Abstract
CDK2 activity is regulated by phosphorylation/dephosphorylation, subcellular localization, cyclin levels, and cyclin dependent kinase inhibitors (CKIs). Using Xenopus egg extracts, we find that degradation of Xic1, a Xenopus p21cip1/p27kip1 family member, is coupled to initiation of DNA replication. Xic1 turnover requires the formation of a prereplication complex (pre-RC). Additionally, downstream initiation factors including CDK2, Cdc7, and Cdc45, but not RPA or DNA polymerase α, are necessary for activating the degradation system. Xic1 degradation is attenuated following completion of DNA replication. Unlike degradation of p27kip1 in mammalian cells, CDK2 activity is not directly involved in Xic1 degradation and interactions between Xic1 and CDK2/cyclin E are dispensable for Xic1 turnover. Interestingly, a C-terminal region (162–192) of Xic1 is essential and apparently sufficient for triggering Xic1 ubiquitination prior to degradation. These observations demonstrate that a direct link exists between DNA replication and CKI degradation.
Keywords: CDK inhibitor, Xic1, initiation of DNA replication, CDK2/cyclin E
Cell cycle transitions are driven by the activity of a set of distinct cyclin-dependent kinases (CDKs). To ensure genome integrity, CDKs are only activated during appropriate windows of the cell cycle. CDK activity is post-translationally regulated at multiple levels, including phosphorylation/dephosphorylation and association with positively regulating cyclins and negatively regulating CKIs (Morgan 1997; Sherr and Roberts 1999).
Although the complete molecular details of its functions are not clear, CDK2 activity is essential for the initiation of DNA replication and progression through S phase in metazoans (Morgan 1997). Initiation of DNA replication involves the stepwise assembly of a set of initiation factors at origins of replication (Stillman 1996; Dutta and Bell 1997). The assembly process is under strict control ensuring the genome is replicated only once in each cell cycle (Diffley 1996). In G1, prereplication complexes (pre-RCs), composed of ORC, Cdc6, Cdt1, and MCM, are assembled on chromatin (Coleman et al. 1996; Hua et al. 1997; Maiorano et al. 2000). At the G1/S transition, Cdc45 is loaded onto chromatin to form a pre-initiation complex (pre-IC) (Zou and Stillman 1998). This requires the activity of at least two kinases, CDK2/cyclin E and Cdc7/Dbf4 (Owens et al. 1997; Mimura and Takisawa 1998; Zou and Stillman 1998, 2000). Subsequently, RPA, which facilitates DNA unwinding, and DNA polymerase α, which is responsible for synthesis of RNA–DNA primers, associate with chromatin (Tanaka and Nasmyth 1998; Walter and Newport 2000). Following this, a proliferating cell nuclear antigen (PCNA)-dependent polymerase switch step is necessary for the loading of DNA polymerase δ, which then synthesizes DNA in a processive manner (Tsurimoto and Stillman 1989; Lee and Hurwitz 1990; Burgers 1991; Lee et al. 1991; Podust et al. 1992). In somatic cells, replication initiation is asynchronous. Recent studies in yeast indicate that Cdc7, Cdc45, and S phase CDKs (e.g., Cdk1/Clb5) are all required for the activation of late origins (Bousset and Diffley 1998; Donaldson et al. 1998a,b; Tercero et al. 2000). Additionally, recent studies reveal that CDK2/cyclin E is involved in the duplication of centrosomes (Hinchcliffe et al. 1999; Lacey et al. 1999; Matsumoto et al. 1999; Meraldi et al. 1999) and the replication-dependent transcription of histone genes during S phase (Ma et al. 2000).
Genetic and biochemical data in budding yeast indicate that an inhibitor of the S phase CDK, SIC1, is degraded at the G1/S transition through the ubiquitin-proteasome pathway (Sheaff and Roberts 1996; Deshaies 1997). Abrogation of the degradation prevents entry into S phase (Schwob et al. 1994; Schneider et al. 1996; Tyers 1996). Ubiquitination and subsequent degradation of SIC1 requires CDC34 and the SCFcdc4 complex including CDC53, Skp1, CDC4, and Rbx1/Roc1/Hrt1(Skowyra et al. 1997; Verma et al. 1997a,b; Kamura et al. 1999; Seol et al. 1999; Skowyra et al. 1999). A feature of this degradation system is that it is phosphorylation-dependent (Skowyra et al. 1997; Verma et al. 1997a; Nishizawa et al. 1998). It has been proposed that G1 CDKs phosphorylate SIC1, thereby, generating a substrate that is recognized by the F-box protein, CDC4 (Tyers 1996). Recent work shows that ubiqutination and degradation of p27kip1 also require components of the mammalian SCF complex (Tsvetkov et al. 1999; Nakayama et al. 2000). In S phase, p27kip1 turnover is dependent on phosphorylation on threonine 187 (Thr 187) by CDK2 and the interaction between p27kip1 and CDK2 (Vlach et al. 1997; Montagnoli et al. 1999; Nguyen et al. 1999; Tsvetkov et al. 1999; Malek et al. 2001). However, p27kip1 can also be degraded independently of T187 phosphorylation, which apparently occurs in G1 phase of the cell cycle (Malek et al. 2001). Although there have been efforts to understand the downstream factors required for the degradation of CKIs, little is known about the regulators that initiate this during the cell cycle.
It has been shown that Xic1, a member of the p21cip1/p27kip1 CKI family in Xenopus, is degraded in a nucleus and Cdc34 dependent process by the ubiquitin–proteasome pathway in Xenopus extracts (Yew and Kirschner 1997; Swanson et al. 2000; Chuang and Yew 2001). In this study, we have investigated the cell cycle regulated degradation of Xic1. Specifically, we have characterized the relationship between initiation of DNA replication and degradation of Xic1 using extracts derived from Xenopus eggs. Our results indicate that Xic1 degradation is coupled to initiation of DNA replication. The degradation of Xic1 requires the assembly of pre-RCs. Furthermore, initiation factors such as CDK2, Cdc7 and Cdc45, but not RPA or DNA polymerase α, are necessary for Xic1 turnover. These observations indicate that during initiation a Xic1 degradation signal is generated after the recruitment of Cdc45 to chromatin but before the association of RPA. In addition, we have found that Xic1 is stable following completion of DNA replication. Our analysis has revealed that CDK2/cyclin E activity is not directly involved in Xic1 turnover. The conserved CDK phosphorylation site in the C terminus of Xic1 is not essential, and the interactions between Xic1 and CDK2/cyclin E are dispensable for Xic1 elimination. Importantly, we have found that a region (162–192) in the C-terminal part of Xic1 may trigger Xic1 ubiquitination, probably through its interaction at the replication fork.
Results
Xic1 is degraded in egg extracts in a DNA-dependent manner
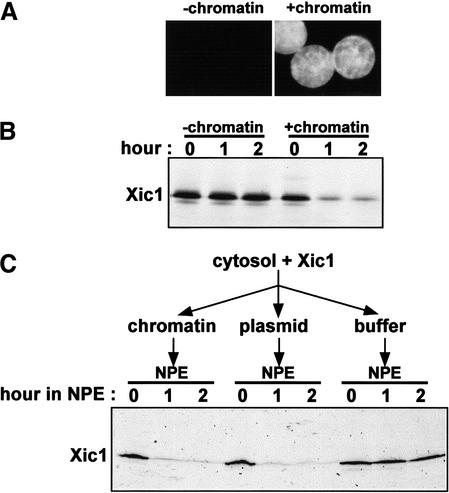
To investigate the regulation of Xic1 during the cell cycle, we used Xenopus egg extracts and in vitro translated [35S]methionine-labeled Xic1. When purified membrane vesicles and demembranated sperm chromatin are added to egg cytosol, nuclei form (Newport 1987; Sheehan et al. 1988). Following nuclear assembly, a single, complete round of semiconservative DNA replication takes place (Blow and Laskey 1986; Newport 1987). When isolated sperm chromatin was incubated with egg cytosol and membranes in the presence of a trace amount of labeled Xic1, nuclei formed (Fig. 1A) and, as reported previously (Yew and Kirschner 1997; Swanson et al. 2000; Chuang and Yew 2001), Xic1 was degraded more than 80% within 1 h (Fig. 1B). Xic1 translated in a bacterial lysate (Novagen, EcoPro transcription/translation system) was also degraded in the egg extract under the same conditions (data not shown). Importantly, when sperm chromatin was not added to the egg extract, nuclei were not assembled (Fig. 1A) and Xic1 was stable (Fig. 1B). Thus, Xic1 is degraded in a nucleus-dependent manner in complete Xenopus egg extracts.
Figure 1.
Degradation of Xic1 in Xenopus egg extracts requires chromatin and a nuclear environment. (A) Nuclei assembled in extracts with in vitro translated Xic1 were stained with Hoechst and visualized by fluorescence microscopy. (B) Radiolabeled Xic1 was incubated with extracts with or without nuclei for the indicated times. Xic1 remaining in extracts was analyzed by SDS-PAGE followed by autoradiography. (C) Radiolabeled Xic1 was incubated with cytosol supplemented with 3000/μL sperm chromatin, 5 ng/μL pBluescript plasmid DNA, or ELB buffer. Subsequently, 2 volumes of NPE was added. Reactions were terminated at indicated times after the addition of NPE and Xic1 was visualized as in B.
To further investigate this nucleus-dependent Xic1 degradation, we asked whether the assembly of a nuclear envelope around sperm chromatin is a prerequisite for Xic1 degradation. To address this issue, we used a nucleus-free DNA replication system (Walter et al. 1998), which does not require the addition of a purified membrane fraction. To do this, sperm chromatin, plasmid pBluescript DNA, or ELB buffer was incubated with egg cytosol containing labeled Xic1 for 30 min. Subsequently, 2 volumes of a concentrated solution of nuclear proteins, NPE, was added to each reaction. As shown in Figure 1C, when NPE was added, Xic1 was degraded efficiently in the presence of sperm chromatin or plasmid DNA. However, in the absence of DNA, Xic1 was stable (Fig. 1C). These observations demonstrate that the nuclear envelope is not essential for Xic1 degradation. Rather, DNA within nuclei or DNA in a nuclear-like environment (NPE) is required for triggering Xic1 degradation.
Xic1 degradation requires chromatin-bound MCM7
Because CKI degradation typically occurs at the onset of S phase we investigated the relationship between DNA replication and Xic1 degradation. Initiation of DNA replication can be separated into two distinct steps. In the first step, ORC, Cdc6, Cdt1, and MCM proteins sequentially bind to chromatin to assemble pre-RCs (Coleman et al. 1996; Hua et al. 1997; Maiorano et al. 2000). This step does not depend on CDK2. In the second step, downstream initiation factors are recruited to pre-existing pre-RCs to form pre-ICs and activate DNA replication. This second step requires the activity of CDK2/cyclin E and Cdc7/Dbf4 kinases (Stillman 1996; DePamphilis 1998; Leatherwood 1998). In Xenopus extracts, pre-RCs form on DNA added to cytosol alone. These preformed pre-RCs are then activated to initiate replication when either membranes are added to induce nuclear formation or when NPE is added to the cytosol (Walter et al. 1998).
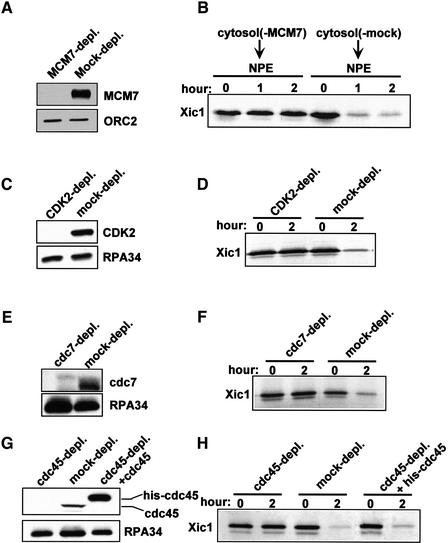
To examine a potential role for replication components in Xic1 degradation we inhibited pre-RC formation by removing MCM from cytosol prior to addition of DNA. To do this, cytosol was treated with protein A beads coupled with either MCM7 antiserum or pre-immune serum. Western blotting results showed that more than 99% of MCM7 was removed from the cytosol treated with MCM7 antiserum, while in the pre-immune serum treated cytosol MCM7 was present at normal levels (Fig. 2A). Sperm chromatin and labeled Xic1 were added to the treated cytosol for 30 min to allow the assembly of pre-RCs. Subsequently, two volumes of NPE was added to each reaction. Xic1 stability and DNA replication were then measured.
Figure 2.
Xic1 degradation requires the association of MCM7 with chromatin, CDK2, Cdc7, and Cdc45. (A) Western blots of MCM7 and ORC2 in the MCM7-depleted or the mock-depleted cytosol. (B) Chromatin and Xic1 were mixed with the treated cytosol depicted in A for 30 min. Subsequently, 2 volumes of untreated NPE was added to each reaction. Xic1 degradation was measured after the addition of NPE. (C,E,G) Western blots of CDK2, Cdc7, Cdc45, and RPA in CDK2-, Cdc7-, and Cdc45-depleted and mock-depleted cytosol. Recombinant histidine-tagged Cdc45 protein (75 ng/μL) was added to rescue activities of the Cdc45-depleted extract. (D,F,H) Xic1 degradation was determined in the treated extracts containing nuclei.
As expected, Xic1 degradation (Fig. 2B) occurred efficiently in the mock-depleted extract. However, when MCM7 was depleted from cytosol, both Xic1 degradation (Fig. 2B) and DNA replication (data not shown) were inhibited. Removal of MCM7 from cytosol prevents the functional assembly of pre-RCs on chromatin. Although MCM proteins are also present in NPE, an activity in NPE, probably CDK2/cyclin E, blocks further binding of MCM to chromatin and pre-RCs are not formed after the addition of NPE (Hua et al. 1997; Walter et al. 1998). Taken together, these data indicate that chromatin bound MCM7 (but not free MCM7) is required for activating the Xic1 degradation system. As MCM loading is presumably the last step of pre-RC assembly on DNA, it is clear that pre-RC formation is a prerequisite for triggering Xic1 degradation.
Xic1 degradation requires CDK2, Cdc7, and Cdc45
The data above demonstrate that pre-RCs are essential for activating Xic1 degradation. To determine whether downstream initiation factors are also required, CDK2, Cdc7, or Cdc45 was depleted from extracts, and then Xic1 degradation was assayed. Depletion of CDK2 (Fig. 2C), Cdc7 (Fig. 2E), or Cdc45 (Fig. 2G) did not affect the kinetics or efficiency of nuclear formation (data not shown). However, Xic1 was stable in each of the depleted extracts (Fig. 2D,F,H). Addition of recombinant histidine-tagged Cdc45 to the Cdc45-depleted extract restored degradation of Xic1 (Fig, 2H). These results indicate that CDK2, Cdc7, and Cdc45 are all necessary for activating degradation of Xic1. Loading of Cdc45 onto chromatin requires MCM, CDK2, and Cdc7 (Owens et al. 1997; Mimura and Takisawa 1998; Walter and Newport 2000) and has been proposed to be the critical switch for converting a pre-RC to a pre-IC (Zou and Stillman 2000). Therefore, our data strongly indicate that the formation of pre-ICs is necessary for activating Xic1 turnover.
RPA and DNA polymerase α are not required for Xic1 degradation
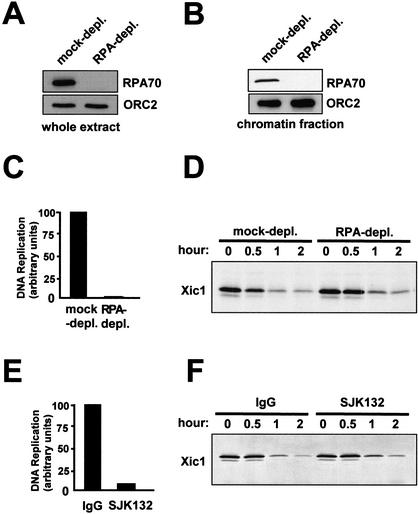
When Cdc45 associates with pre-RCs to form pre-ICs, the MCM helicase complex is activated and begins moving along and unwinding DNA. This movement and unwinding occurs even in the absence of RPA (Walter and Newport 2000). However, if RPA is present, it associates with the unwound DNA generated by MCM to form a stable single-stranded DNA (ssDNA) complex. To determine whether MCM activation alone was sufficient for Xic1 turnover, or whether an RPA–ssDNA complex was needed, we assayed Xic1 degradation in the absence of RPA. To do this, RPA was immunodepleted from cytosol (Fig. 3A). Treated cytosol was then used to form nuclei in the presence of labeled Xic1. No RPA was observed to bind to chromatin in the depleted extract (Fig. 3B). As shown in Figure 3C, removal of RPA reduced DNA replication more than 98.5%. However, Xic1 degradation still occurred very efficiently in these RPA-depleted extracts (Fig. 3D). This result shows that Xic1 degradation is activated at an intermediate initiation step, which occurs after cdc45 forms pre-ICs but before generation of an RPA–ssDNA complex by active MCM helicase.
Figure 3.
RPA and pol α are not required for Xic1 degradation. (A) Western blots of RPA and ORC2 in the RPA-depleted or the mock-depleted extract. (B) Chromatin-bound RPA and ORC2 in the RPA-depleted or the mock-depleted extract containing nuclei. (C) DNA replication in the RPA-depleted or mock-depleted extracts containing nuclei. (D) Xic1 degradation in extracts depicted in C. (E) DNA replication in the extract containing nuclei with 1 mg/mL monoclonal antibody SJK132 or 1 mg/mL nonspecific mouse IgG. (F) Xic1 degradation in the extracts depicted in E.
In the absence of RPA, DNA polymerase α cannot load onto chromatin (Walter and Newport 2000). As such, one would expect that DNA polymerase α is not required for activation of Xic1 degradation. To determine whether this is the case, SJK132, an inhibitory monoclonal antibody against DNA polymerase α was employed. As shown in Figure 3E, SJK132 antibody inhibited DNA replication to 91% of control. However, Xic1 was still degraded under these conditions (Fig. 3F). Similar results were obtained when DNA polymerase α was immunodepleted from extracts (data not shown). Therefore, although initiation is required to activate Xic1 degradation, ongoing DNA replication is not.
Xic1 degradation is attenuated after the completion of DNA replication
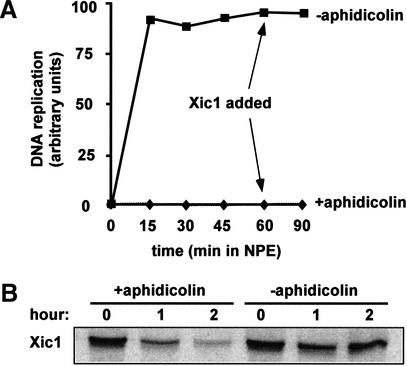
MCM and Cdc45 remain associated with DNA during replication and then dissociate following completion of replication (Aparicio et al. 1997; Mimura et al. 2000). If a MCM–Cdc45 complex is key to the Xic1 degradation pathway, it would be expected that Xic1 degradation would persist during replication and then attenuate once MCM and Cdc45 dissociate from DNA at the end of replication. To address this issue, cytosol was incubated with sperm chromatin for 30 min followed by the addition of 2 volumes of NPE with or without a replication elongation inhibitor, aphidicolin. Aphidicolin inhibits the activity of DNA replication polymerases but still allows initiation to occur. Sixty minutes after the addition of NPE, labeled Xic1 was added to each reaction. As shown in Figure 4A, in the absence of aphidicolin, DNA replication was complete before Xic1 addition. In this reaction added Xic1 was stable (Fig. 4B). The stabilization of Xic1 after DNA replication was not due to the inactivation of extracts, as Xic1 continued to be degraded in the reaction containing aphidicolin (Fig. 4B). These results show that following the dissociation of MCM and Cdc45 from DNA at the end of replication, the Xic1 degradation pathway is inactivated. Thus, activation and inactivation of both the Xic1 degradation system and MCM helicase activity are temporally coincident, suggesting that a mechanistic link exists between these two processes at the molecular level.
Figure 4.
Xic1 is stable after the completion of DNA replication. (A) Chromatin was incubated with cytosol for 30 min followed by the addition of 2 volumes of NPE with aphidicolin or without aphidicolin. DNA replication was measured in both reactions. (B) To the reactions depicted in A Xic1 was added after 60 min incubation in NPE. Xic1 degradation was examined in both reactions. Time points are hours after Xic1 addition.
CDK2 activity is not directly required for Xic1 degradation
Depletion of CDK2 blocks Xic1 degradation (Fig. 2D), indicating that CDK2 is necessary for Xic1 turnover. CDK2 could act directly on Xic1 by phosphorylation. Alternatively, if initiation of replication activates Xic1 degradation, then the role of CDK2 in Xic1 degradation could be indirect and mediated through its role in the initiation process. For example, it is possible that the role of CDK2 is to simply facilitate the recruitment of Cdc45 to pre-RC, and it is this recruitment that activates Xic1 degradation (Fig. 2H).
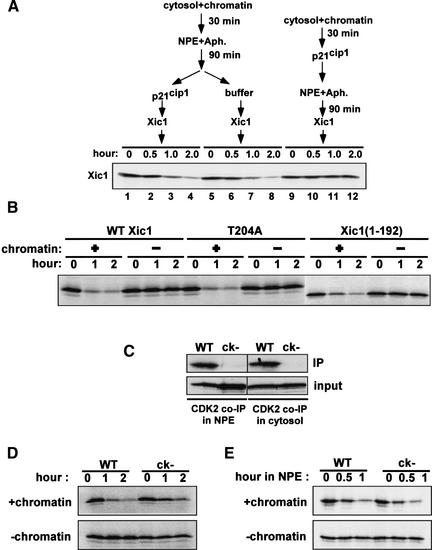
To examine the direct involvement of CDK2/Cyclin E in Xic1 degradation, we first determined whether phosphorylation of Xic1 by CDK2 is required for Xic1 turnover. Because CDK2 is required for loading of Cdc45 at pre-RCs, we determined the stability of Xic1 after initiation (post Cdc45 loading). To do this, replication was initiated in the presence of aphidicolin by first incubating chromatin in cytosol and then adding NPE. After 90 minutes in NPE, human p21cip1, which unlike Xic1 is stable in egg extracts (Z. You and J. Newport, unpubl.), was added to the reaction to inhibit CDK2 activity. Following this, labeled Xic1 was added and its stability measured. To our surprise, in the presence of p21cip1, Xic1 was destroyed at the same rate as a control reaction lacking p21cip1 (Fig. 5A, lanes 1–4 and 5–8). In contrast, if CDK2 activity was inhibited by p21cip1 prior to initiation of replication (p21cip1 added before NPE addition), Xic1 was not degraded (Fig. 5A, lanes 9–12). These results suggest that the role of CDK2 in Xic1 degradation is indirect and mediated through its role in activating initiation.
Figure 5.
CDK2 is not directly involved in Xic1 degradation. (A) Chromatin was incubated with cytosol for 30 min followed by the addition of 2 volumes of NPE with aphidicolin. Ninety minutes after the addition of NPE, 1.2 μM p21cip1 (lanes 1–4) or buffer (lanes 5–8) was added. Subsequently, Xic1 was added and its stability was determined. In another reaction (lanes 9–12), the same amount of p21cip1 was added into the reaction before the addition of NPE. (B) Degradation assays of wild-type Xic1 and its mutants, T204A and Xic1(1–192), in extracts with or without nuclei. (C) Coimmunoprecipitation of labeled wild-type Xic1 and Xic1(ck−) in cytosol or NPE with anti-CDK2 antibody. (WT) Wild-type Xic1; (ck−) Xic1(ck−). (D) Degradation assays of Xic1 and Xic1(ck−) in complete extracts in the presence or absence of chromatin. (E) Degradation assays of Xic1 and Xic1(ck−) in the nucleus-free replication system in the presence or absence of chromatin.
Further evidence that Xic1 degradation is independent of direct phosphorylation by CDK2 is the observation that addition of the CDK2 kinase inhibitior, 6-DMAP, to extracts following initiation of replication does not block Xic1 degradation (data not shown). Moreover, Xic1 mutants lacking the CDK2 phosphorylation consensus site are degraded normally. Substitution of Thr 204 with alanine has no affect on the kinetics of Xic1 turnover (Fig. 5B; Chuang and Yew 2001). Similarly, a Xic1 deletion mutant [Xic1(1–192)] lacking the C-terminal 18 amino acids, which includes the CDK phosphorylation consensus site, is also degraded in a nucleus-dependent manner (Fig. 5B). Together, these results strongly suggest that Xic1 degradation is independent of phosphorylation by CDK2.
Interactions between Xic1 and CDK2/cyclin E are dispensable for Xic1 degradation
Next, we examined whether interactions between Xic1 and CDK2/cyclin E, rather than phosphorylation, are essential for Xic1 degradation. As shown in Figure 5A (lanes 1–4), addition of competing concentrations of p21cip1 after initiation did not inhibit Xic1 degradation, suggesting that interactions between Xic1 and CDK2/cyclin E might not be essential. To examine this further, a Xic1 mutant, Xic1(ck−), was generated. In Xic1(ck−), Arg 33 and Leu 35 in the conserved cyclin-binding motif, and Pro 65 and Pro 67 within the CDK binding motif are replaced with alanines. Coimmunoprecipitation experiments using a CDK2-specific antibody showed that wild-type Xic1 bound to CDK2/cyclin E efficiently in both NPE and cytosol (Fig. 5C). However, mutation of both the CDK and cyclin-binding sites in Xic1(ck−) completely abolished the association of Xic1 with CDK2/cyclin E in both NPE and cytosol (Fig. 5C).
Degradation of Xic1 and Xic1(ck−) was determined in extracts in the presence and in the absence of nuclei. In the absence of nuclei, both wild-type Xic1 and Xic1(ck−) were stable (Fig. 5D, bottom), indicating that nonspecific degradation of the mutant did not occur. Destruction of both CDK and cyclin-binding sites in Xic1 only partially inhibited Xic1 turnover (Fig. 5D, top). In the presence of aphidicolin, wild-type Xic1 and Xic1(ck−) were degraded to the same degree as seen in untreated extracts albeit with slower kinetics (see Fig. 5A; data not shown).
The modest inhibition of degradation observed with the Xic1(ck−) mutant might suggest that CDK2/cyclin E interactions are, at least partially, involved in Xic1 degradation. However, it has been observed recently that interactions between Xic1 and CDK2/cyclin E facilitate transport of Xic1 into the nucleus (Chuang and Yew 2001). Therefore, disruption of these interactions could slow nuclear import of Xic1, which in turn would appear as an inhibition of Xic1 degradation. Indeed, we found that both wild type and mutant Xic1(ck−) were degraded at the same rate in the nucleus-free replication system that does not require nuclear transport (Fig. 5E). Together, our results indicate that interactions between Xic1 and CDK2/cyclin E are not essential for the initiation-coupled degradation of Xic1.
A C-terminal region (162–192) in Xic1 is essential and sufficient for triggering Xic1 ubiquitination
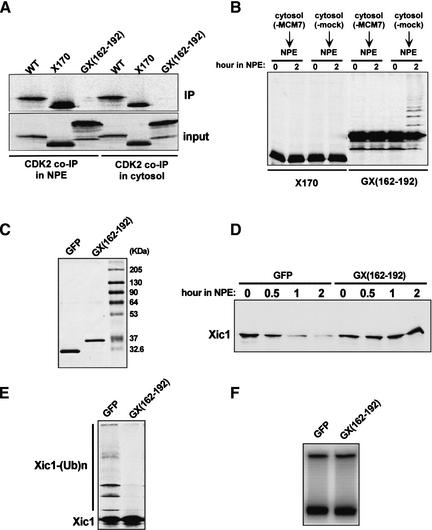
Previous observations have demonstrated that three lysine residues (Lys 180, Lys 182, Lys 183) in the C-terminal part of Xic1 are needed for Xic1 degradation (Chuang and Yew 2001). Interestingly, we have found that a C-terminal region in Xic1 is both necessary and sufficient for triggering Xic1 ubiquitination. A C-terminal 40-amino-acid truncation mutant of Xic1, Xic1(1–170), associates with CDK2 in both NPE and cytosol (Fig. 6A). However, this mutant is neither ubiquitinated nor degraded (Fig. 6B). This indicates that the C-terminal part of Xic1 is essential for its ubiquitination and degradation. To test this possibility we constructed a fusion protein consisting of this C-terminal segment (162–192) fused to the fluorescent protein GFP. This fusion protein did not interact with CDK2 (Fig. 6A). Importantly, upon addition to extract, the fusion was modified in a pre-RC dependent generating a ladder of slow migrating forms (Fig. 6B). GFP protein was not modified in the extract in this way (data not shown). That this ladder of slow migrating proteins is the result of ubiquitination is supported by the observation that ubiquitin aldehyde, an inhibitor of de-ubiquitination, increased the fraction of protein in the ladder (data not shown).
Figure 6.
A C-terminal region (162–192) of Xic1 is essential for its ubiquitination and degradation. (A) Coimmunoprecipitation of labeled Xic1, Xic1(1–170), and GFP–Xic1(161–192) in cytosol or NPE with anti-CDK2 antibody. (WT) Wild-type Xic1; (X170) Xic1(1–170); [GX(162–192)] GFP–Xic1(162–192). (B) Degradation assays of Xic1(1–170) and GFP–Xic1(162–192) in the nucleus-free replication system in the presence or absence of pre-RC. Reactions were assembled as depicted in Figure 2B. Chromatin and Xic1(1–170) or GFP–Xic1(162–192) were mixed with MCM7-depleted or mock-depleted cytosol for 30 min. Subsequently, 2 volumes of untreated NPE was added to each reaction. Reactions were terminated at the designated times after the addition of NPE. (C) Bacterially expressed and purified recombinant GFP and GFP–Xic1(162–192). (D) Degradation assays of Xic1 in the nucleus-free replication system containing 100 ng/μL of GFP or GFP–Xic1(162–192). (E) Same as depicted in D except 100 μM MG132 was added to each reaction. (F) DNA replication in the extracts as depicted in D.
These results suggest that the C-terminal region may facilitate Xic1 ubiquitination prior to its degradation. To determine if this is true, we tested whether the addition of bacterially expressed GFP–Xic1(162–192) could competitively inhibit Xic1 ubiquitination and degradation in a dominant-negative manner. Indeed, we found that GFP–Xic1(162–192), but not GFP (Fig. 6C), inhibited both degradation (Fig. 6D) and ubiquitination (Fig. 6E) of full-length Xic1. In this experiment the concentration of Xic1 is low relative to CDK2. Therefore, neither GFP–Xic1(162–192) nor GFP alone had any effect on DNA replication (Fig. 6F). Taken together, these results strongly suggest that the region (162–192) in Xic1 facilitates Xic1 ubiquitination prior to its degradation. The fact that GFP–Xic1(162–192) is degraded slowly suggests that additional sequences are required for efficient Xic1 proteolysis after ubiquitination.
S phase-specific association of GFP–Xic1(162–192) with chromatin requires Cdc45
The coupling of Xic1 degradation and initiation of DNA replication led us to consider whether Xic1 is ubiquitinated and/or degraded on chromatin. While this article was in preparation, Furstenthal et al. (2001) reported evidence from which it was concluded that Xic1 is recruited to chromatin through interaction with CDK2/cyclin E. In contrast, our observation that Xic1(ck−), which is deficient in CDK2/cyclin E binding, is still degraded with the same kinetics as Xic1 (Fig. 5E) suggests that other site(s) in Xic1 are required for this association.
As shown above, the C-terminal region (162–192) of Xic1 is required for Xic1 ubiquitination and degradation. In addition, fusion of GFP with this 31-amino-acid segment is sufficient to trigger the pre-RC-dependent ubiquitination of the fusion protein. Therefore, this segment seems to be a possible candidate for the site that recruits Xic1 to chromatin.
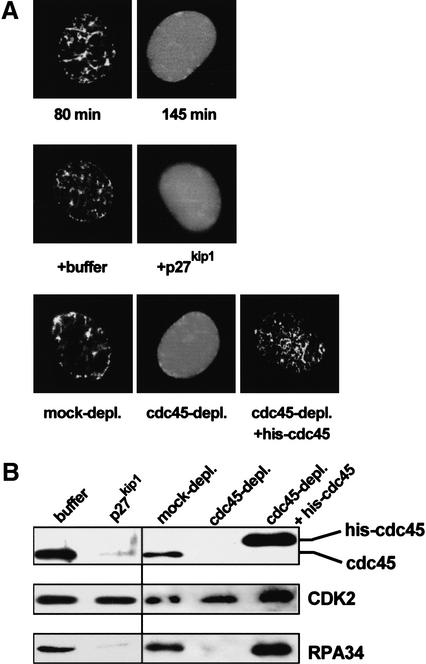
To investigate this possibility, we examined the association of GFP–Xic1(162–192) with chromatin in nuclei by fluorescence microscopy. We found that GFP–Xic1(162–192), but not GFP (data not shown), was imported into nuclei efficiently. Further, GFP–Xic1(162–192) protein associated with chromatin during DNA replication (Fig. 7A, top, 80 min). Following completion of replication it dissociated from DNA (Fig. 7A, top, 145 min), consistent with the observation that Xic1 degradation occurs during replication and then attenuates after completion of replication (Fig. 4). Further, when initiation of replication is blocked by addition of p27kip1, GFP–Xic1(162–192) failed to associate with DNA (Fig. 7A, middle). Importantly, depletion of Cdc45 also inhibited association of GFP–Xic1(162–192) protein with chromatin, and this inhibition was rescued by addition of recombinant Cdc45 to the depleted extract (Fig. 7A, bottom). Together these results show that association of GFP–Xic1(162–192) with chromatin requires Cdc45 and initiation of replication and that the fusion protein, like Cdc45, remains bound to chromatin during the course of replication.
Figure 7.
Chromatin association of GFP–Xic1(162–192) requires Cdc45. (A, top) Sperm chromatin was incubated with cytosol and membranes for 10 min. After this, recombinant GFP–Xic1(162–192) (10 ng/μL) was added to the reaction. GFP signal and chromatin signal (stained with Hoeschst, data not shown) were detected during replication (80 min) and after replication (145 min) by fluorescence confocal microscopy (Nikon, PCM200 confocal microscope). (Middle) Reactions were assembled as depicted in A except buffer or p27kip1 was added to cytosol. GFP and chromatin signals were examined after 80 min incubation. (Bottom) Reactions were assembled as depicted in A except cytosol was pretreated with either Cdc45 antiserum or pre-immune serum coupled beads. In another reaction, histidine-tagged Cdc45 was added to Cdc45-depleted cytosol. (B) Effects of p27kip1 addition or Cdc45 depletion as depicted in A on the chromatin association of CDK2, Cdc45, and RPA.
Interestingly, in the absence of Cdc45, the level of CDK2 bound to chromatin was normal (Fig. 7B). Similarly, chromatin-bound level of CDK2 was normal in the presence of p27kip1. However, despite normal CDK2 levels, GFP–Xic1(162–192) failed to bind to chromatin. This observation is consistent with data showing that GFP–Xic1(162–192) does not interact with CDK (Fig. 6A). Taken together, our results suggest that the C-terminal region (162–192) of Xic1, rather than the N-terminally located CDK and cyclin-binding sites, directly facilitates the association of Xic1 with chromatin during DNA replication and that this interaction may facilitate ubiquitination of Xic1 prior to its degradation.
Discussion
A Xic1 degradation signal is generated during initiation of DNA replication
Regulated protein degradation is essential for multiple steps during the cell cycle including entry into mitosis, sister chromatid separation, and initiation of DNA replication (King et al. 1996). Because of its irreversible nature, proteolysis is under tight regulatory control ensuring that it occurs only during appropriate stages of the cell cycle. Our results indicate that in Xenopus egg extracts, degradation of the CKI, Xic1, is dependent on initiation of DNA replication. Degradation of Xic1 requires not only the assembly of pre-RCs on chromatin, but also the origin activating kinases CDK2 and Cdc7, as well as the initiation protein Cdc45 (Fig. 2). The requirement for CDK2 and Cdc7 may be indirect in that loading of Cdc45 onto chromatin during initiation requires the activity of both of these kinases (Owens et al. 1997; Mimura and Takisawa 1998; Walter and Newport 2000). Supporting this conclusion is the observation that inhibition of CDK2 activity following initiation has little effect on Xic1 degradation (Fig. 5A, see below). Recently, Furstenthal et al. (2001) proposed that Xic1 is degraded in a pre-RC-dependent manner prior to initiation of DNA replication. However, based on our results, we believe that Xic1 degradation is a post-initiation event.
During initiation of replication the MCM helicase complex is activated and then RPA and DNA polymerase α are sequentially loaded onto DNA (Walter and Newport 2000). Strikingly, depletion of either of these two late initiation proteins had little, if any, effect on activation of the Xic1 degradation system (Fig. 3). These results demonstrate that initiation-coupled Xic1 degradation occurs following recruitment of Cdc45 to generate pre-ICs and activation of the MCM helicase. Following completion of DNA replication, MCM and Cdc45 dissociate from chromatin (Aparicio et al. 1997; Mimura et al. 2000). Our data suggest that it is this dissociation that causes attenuation of the Xic1 degradation pathway (Fig. 4). Overall, our results are consistent with a model whereby Cdc45-dependent activation of the MCM helicase late in the initiation sequence activates the Xic1 degradation system. Physical interaction between Xic1 and Cdc45 have not been observed (data not shown), suggesting that MCM activation is directly responsible for activation of the degradation pathway.
It has been shown that Xic1 is degraded by the proteasome complex in frog egg extracts (Yew and Kirschner 1997). We have found that neither calpain inhibitors (NCO-700 and EST) nor caspase inhibitors have any obvious effects on Xic1 turnover (data not shown). Additionally, the ATM/ATR checkpoint kinase inhibitor, caffeine, had no effect on Xic1 degradation (data not shown), suggesting that Xic1 degradation is not simply a consequence of the activation of cell cycle checkpoints.
CDK2 is not directly involved in Xic1 degradation
Our results indicate that CDK2 is not directly involved in Xic1 turnover. This conclusion is supported by the following observations. (1) The conserved CDK phosphorylation site in Xic1 is not necessary for Xic1 turnover (Fig. 5B). (2) CDK2 activity is not required for Xic1 degradation after initiation of DNA replication (Fig. 5A). (3) Disruption of binding interactions between Xic1 and CDK2/cyclin E in Xic1(ck−) did not affect the kinetics of Xic1 turnover (Fig. 5E). (4) A Xic1 mutant, Xic1(1–170), associates with CDK2/cyclin E, but is not degraded (Fig. 6B). (5) A C-terminal region (162–192) of Xic1, which does not interact with CDK2, is essential and apparently sufficient for triggering Xic1 ubiquitination and association with chromatin (Figs. 6 and 7). Taken together, these results strongly suggest that although Xic1 may interact with chromatin through CDK2/cyclin E, this interaction is not essential for Xic1 degradation. Rather, because both Xic1 degradation (Fig. 2H) and the association of GFP–Xic1(162–192) (Fig. 7A) with chromatin requires Cdc45, it is likely that the role of CDK2/cyclin E in Xic1 degradation is indirect and occurs through its role in loading Cdc45 onto chromatin during initiation.
A C-terminal region (162–192) in Xic1 is essential for Xic1 ubiquitination and subsequent degradation
Degradation assays for Xic1(1–192) (Fig. 5B) and Xic1(1–170) (Fig. 6B) indicate that the C-terminal region (170–192) in Xic1 is essential for its destruction. Interestingly, fusion of the region (162–192) in Xic1 with GFP triggers ubiquitination of the fusion protein in a pre-RC-dependent manner (Fig. 6B). Moreover, excess GFP–Xic1 (162–192) has a dominant-negative effect inhibiting both Xic1 ubiquitination and degradation (Fig. 6D,E). Fluorescence experiments show that this 31-amino-acid segment recruits GFP–Xic1(162–192) to chromatin in a Cdc45-dependent manner (Fig. 7A). Taken together, these observations strongly suggest that the C-terminal region (162–192) of Xic1 facilitates its association with chromatin and subsequent ubiquitination.
This C-terminal region contains a PCNA-binding site (Su et al. 1995). However, it is unlikely that interaction of this region with PCNA contributes to Xic1 degradation as p21cip1 and Xic1 chimeric proteins, in which the PCNA binding sites are exchanged, associate with PCNA but are not degraded (Z. You and J. Newport, unpubl.).
It appears that negative charges on three lysine residues (Lys 180, Lys 182, Lys 183) in the C-terminal region are necessary for Xic1 degradation. Mutation of these lysine residues to alanine abrogated Xic1 degradation. However, Xic1 degradation was retained when these lysine residues were mutated to arginine (data not shown; Chuang and Yew 2001). It is possible that the negative charges of these three lysine residues play a role in the interaction between the C-terminal region (162–192) in Xic1 and an unknown factor that facilitates ubiquitination of Xic1.
Possible roles of the initiation coupled degradation of Xic1
There are several reasonable explanations for the existence of an initiation-coupled Xic1 degradation system. Firstly, because replicon size is established by the distance between adjacent pre-RCs it is essential that a sufficient number of pre-RCs be formed along the chromosome during G1 of the cell cycle, when CDK2 activity is low. If an insufficient number of pre-RCs form during G1, the average replicon size would be very large and, as a result, S phase would be excessively long. Therefore, a process that ensures formation of a threshold number of pre-RCs during G1 could contribute to defining the duration of S phase. An initiation-dependent Xic1 degradation system may contribute to defining this threshold number. For example, it is possible that once a threshold number of pre-RCs have formed, a small subset of these might become activated by trace amounts of active CDK2. This, in turn, would cause Xic1 degradation, thereby, initiating an autocatalytic loop resulting in activation of the remaining CDK2 and suppression of further pre-RC formation (Dahmann et al. 1995; Hua et al. 1997). Such a loop could also serve to activate CDK2 rapidly at the G1/S transition. During G1 the CDK2/cyclin E complex remains inactive because of the presence of Xic1. However, once the concentration of CDK2/cyclin E exceeds Xic1 levels because of the synthesis of cyclin E, some pre-RCs would be activated and initiation-coupled degradation of Xic1 would ensure rapid activation of all CDK2/cyclin E kinase present.
Alternatively, coordination of replication initiation with degradation of a CDK inhibitor may ensure uninterrupted CDK2 activity for the duration of S phase. Though CDK2 may be subsequently regulated by other mechanisms, such as phosphorylation, regulation at these other levels would be rapid and reversible in nature. For instance, in somatic cells of metazoans, such regulation during S phase would allow the CDK2-dependent duplication of centrosomes (Hinchcliffe et al. 1999; Lacey et al. 1999; Matsumoto et al. 1999; Meraldi et al. 1999) as well as the replication-dependent transcription of histone genes in S phase (Zhao et al. 2000). Furthermore, we believe that the initiation-coupled degradation system may also provide a mechanism to block re-replication in S phase attributable to the accumulation of active CDK2, which is inhibitory for de novo pre-RC assembly (Dahmann et al. 1995; Hua et al. 1997).
Recent work reveals that mammalian p27kip1 is degraded by two distinct pathways (Malek et al. 2001). One pathway is dependent on the phosphorylation of T187 by CDK2 whereas the other is T187 independent. The phosphorylation-independent pathway is active in cultured mouse embryonic fibroblasts (MEFs) 12–19 h after serum stimulation. About 5% of MEFs entered S phase 16 h after serum stimulation (Malek et al. 2001). We suspect that the initiation-coupled degradation of Xic1 may mimic this T187 independent p27kip1 degradation pathway. Like Xic1, p27kip1 degradation by this pathway depends on components of the SCF complex (Malek et al. 2001). Also like Xic1, p27kip1 can be degraded in the nucleus (Rodier et al. 2001).
In egg extracts, when initiation of replication is blocked, Xic1 is stable even if high CDK2 activity is present. Therefore, the CDK2 phosphorylation-dependent p27kip1 degradation pathway present in somatic cells appears to be absent from egg extracts. The absence of a phosphorylation-dependent pathway in embryos could be due to cyclin dependent changes in CDK2 substrate specificity. In somatic cells CDK2-associated cyclin E is degraded shortly after S phase starts and is replaced by cyclin A. In contrast, in early frog embryos, cyclin E is not degraded during the cell cycle and cyclin A is exclusively associated with Cdc2 and is not required for DNA replication (Walker and Maller 1991; Howe and Newport 1996; Strausfeld et al. 1996). Therefore, if phosphorylation-dependent Xic1 degradation occurs through the action of CDK2/cyclin A kinase, this pathway should be absent from embryos.
Materials and methods
Preparation of Xenopus egg extracts
Interphase cytosol, demembranated sperm chromatin, and purified membranes were prepared essentially as described previously (Smythe and Newport 1991). NPE was prepared by isolating nuclei assembled in low-speed egg extract in the presence of 3.3 μg/mL nocodazole, according to a detailed protocol (Walter et al. 1998).
Plasmids, mutagenesis, and in vitro transcription and translation
Open reading frames from cDNAs encoding WT Xic1, its mutants, and GFP–Xic1(162–192) were subcloned into pET28a(+) (Novagen) at BamHI and EcoRI sites. Mutations were generated using a standard PCR-based oligonucleotide-directed mutagenesis protocol. Sequences of the primers are available on request. All of the mutants were verified by sequencing.
Messager RNAs of Xic1, its deletion and point mutation mutants, and GFP–Xic1(162–192) were in vitro-transcribed with T7 RNA polymerase (Ambion, mMESSAGE mMACHINE kit). After the DNA templates were digested, mRNAs were recovered and translated in nuclease-treated rabbit reticulocyte lysate (Promega) or the Escherichia coli S30 extract (Novagen, EcoPro T7 System) in the presence of [35S]methionine.
Inhibitors, antibodies, and recombinant proteins
Aphidicolin (Sigma, A0781) and nocodazole (Sigma, M1404) were dissolved in DMSO and used at 50 μg/mL, 3.3 μg/mL, respectively.
MCM7 and Cdc7 antibodies were generously provided by Dr. Li Sun. These antibodies were raised in rabbits against the C-terminal 176 amino acids of Xenopus MCM7 protein and the full length of Xenopus Cdc7 protein, respectively. CDK2, ORC2, RPA, and Cdc45 antibodies were obtained as described previously (Walter and Newport 2000; Fang and Newport 1991). Monoclonal antibody against human DNA polymerase α, SJK132, was purified from cell culture supernatant (ATCC no. CRL-1640).
Recombinant histidine-tagged Cdc45 was used to rescue the activities of the Cdc45-depleted extract at 75 μg/mL (Walter and Newport 2000). Recombinant histidine-tagged human p21cip1 was described previously (Guadagno and Newport 1996). Recombinant GST-fused human p27kip1 and histidine-tagged GFP, GFP–Xic1(162–192) were expressed in E. coli and affinity purified from glutathione sepharose 4B column (Amersham Phamarcia Biotech AB) and Ni-NTA agarose column (QIAGEN), respectively.
Immunodepletion, immunoprecipitation, and chromatin binding assay
To remove specific proteins from egg cytosol, three consecutive immunodepletions were carried out as described previously (Walter and Newport 1999). Briefly, protein A sepharose 4B fast flow beads were incubated with specific antisera or the corresponding pre-immune sera for 30 min at room temperature. After washing 3 times with ELB (250 mM sucrose, 2.5 mM MgCl2, 50 mM KCl, 10 mM HEPES, 1 mM DTT at pH 7.7), the beads were split into three aliquots of equal volume. Extracts were then consecutively incubated with the three bead aliquots in the presence of 3.3 μg/mL nocodazole at 4°C for 1 h with constant rotation. The mixture was then centrifuged (5000 rpm, 2 min) to remove beads. The ratio of the volume of beads, antibody, and cytosol was optimized for the depletion of each protein. Western blotting results showed the protein levels of each specific factor were reduced to <1% after depletion.
Immunoprecipitation was done by first incubating 10 μL protein A beads with 30 μL of a CDK2 antiserum or its pre-immune serum for 30 min; after washing 3 times with ELB, beads were then incubated with 20 μL cytosol or NPE containing labeled Xic1, Xic1(ck−), Xic1(1–170), GFP–Xic1(162–192) for 1 h at room temperature with constant rotation. Subsequently, beads were pelleted by low-speed centrifugation (5000 rpm, 2 min) and washed 5 times with washing buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% Tween 20 at pH 7.5). The bound proteins were then analyzed by SDS-PAGE followed by autoradiography.
Chromatin binding assays were essentially performed as described previously (Walter and Newport 1999). In Figure 3B, nuclei formed in RPA-depleted or mock-depleted cytosol were isolated by centrifugation through a 0.5 M sucrose ELB cushion and then soloubilized with 0.6% Trition X-100 in ELB. Chromatin fractions were then isolated by centrifugation through a 0.5 M sucrose ELB cushion. Chromatin-bound proteins were then subjected to Western blot analysis.
Xic1 degradation and DNA replication assays
Before use, cytosol was supplemented with 3.3 μg/mL nocodazole and an ATP regenerating system (2 mM ATP, 20 mM phosphocreatine, 5 μg/mL creatine phosphokinase). The ATP regenerating system was also added to NPE. Typical reactions in various replication systems were performed as follows. Variations of the reactions are specified in Figure legends 2–6.
In cytosol + membrane
Cytosol (20 μL) was mixed with purified membranes (2 μL) and sperm chromatin (3000/μL final concentration from a 50,000/μL stock). Next, 1 μL of labeled Xic1 was added, and reactions were generally split into two parts. One part was used for determining Xic1 degradation. Aliquots (3 μL) were taken at appropriate time points and the reactions were stopped by adding 10 μL SDS sample buffer. Proteins were separated by 12% SDS-PAGE followed by analysis with PhosphorImager (Molecular Dynamics) or autoradiography. To the other half of the reaction ∼0.1 μCi/μL [α-32P]dATP was added to measure DNA replication. Again, 3 μL aliquots were withdrawn and quenched at appropriate time points by adding 5 μL stop solution (8 mM EDTA, 0.13% phosphoric acid, 10% ficoll, 5% SDS, 0.2% bromophenol blue, 80 mM Tris at pH 8.0). DNA replication was analyzed using 0.8% agarose gel electrophoresis and quantified after PhosphorImaging.
In cytosol → NPE
Sperm chromatin (3000 sperms/μL) or pBluecript plasmid DNA (5 ng/μL) and 1 μL of labeled Xic1 were incubated with 6 μL of membrane-free cytosol for 30 min at room temperature. Subsequently, 12 μL of NPE was added. As described above, reaction samples were split in half and Xic1 degradation and DNA replication were measured in parallel.
Acknowledgments
We thank Dr. James Maller for providing Xic1 cDNA and Dr. Tony Hunter for providing p27kip1 expression vector. We also thank Drs. Johannes Walter, Matthew Michael, Ryuji Yamagochi, Helen Hua, Li Sun, and Jianghuai Xu for helpful advice and technical assistance. This work was supported by NIH grant (GM44656) to J.N.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jnewport@ucsd.edu; FAX (858) 822-3531.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.985302.
References
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Bousset K, Diffley JF. The Cdc7 protein kinase is required for origin firing during S phase. Genes & Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J Biol Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- Chuang LC, Yew PR. Regulation of nuclear transport and degradation of the Xenopus cyclin-dependent kinase inhibitor, p27Xic1. J Biol Chem. 2001;276:1610–1617. doi: 10.1074/jbc.M008896200. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Diffley JF, Nasmyth KA. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML. Initiation of DNA replication in eukaryotic chromosomes. J Cell Biochem Suppl. 1998;30–31:8–17. [PubMed] [Google Scholar]
- Deshaies RJ. Phosphorylation and proteolysis: Partners in the regulation of cell division in budding yeast. Curr Opin Genet Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- Diffley JF. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes & Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Donaldson AD, Fangman WL, Brewer BJ. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes & Dev. 1998a;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998b;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- Fang F, Newport JW. Evidence that the G1–S and G2–M transitions are controlled by different cdc2 proteins in higher eukaryotes. Cell. 1991;66:731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- Furstenthal L, Swanson C, Kaiser BK, Eldridge AG, Jackson PK. Triggering ubiquitination of a CDK inhibitor at origins of DNA replication. Nat Cell Biol. 2001;3:715–722. doi: 10.1038/35087026. [DOI] [PubMed] [Google Scholar]
- Guadagno TM, Newport JW. Cdk2 kinase is required for entry into mitosis as a positive regulator of Cdc2–cyclin B kinase activity. Cell. 1996;84:73–82. doi: 10.1016/s0092-8674(00)80994-0. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2–cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Howe JA, Newport JW. A developmental timer regulates degradation of cyclin E1 at the midblastula transition during Xenopus embryogenesis. Proc Natl Acad Sci. 1996;93:2060–2064. doi: 10.1073/pnas.93.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Yan H, Newport J. A role for Cdk2 kinase in negatively regulating DNA replication during S phase of the cell cycle. J Cell Biol. 1997;137:183–192. doi: 10.1083/jcb.137.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherwood J. Emerging mechanisms of eukaryotic DNA replication initiation. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- Lee SH, Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci. 1990;87:5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kwong AD, Pan ZQ, Hurwitz J. Studies on the activator 1 protein complex, an accessory factor for proliferating cell nuclear antigen-dependent DNA polymerase delta. J Biol Chem. 1991;266:594–602. [PubMed] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes & Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D, Moreau J, Méchali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- Malek NP, Sundberg H, McGrew S, Nakayama K, Kyriakides TR, Roberts JM, Kyriakidis TR. A mouse knock-in model exposes sequential proteolytic pathways that regulate p27Kip1 in G1 and S phase. Nature. 2001;413:323–327. doi: 10.1038/35095083. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2–cyclin A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Masuda T, Matsui T, Takisawa H. Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells. 2000;5:439–452. doi: 10.1046/j.1365-2443.2000.00340.x. [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Fiore F, Eytan E, Carrano AC, Draetta GF, Hershko A, Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes & Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama T, Ishida N, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vitro: Stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Gitig DM, Koff A. Cell-free degradation of p27(kip1), a G1 cyclin-dependent kinase inhibitor, is dependent on CDK2 activity and the proteasome. Mol Cell Biol. 1999;19:1190–1201. doi: 10.1128/mcb.19.2.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M, Kawasumi M, Fujino M, Toh-e A. Phosphorylation of sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol Biol Cell. 1998;9:2393–2405. doi: 10.1091/mbc.9.9.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens JC, Detweiler CS, Li JJ. CDC45 is required in conjunction with CDC7/DBF4 to trigger the initiation of DNA replication. Proc Natl Acad Sci. 1997;94:12521–12526. doi: 10.1073/pnas.94.23.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust VN, Georgaki A, Strack B, Hübscher U. Calf thymus RF-C as an essential component for DNA polymerase delta and epsilon holoenzymes function. Nucleic Acids Res. 1992;20:4159–4165. doi: 10.1093/nar/20.16.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier G, Montagnoli A, Di Marcotullio L, Coulombe P, Draetta GF, Pagano M, Meloche S. p27 cytoplasmic localization is regulated by phosphorylation on Ser10 and is not a prerequisite for its proteolysis. EMBO J. 2001;20:6672–6682. doi: 10.1093/emboj/20.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Yang QH, Futcher AB. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- Schwob E, Böhm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes & Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaff RJ, Roberts JM. End of the line: Proteolytic degradation of cyclin-dependent kinase inhibitors. Chem Biol. 1996;3:869–873. doi: 10.1016/s1074-5521(96)90174-x. [DOI] [PubMed] [Google Scholar]
- Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes & Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin–ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Smythe C, Newport JW. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Strausfeld UP, Howell M, Descombes P, Chevalier S, Rempel RE, Adamczewski J, Maller JL, Hunt T, Blow JJ. Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopus egg extracts. J Cell Sci. 1996;109:1555–1563. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- Su JY, Rempel RE, Erikson E, Maller JL. Cloning and characterization of the Xenopus cyclin-dependent kinase inhibitor p27XIC1. Proc Natl Acad Sci. 1995;92:10187–10191. doi: 10.1073/pnas.92.22.10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson C, Ross J, Jackson PK. Nuclear accumulation of cyclin E/Cdk2 triggers a concentration-dependent switch for the destruction of p27Xic1. Proc Natl Acad Sci. 2000;97:7796–7801. doi: 10.1073/pnas.97.14.7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Labib K, Diffley JF. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 2000;19:2082–2093. doi: 10.1093/emboj/19.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, alpha and delta. EMBO J. 1989;8:3883–3889. doi: 10.1002/j.1460-2075.1989.tb08567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov LM, Yeh KH, Lee SJ, Sun H, Zhang H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997a;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Verma R, Feldman RM, Deshaies RJ. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997b;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DH, Maller JL. Role for cyclin A in the dependence of mitosis on completion of DNA replication. Nature. 1991;354:314–317. doi: 10.1038/354314a0. [DOI] [PubMed] [Google Scholar]
- Walter J, Newport J. The use of Xenopus laevis interphase egg extracts to study genomic DNA replication. In: Cotterill S, editor. Eukaryotic DNA replication: A practical approach. Oxford, UK: Oxford University Press; 1999. pp. 201–221. [Google Scholar]
- ————— Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Yew PR, Kirschner MW. Proteolysis and DNA replication: The CDC34 requirement in the Xenopus egg cell cycle. Science. 1997;277:1672–1676. doi: 10.1126/science.277.5332.1672. [DOI] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes & Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- ————— Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p–Dbf4p kinase. Mol Cell Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]