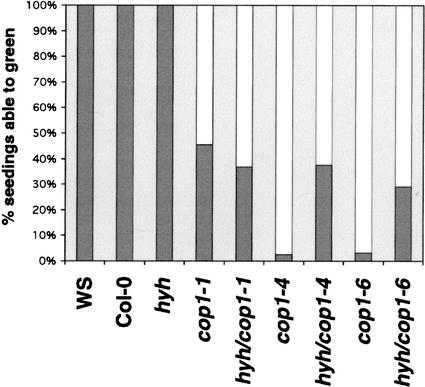
In an effort to examine a potential genetic interaction between hyh and cop1, we generated plants homozygous for both hyh and each of the three viable cop1 alleles, cop1-1, cop1-4, and cop1-6. The double-mutant seedlings are indistinguishable from the respective cop1 mutants when grown in darkness, suggesting that the hyh mutation is unable to suppress the dark phenotypes of cop1. However, we were able to detect suppression of a light-dependent cop1 phenotype, the COP1-dependent block of greening. Dark-grown wild-type seedlings transferred to light readily begin photomorphogenic development with production of photosynthetic pigments and expansion and differentiation of the cotyledons. However, if the dark-grown cop1 seedlings are transferred to the light, a significant portion of the seedlings are unable to green and will bleach out and die. This COP1-dependent block of greening phenotype follows an allelic series and becomes more pronounced the longer the seedlings have been grown in the dark (Ang and Deng 1994). We found that a significant percentage of hyh/cop1-4 (37.7%) and hyh/cop1-6 (29.1%) seedlings were able to green when germinated in the dark for 4 d and then transferred to light for an additional 4 d. In contrast, no dramatic difference was observed between cop1-1 and cop1-1/hyh seedlings, suggesting that hyh acts as an allele-specific suppressor of this cop1 phenotype (Fig. 7). This allele-specific suppression indicates a genetic interaction between hyh and cop1 and supports the notion of a direct physical interaction between the two proteins. Thus, our evidence places HYH in the same regulatory hierarchy as HY5.
Figure 7.
The hyh mutation suppresses the block-of-greening phenotype of cop1 alleles. Seedlings were germinated in the dark for 4 d and then transferred to constant white light for 4 d. Seedlings with green cotyledons and/or true leaves were scored as able to green, and bleached seedlings were scored as unable to green (n = >109).
HYH and HY5 interact and bind DNA together in vitro
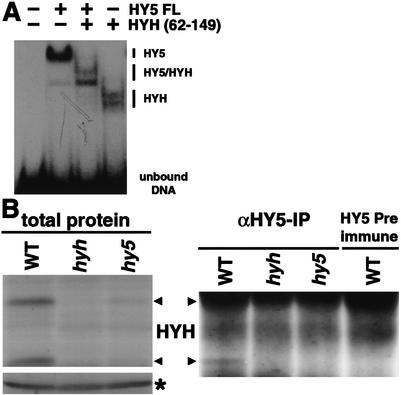
Considering their similar role in light-regulated development and the high homology between the DNA-binding basic domains of HYH and HY5 (21 acids are identical), it could be expected that these two proteins recognize the same or similar DNA sequences. HY5 has previously been shown to bind the G-box motif commonly found in light-regulated promoters (Chattopadhyay et al. 1998). To examine the ability of HYH to bind DNA, we used recombinant HYH and HY5 proteins in electrophoretic mobility shift assays (EMSA) with the G-box sequence from the RBCS promoter (Chattopadhyay et al. 1998). We found that full-length HYH binding to the G-box is comparable to that of HY5 (data not shown). However, because the mobility shifts observed for HY5 and HYH homodimers are very close, we were unable to determine whether the proteins are able to form heterodimers upon mixing. To increase the resolution of the assay, we expressed a truncated version (amino acids 62–149) of HYH containing the bZIP domain. Pre-existing homodimeric complexes were dissociated by incubation of full-length HY5, HYH62–149, and a mixture of HY5 and HYH62–149 at 50°C for 5 min prior to addition to the DNA. As shown in Figure 8A, HY5 and HYH62–149 form discrete protein–DNA complexes that are readily resolved on the gel. Interestingly, when HY5 and HYH62–149 are mixed together, a complex with intermediary mobility can be detected. Because the DNA probe contains only one copy of the G-box motif, our result suggests the formation of HY5/HYH62–149 heterodimers when the proteins are incubated together. Furthermore, when mixing the HY5 and HYH62–149 proteins, the HY5/HYH62–149 heterodimeric complex constitutes the clear majority of protein binding to DNA, suggesting that the HY5/HYH62–149 heterodimer complex is preferentially formed in vitro.
Figure 8.
HYH interacts with HY5 on DNA and in vivo. (A) Gel retardation using recombinant HY5 and HYH protein binding to the G-box of the RBCS promoter. Approximately 40 ng each of the HY5 and HYH protein were mixed and incubated at 50°C for 5 min to dissociate pre-existing dimers prior to addition to radioactively labeled probe. The protein–DNA complexes were resolved on a 4% native 0.5× TBE polyacrylamide gel. (B) HY5 immunoprecipitation of 3-DAG wild-type, hyh, and hy5-ks50 seedlings grown in white light. (Left panel) Total protein extract, (*) a cross-reacting protein band serving as loading control, and (right panel) the anti-HY5 precipitate following immunodetection with HYH antibodies. The right lane shows the HY5 preimmune precipitate from wild-type extract.
HYH and HY5 interact in vivo
To examine whether a complex containing both HYH and HY5 protein can be detected in vivo, we performed coimmunoprecipitation experiments using antibodies against HY5. As shown in Figure 8B, the HYH proteins are readily detectable in 3-DAG wild-type seedlings and present at reduced levels in hy5 seedlings. After immunoprecipitation with HY5 antibodies (Fig. 8B), we found that the HYH proteins, in particular the 21-kD band, are clearly precipitated from the wild-type extract, whereas no protein is detected from the hy5 and hyh extracts, as expected. Because the ∼27-kD HYH protein is largely masked by the IgG light chain, it is not feasible to judge its abundance in the immunoprecipitates. The absence of HYH protein in immunoprecipitates from hy5 seedlings suggests that the presence of HY5 protein is required for the immunoprecipitation of HYH protein with HY5 antibodies. The ability of HY5 antibodies to precipitate HYH protein appear to be specific because no HYH protein is detected in the HY5 preimmune precipitate from the wild-type extract (Fig. 8B). The result therefore indicates that precipitation of the ∼21-kD HYH protein requires the presence of HY5 protein, suggesting that HYH and HY5 are physically associated in vivo.
Overlapping genome expression profiles in hyh and hy5 seedlings
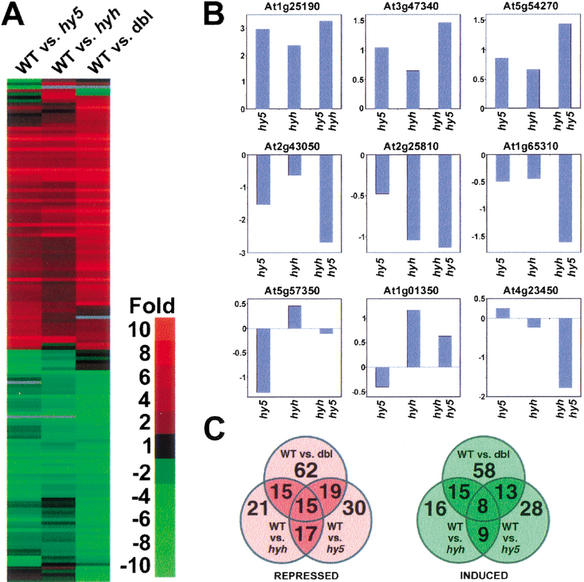
To further investigate the functional relevance of the observed physical interaction between HYH and HY5, we compared the expression profiles in blue-light-grown wild-type seedlings with that of hyh, hy5, and hyh/hy5 mutant seedlings using an EST microarray. The microarray contains a total of 9216 ESTs, corresponding to an estimated 6200 unique genes (Ma et al. 2001). Of the 9216 ESTs, 292, or 3.2%, were ≥twofold up- or down-regulated compared with wild type in at least one of the three mutant strains. The 292 EST entries with twofold or more differential regulation correspond to 149 unique genes (http://www.plantgenomics.biology.yale.edu). The cluster analysis (Fig. 9A) shows that the differential expression (as indicated by color intensity of the bands) is weakest in wild-type versus hyh and strongest in the wild-type versus hyh/hy5, corresponding well with the observed phenotype.
Figure 9.
A functional overlap between HYH and HY5. (A) Hierarchical clustering display of the expression ratios from 6-DAG wild type (WT) seedlings versus hy5-ks50, hyh, or hyh/hy5-ks50 (dbl) seedlings, all grown in blue light. The 149 genes (http://www.plantgenomics.biology.yale.edu/) that showed at least twofold differential change in at least one of the three sample pairs were included for comparison. (B) The differential expression of nine different genes in the hy5, hyh, and hy5/hyh experiments. See text for predicted function and domains of the nine displayed genes. The ratio of wild-type/mutant is expressed as log2(ratio). (C) Venn diagrams of the number of differentially expressed genes that showed twofold or higher expression in wild-type (repressed-red) or lower (induced-green) compared with respective mutant. The numbers in the overlapping areas indicate the shared number of genes showing twofold or higher differential expression in either two or three sample pairs.
Of the 149 genes with twofold or more differential regulation in at least one of the three mutant strains, 76 genes had higher expression in wild type than in respective mutant (repressed), and 73 genes had higher expression in the mutants compared with wild type (induced). The vast majority of the 149 genes, 135 (90.6%), were qualitatively similarly regulated in all three mutant strains, suggesting an extensive overlap between the expression profiles of hyh and hy5. The expression profiles for some of the representative genes are shown in Figure 9B. For example, Arabidopsis gene At1g25190 (encoding an F-box protein) showed the highest fold repression in all three mutant strains with a ratio of 9.6, 7.9, and 5.1 in the hyh/hy5, hy5, and hyh mutants, respectively. Other repressed genes include At3g47340 (encoding asparagine synthase) and genes encoding chloroplast-targeted proteins such as Lhcb3 (At5g54270), consistent with the specific role of HYH and HY5 in light-dependent G-box-mediated transcription. On the other hand, several of the genes that were expressed at higher levels in the mutant strains encode proteins affecting hypocotyl elongation. These include At2g43050 and At1g65310, which encode a putative pectinesterase and a xyloglucan endotransglycosylase, respectively, as well as tonoplast-associated protein such as the putative aquaporin At2g25810 (Fig. 9B), consistent with the hypocotyl phenotype seen in the hyh and hy5 mutants.
However, the gene expression profile overlap was not complete, and the hy5 and hyh mutations had opposite effects on the expression of 11 different genes (7.3% of the 149 genes), suggesting that HYH and HY5 also have distinct or opposite roles toward certain target genes. The genes that are regulated oppositely in the hyh and hy5 mutants include At5g57350, which encodes the plasma membrane proton pump ATPase 3, and the RING finger protein-encoding genes At1g01350 and At4g23450 (Fig. 9B).
Figure 9C shows Venn diagrams of the genes with twofold or higher difference in expression in each of the three mutant strains. The expression profiles of hyh and hy5 seedlings showed an almost complete (17/21) overlap in the genes repressed by the mutations. The apparent overlap was slightly less for the genes expressed more highly in the mutants. Of the 16 genes with more than twofold expression in the hyh mutants, only nine genes also show twofold or higher expression in the hy5 mutant. However, the remaining five genes all showed higher expression in the hy5 mutant than in the wild type but fell short of the twofold threshold. Taken together, the high degree of overlap in the expression profiles of hyh, hy5, and hyh/hy5 seedlings and the finding that the fold of differential expression is lowest in the hyh mutant and highest in the hyh/hy5 mutant strongly support the notion that HYH and HY5 predominantly act in concert on the same set of genes under blue light.
Discussion
This study identifies HYH as a target of COP1-mediated protein degradation in the dark. We show that HYH mediates light-dependent transcription and revealed a functional relationship between HYH and HY5.
The interaction with the WD40 domain of COP1 results in COP1-dependent degradation of HYH
We identified HYH in a yeast two-hybrid screen using full-length COP1 protein as a bait. We characterized the interaction between HYH and COP1 in yeast and found that the interactions require both the WD40 domain in COP1 and a functional COP1-interacting motif in HYH. Additional and independent support for the interaction between HYH and COP1 was obtained using GFP and BFP fusions of the proteins in living plant cells. Furthermore, the allele-specific ability of an hyh mutation to suppress the block-of-greening phenotype in viable cop1 alleles shows a genetic interaction between the two genes, thus providing genetic support for the interaction.
Previous work suggested that the interaction between COP1 and HY5 resulted in COP1-dependent degradation of HY5 in the dark (Osterlund et al. 2000). In an effort to analyze a potential functional role of the interaction between COP1 and HYH, we assayed the levels of HYH protein upon transfer to darkness. As with HY5, we found that the level of endogenous HYH protein is dramatically decreased after 24 h in the dark. Our results suggest that the decrease occurs on a posttranscriptional level because a similar decrease in HYH protein levels was seen in seedlings overexpressing the HYH protein from a heterologous promoter. Similar to HY5, the reduction of HYH protein levels requires the activity of the COP/DET/FUS proteins, and no reduction of HYH protein is seen in cop/det/fus mutants after 20 h in darkness. Finally, we show a direct correlation between HYH interaction with amino-acid-substituted COP1 proteins in yeast two-hybrid assays and the ability of the same substituted COP1 proteins to mediate dark-dependent degradation of HYH. This strongly supports the notion that the interaction between COP1 and HYH, like the interaction between COP1 and HY5, results in a direct COP1-mediated degradation of HYH in the dark.
HYH mediates light-dependent transcription and shows a functional overlap with HY5
Since both HYH and HY5 are regulated by COP1, we have tried to determine the individual contributions as well as the functional interplay between these two proteins. HYH is a close homolog of HY5, and both proteins are members of the large bZIP family of transcription factors. HY5 has previously been shown to be required for accumulation of anthocyanin pigments, and deficiency of HY5 results in suppression of the inhibition of hypocotyl elongation in all light conditions, suggesting that HY5 acts downstream of each of the photoreceptors.
We found that hyh plants flower early. However, no major effect on anthocyanin or chlorophyll accumulation was detected in white light. Furthermore, unlike hy5 mutant seedlings, the hypocotyl phenotype of hyh seedlings is restricted to blue-light-specific inhibition of hypocotyl elongation. This suggests that, although HYH is expressed at high levels in white, red, and blue light, HYH predominantly acts as an essential positive regulator of light signals mediated by the blue-light photoreceptors CRY1 and CRY2. HYH is thus the first example of a transcription factor specifically affecting blue-light signaling. Interestingly, the simultaneous loss of both HYH and HY5 proteins in blue-light-grown seedlings has an additive effect on hypocotyl length. This suggests that the two proteins have an overlapping yet nonredundant function in the transcriptional response to blue-light signaling.
The functional overlap between HYH and HY5 in blue-light signaling was further characterized in microarray experiments. The expression profiles of hyh and hy5 seedlings showed an extensive overlap: 135 of the 149 genes showing differential expression were similarly regulated in all three experiments. Only 11 genes (7.3%) showed distinct regulation in the hyh and hy5 mutants. This high level of functional overlap between HYH and HY5 in blue-light-dependent transcription stands in stark contrast to the phenotypic differences seen in white-light-grown hyh and hy5 seedlings. In white light, where light signals are transduced by both phytochromes and cryptochromes, only hy5 gives a clear phenotype. This suggests that the HYH and HY5 proteins differ in their abilities to respond to phytochrome-mediated signals. However, because overexpressed HYH protein is able to compensate for the much broader phenotype of hy5 mutant seedlings in white light, we conclude that elevated levels of HYH can overcome the apparent lower sensitivity of HYH to phytochrome signaling and regulate expression of the same set of genes as HY5 in white light.
Therefore, HYH has a distinct role in blue-light-dependent transcription, where it regulates a set of genes overlapping that of HY5. Although HYH and HY5 differ in their ability to respond to different photoreceptors, the finding that increased levels of HYH can partially suppress the white-light phenotype of hy5 suggests that the two proteins can act on the same set of genes in other light conditions also.
The mechanistic relationship between HYH and HY5 in light-regulated transcription
We found that the HYH and HY5 proteins interact in vivo and that they readily form G-box-binding heterodimers, suggesting that HYH and HY5 can act together. We have previously shown that HY5 interacts well with the K422E-, R465E-, and E592R-substituted COP1 proteins in yeast. However, only seedlings expressing COP1-E592R were able to degrade HY5 to the same extent as wild-type COP1, whereas COP1-K422E and -R465E had dramatically reduced, but clearly detectable, levels of HY5 protein after 20 h in darkness (Holm et al. 2001). If the HY5 and HYH proteins exist as heterodimers in vivo, it is conceivable that the degradation of HY5 would be slower in seedlings where the COP1 protein is unable to recognize the HYH heterodimer partner. This result would be consistent with the notion that the HY5 and HYH proteins exist in an equilibrium of homo- and heterodimers.
Our results show that although HYH transcription is maintained at wild-type levels in hy5 seedlings, the HYH protein level is dramatically reduced. This suggest that the presence of the HY5 protein is important for the accumulation of HYH protein. It is possible that HY5 directly enhances the stability of the HYH protein by allowing formation of more stable heterodimers. However, an alternative possibility is that HY5 indirectly affects HYH protein levels by regulating factors involved in the translation or stability of the HYH protein. If an equilibrium exists between HYH and HY5 homo- and heterodimers, the relative strength of hy5 and hyh single mutants would suggest that HY5 homodimers could mediate a large portion of the HY5/HYH heterodimer function in the hyh mutant, but the HYH homodimer is only able to partially compensate for the loss of both HY5 homodimers and HY5/HYH heterodimers in an hy5 mutant. The fact that hyh shows a largely blue-light-specific phenotype would suggest that the HYH homodimer and HY5/HYH heterodimer play an essential role in blue-light-mediated gene expression that can not be substituted by the HY5 homodimer.
COP1 regulation of light-dependent transcription
The finding that degradation of HYH is impaired in cop1, det1, and cop10 mutants as well as in the cop/det/fus mutant lacking the COP9 signalosome, gives support for the notion that the COP/DET/FUS proteins mediate degradation of a set of key transcription factors. The very low expression of HYH in the dark, together with the inability of the hyh mutation to suppress the phenotype of dark-grown cop1 seedlings, suggests that HYH mainly functions in the light.
The current model proposes that COP1 accumulates in the nucleus in the dark and interacts with light-dependent transcription factors, and that this interaction targets these factors for proteasome-mediated degradation with the involvement of the COP9 signalosome, COP10, and possibly DET1. Light decreases the nuclear abundance of COP1 and thereby relieves the repression of the transcription factors. Although the cop/det/fus mutants show striking phenotypes in darkness, light-grown null mutants in all 11 loci are adult lethal and the viable cop1 alleles are stunted, flower early, and are deficient in light-dependent physiological responses such as the shade-avoidance response and the end-of-day far-red response. The role of COP1 in the light suggests a more dynamic interplay between the positive signals from the photoreceptors and repressive activity of COP1. Light signals from the photoreceptors decrease the nuclear levels of COP1; however, some COP1 protein is likely to remain in the nucleus, allowing fine tuning of the transcriptional response.
The positive signals from the photoreceptors received by downstream transcription factors are thus balanced by the ability of the COP1 to target some of these transcription factors (including HY5 and HYH) for degradation, allowing a very dynamic control of the transcriptional output. Because much of the light signaling appears to act directly on transcription factors, future work will be directed at identifying more of these downstream factors so that the mechanisms of their activation as well as the regulatory interplay between them can be revealed.
Materials and methods
Biological materials and growth conditions
The Escherichia coli strain BNN123 was used for library excision, DH5α for subcloning, and BL21 DE3 for expression of recombinant protein. The Agrobacterium strain GV3101 pMP90 was used for plant transformation. The yeast strain Y190 (Kim et al. 1997) was used for the two-hybrid screen and for the two-hybrid assays. The hy5-ks50 (Oyama et al. 1997), hyh, and cop10-1 alleles are in the Wassiljevskaja ecotype as are the cop1-5 plants overexpressing wild type and K422E-, R465E-, W467A-, K550E-, and E592R-substituted COP1 proteins. The cop9-1, det1-1, cop1-1, cop1-4, and cop1-6 alleles as well as the HYH-OE plants overexpressing HYH are in the Col-0 ecotype. The HYH-OE/hy5-215 plants were generated by crossing HYH-OE into the Col-0 allele hy5-215. The HYH cDNA (accession no. AF453477) was identical to portions of the predicted gene At3g17610 (The Arabidopsis Genome Initiative 2000), which is predicted to contain five exons. However, the cDNA differs from the prediction in that it contains 51 nucleotides 5′ of the ATG and that the third exon was the last exon but extends 151 nucleotides, including an in-frame stop codon 3′. The COP1 cDNA for transgenic expression under control of the 35S cauliflower mosaic virus 35SCaMV promoter and terminator was cloned into pPZP222 (Hajdukiewicz et al. 1994). The HYH cDNA was cloned into pBAR-A (DDBJ/EMBL/GenBank accession no. AJ251013) for expression under control of the full-length 35S CaMV promoter and the nopaline synthase terminator. The 35S::COP1 and HYH-OE constructs were transformed into cop1-5 (Deng et al. 1992) and Col-0 plants, respectively, by the floral dip method (Clough and Bent 1998). Homozygous lines were then established from primary transformants. At least two independent lines with similar HYH and COP1 protein levels, respectively, were selected and used for detail analysis. Seeds were surface-sterilized, put on Murashige Skoog plates containing 1% sucrose, and cold-treated at +4°C for 72 h prior to germination. The Arabidopsis growth conditions have been described previously (Hsieh et al. 2000). The intensities of continuous light sources are: white light (173, 25, 2.2 μmole/sec per m2), blue light (21.6, 2.6, 0.13 μmole/sec per m2), red light (115, 19, 2.4 μmole/sec per m2), and far-red light (145, 22.5, 2.9 μmole/sec per m2). Unless specifically stated, the highest light intensities for each light regime were used for the experiments.
Yeast two-hybrid methods and onion experiments
The λACT cDNA expression library (Arabidopsis Biological Resource Center no. CD4-22) was converted into a pACT library according to Durfee et al. (1993). The two-hybrid screen was performed essentially as described in Kim et al. (1997). The β-galactosidase assays were performed as previously described (Holm et al. 2001). The pRTL2–S65TGFP–HYH and pRTL2–BFP–COP1 constructs, containing versions of the green fluorescent protein (GFP) that have optimum excitations by blue and UV light and have maximum emission for green and blue light, respectively (von Arnim et al. 1998), were introduced into onion epidermal cells by particle bombardment as previously described (Ang et al. 1998). After bombardment, the onion cell layers were incubated at 22°C for 48 h in darkness. The cell layers were then mounted in water and examined by epifluorescence microscopy as previously described (Ang et al. 1998).
Chlorophyll and anthocyanin measurements
The chlorophyll and anthocyanin measurements were performed essentially as described (Chory et al. 1991, 1998). Briefly, for the chlorophyll measurements, 4-DAG seedlings were harvested in darkness, weighed, frozen in liquid nitrogen, and ground to a fine powder. Total chlorophyll was extracted into 80% acetone, and chlorophyll a and b content were calculated using MacKinney's specific absorption coefficients in which chlorophyll a = 12.7(A663) − 2.69(A645) and chlorophyll b = 22.9(A645) − 4.48(A663). The total specific chlorophyll content is expressed as micrograms of chlorophyll per gram seedlings.
For the anthocyanin determinations, 3-DAG seedlings were weighed, frozen in liquid nitrogen, and ground, and total plant pigments were extracted overnight in 0.3 mL of 1% HCl in methanol. After addition of 0.2 mL of H20, chlorophyll was extracted with an equal volume of chloroform. The quantity of anthocyanins was determined by spectrophotometric measurements of the aqueous phase (A530 − A657) and normalized to the total fresh weight of tissue used in each sample.
Gel retardation and immunoprecipitation
Recombinant truncated HYH protein with amino acids 62–149 fused to the maltose-binding protein (MBP) in the pMAL-c2X vector (New England Biolabs) was expressed in bacteria and purified with amylose resin (Biolabs) according to the manufacturer's protocol. The MBP was cleaved overnight at 4°C with factor Xa (Biolabs); the protease was subsequently inhibited with 10 μM of the specific inhibitor 1,5-Dansyl-Glu-Gly-Arg Chloromethyl Ketone (Calbiochem).
Then ∼40 ng of HYH62–149 protein and 40 ng of purified His-tagged HY5 protein (Osterlund et al. 2000) were incubated at 50°C for 5 min either alone or mixed together in a volume of 4 μL. Two microliters of the heat-treated proteins were then added to 18 μL of buffer (15 mM HEPES, 1 mM EDTA, 6% glycerol, 1 mM DTT, 2 mM MgCl2, and 1 mM ZnSO4) containing 20,000 cpm kinased and annealed oligos 5′-AATTATCT TCCACGTGGCATTATTCC-3′. The proteins were incubated with DNA at 22°C for 10 min prior to separation on a 4% native polyacrylamide gel with 0.5× TBE as buffer. Immunoprecipitations were performed according to a previously published procedure (Serino et al. 1999).
Microarray analysis
Microarray experiments and data analysis were performed according to a previously established protocol (Ma et al. 2001). Briefly, 100 μg of total mRNA from 6-DAG blue-light-grown hyh, hy5, hyh/hy5, and wild-type seedlings was used to synthesize Cy3- and Cy5-labeled cDNA, respectively. Pairwise combinations of wild-type cDNA and an equal amount of respective mutant cDNA were used to simultaneously probe a glass slide containing duplicate arrays in three microarray experiments. Each experiment was repeated with reverse labeling.
Acknowledgments
We are grateful to the Arabidopsis knockout facility, Madison WI, for the hyh T-DNA insertion line and to the Arabidopsis stock center for the CD4-22 yeast two-hybrid library. We thank Casey Dunn, James Sullivan, Haiyang Wang, and Vicente Rubio for critically reading the manuscript. This work was supported by a NIH grant (GM47850) and in part by grants from the Human Frontier Science Program Organization (RG0043/97) and the National Program for Transgenic Plants from China (J99-A-001). M.H. was in part supported by a postdoctoral fellowship from the Wenner-Gren foundation and the Swedish Medical Research Council, and X.W.D. was an NSF Presidential Faculty Fellow.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL xingwang.deng@yale.edu; FAX (203) 432-3854.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.969702.
References
- Ang LH, Deng XW. Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell. 1994;6:613–628. doi: 10.1105/tpc.6.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH. LAF1, a MYB transcription activator for phytochrome A signaling. Genes & Dev. 2001;15:2613–2625. doi: 10.1101/gad.915001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1998;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G β homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes & Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Hudson M, Ni M, Qin M, Quail PH. poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc Natl Acad Sci. 1999;96:5832–5837. doi: 10.1073/pnas.96.10.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW. HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 2000;19:4997–5006. doi: 10.1093/emboj/19.18.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 2001;20:118–127. doi: 10.1093/emboj/20.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh H-L, Okamoto H, Wang M, Ang L-H, Matsui M, Goodman H, Deng X-W. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes & Dev. 2000;14:1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. Protein–protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Chen Z, Zhao H, Deng XW. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. Light: An indicator of time and place. Genes & Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix–loop–helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Ang LH, Deng XW. The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 1999;9:113–118. doi: 10.1016/s0962-8924(99)01499-3. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. Phytochrome-interacting factors. Semin Cell Dev Biol. 2000;11:457–466. doi: 10.1006/scdb.2000.0199. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292:1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- Serino G, Tsuge T, Kwok S, Matsui M, Wei N, Deng XW. Arabidopsis cop8 and fus4 define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell. 1999;11:1967–1979. doi: 10.1105/tpc.11.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh MS, Kim YM, Han SJ, Song PS. REP1, a basic helix–loop–helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell. 2000;12:2061–2074. doi: 10.1105/tpc.12.11.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman JI, Mindrinos MN, Fankhauser C, Richards D, Lutes J, Chory J, Oefner PJ. Cloning of the Arabidopsis RSF1 gene by using a mapping strategy based on high-density DNA arrays and denaturing high-performance liquid chromatography. Plant Cell. 2000;12:2485–2498. doi: 10.1105/tpc.12.12.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Hicks SN, von Arnim AG. Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell. 1999;11:349–364. doi: 10.1105/tpc.11.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S. The Arabidopsis knockout facility at the University of Wisconsin–Madison. Plant Physiol. 2000;124:1465–1467. doi: 10.1104/pp.124.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Stoop-Myer CD, Okamoto H, Coleman JE, Matsui M, Deng XW. The RING finger motif of photomorphogenic repressor COP1 specifically interacts with the RING-H2 motif of a novel Arabidopsis protein. J Biol Chem. 1999;274:27674–27681. doi: 10.1074/jbc.274.39.27674. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene. 1998;221:35–43. doi: 10.1016/s0378-1119(98)00433-8. [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Yamamoto YY, Matsui M, Ang LH, Deng XW. Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell. 1998;10:1083–1094. doi: 10.1105/tpc.10.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Deng XW, Matsui M. Cip4, a new cop1 target, is a nucleus-localized positive regulator of Arabidopsis photomorphogenesis. Plant Cell. 2001;13:399–411. doi: 10.1105/tpc.13.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]