Abstract
Regulation of gene expression in mammals through methylation of cytosine residues at CpG dinucleotides is involved in the development and progression of tumors. Because many genes that are involved in the control of cell proliferation are regulated by members of the E2F family of transcription factors and because some E2F DNA-binding sites are methylated in vivo, we have investigated whether CpG methylation can regulate E2F functions. We show here that methylation of E2F elements derived from the dihydrofolate reductase, E2F1, and cdc2 promoters prevents the binding of all E2F family members tested (E2F1 through E2F5). In contrast, methylation of the E2F elements derived from the c-myc and c-myb promoters minimally affects the binding of E2F2, E2F3, E2F4, and E2F5 but significantly inhibits the binding of E2F1. Consistent with these studies, E2F3, but not E2F1, activates transcription through methylated E2F sites derived from the c-myb and c-myc genes whereas both E2F1 and E2F3 fail to transactivate a reporter gene that is under the control of a methylated dihydrofolate reductase E2F site. Together, these data illustrate a means through which E2F activity can be controlled.
Methylation of cytosine residues in CpG dinucleotide pairs is an important mechanism through which genes can be differentially transcribed in various cell types. CpG dinucleotides and 5-methylcytosine residues are unevenly distributed in the genome. Approximately 1–2% of the human genome consists of clusters of nonmethylated CpG stretches of DNA (CpG islands) that are typically 1–2 kb in length and are usually associated with gene promoters. The remaining CpG dinucleotides are dispersed and predominantly methylated (1, 2). CpG islands are present in nearly all known housekeeping genes and in a number of genes that are expressed in a tissue-dependent fashion (3).
Much recent interest in methylation has focused on the potential for abnormal methylation patterns to effect either activation (hypomethylation) or silencing (hypermethylation) of genes that are important for development, the immune response, and progression and metastasis of tumors (4–8). Hypomethylation of specific protooncogenes such as c-Myc, c-Fos, H-Ras, K-Ras, ERB-A1, and BCL-2 has been observed, particularly in hepatocellular carcinomas and leukemias (5). In certain tumors, aberrant methylation and corresponding decreases in expression have been reported for the tumor suppressor genes von Hippel-Lindau (9), retinoblastoma (pRb) (10–12), BRCA1(13, 14), and the cyclin-dependent kinase inhibitor p16 (15, 16).
The E2F family of transcription factors plays a key role in the regulation of cell growth, apoptosis, and oncogenic transformation by regulating the timely expression of a number of genes involved in these processes (for reviews and references see refs. 17 and 18). E2F is a heterodimeric factor composed of an E2F and a DP family member. Currently, six different E2F family members (E2F1 through E2F6) and three DP proteins (DP1 through DP3) have been identified in mammals. The heterodimerization of E2F and DP subunits is essential for both DNA binding and E2F-site-dependent transactivation because E2F and DP homodimers have minimal affinity for DNA. The carboxyl-terminal regions of E2F1–5 contain potent transactivation domains (no equivalent activity has been identified in E2F6, or in the DP proteins) that can be repressed through the interaction with the pRb family members pRb, p107, and p130 (18).
Because pathways responsible for regulating E2F are aberrant in most human tumors (19–21), a detailed knowledge of the mechanisms that control E2F activity is essential to understand the molecular basis of tumorigenesis. E2F binding sites (T/CTTC/GG/CCGC/G) contain one or two CpG motifs and, therefore, are candidates for regulation by DNA methylation. In this regard, it has been shown that some E2F-regulated genes, such as pRb and p14ARF, are hypermethylated in certain tumor cells and others, like the c-myc gene, are hypermethylated in terminally differentiated K562 cells (10–12, 22, 23). Moreover, a detailed sequence analysis of the CpG island spanning the pRb promoter from retinoblastoma tumors revealed that the E2F binding site is methylated in most cases (12). Nevertheless, little is known regarding the functional consequences of E2F binding-site methylation. Therefore, we explored how methylation of CpG dinucleotides within E2F binding sites regulates the binding of E2F family members, and these studies revealed that CpG methylation differentially regulates the response of distinct E2F elements to different E2F factors.
Materials and Methods
Cell Culture.
X50-7 cells were grown in RPMI 1640 medium supplemented with 10% FBS, penicillin, streptomycin, and glutamine. U2OS, Saos-2, and NIH 3T3 cells were grown in DMEM medium supplemented with 10% FBS, penicillin, streptomycin, and glutamine. All cells were maintained at 37°C in a humidified 5% CO2-containing atmosphere. NIH 3T3 cells were synchronized as described (24).
Methylation and Radiolabeling of Oligonucleotides.
The following double stranded oligonucleotides were used: E2F-dhfr-wt (5′-CTAGAGCAATTTCGCGCCAAACTTG-3′ and 5′-GATCCAAGTTTGGCGCGAAATTCGT-3′), E2F-E2F1-A (5′-CTAGAGCTCTTTCGCGGCAAAAAGGAG-3′ and 5′-GATCCTCCTTTTTGCCGCGAAAGAGCT-3′), E2F-E2F1-B (5′-CTAGAGGATTTGGCGCGTAAAAGTGG-3′ and 5′-GATCCCACTTTTACGCGCCAAATCCT-3′), E2F-CDC2 (5′-CTAGATTTCTTTCGCGCTCTAGCCG-3′ and 5′-GATCCGGCTAGAGCGCGAAAGAAAT-3′), E2F-c-myc-wt (5′-CTAGAGAGGCTTGGCGGGAAAAAG-3′ and 5′-GATCCTTTTTCCCGCCAAGCCTCT-3′), E2F-c-myb-wt (5′-CTAGACAGATTTGGCGGGAGGGGGG-3′ and 5′-GATCCCCCCCTCCCGCCAATCTGT-3′), control (5′-CGCGTAGGCTCAGA-3′), and (5′-GATCTCTGACGGTA-3′), 2xE2Fdhfr(wt) (5′-CGCGTGCAATTTCGCGCCAAACTTGGCAATTTCGCGCCAAACTTGA-3′), and (5′-GATCTCAAGTTTGGCGCGAAATTGCCAAGTTTGGCGCGAAATTGCA-3′) 2xE2Fdhfr(mut) (5′-CGCGTGCAATTGCTCGACCAACTTGGCAATTGCTCGACCAACTTGA -3′) and (5′-GATCTCAAGTTGGTCGAGCAATTGCCAAGTTGGTCGAGCAATTGCA-3′), 2xE2Fdhfr(Met) (5′-CGCGTGCAATTTmeCGmeCGCCAAACTTGGCAATTTmeCGmeCGCCA AACTTGA) and (5′-GATCTCAAGTTTGGmeCGmeCGAAATTGCCAAGTTTGGmeCGmeCG AAATTGCA), 2xE2Fc-myb(wt) (5′-CGCGTCAGATTTGGCGGGAGGGGGACAGATTTGGCGGGAGGGGGAA-3′) and (5′-GATCTTCCCCCTCCCGCCAAATCTGTCCCCCTCCCGCCAAATCTGA-3′), 2xE2Fc-myb(mut) (5′-CGCGTCAGATTTAACGGGAGGGGGACAGATTTAACGGGAGGGGGAA-3′) and (5′-GATCTTCCCCCTCCCGTTAAATCTGTCCCCCTCCCGTTAAATCTGA-3′), 2xE2Fc-myb(Met) (5′-CGCGTCAGATTTGGmeCGGGAGGGGGACAGATTTGGmeCGGGAGGGGGAA-3′) and (5′-GATCTTCCCCCTCCmeCGCCAAATCTGTCCCCCTCCmeCGCCA AATCTGA-3′), 2xE2Fc-myc(wt) (5′-CGCGTGAGGCTTGGCGGGAAAAAGAGAGGCTTGGCGGGAAAAAGAA-3′) and (5′-GATCTTCTTTTTCCCGCCAAGCCTCTCTTTTTCCCGCCAAGCCTCA-3′), 2xE2Fc-myc(Met) (5′-CGCGTGAGGCTTGGmeCGGGAAAAAGAGAGGCTTGGmeCGGGAAAAAGAA-3′) and (5′-GATCTTCTTTTTCCmeCGCCAAGCCTCTCTTTTTCCmeCGCCAAGCCTCA). Oligonucleotides were synthesized by BioSource International (Camarillo, CA). Purification and annealing of complementary oligonucleotides were performed as described (24). The 2x oligonucleotides used for reporter assays were synthesized by using 5-methyl cytosine at the indicated positions. Double stranded oligonucleotides (100 ng) used for gel shift assays were methylated in vitro with Sss I (New England Biolabs) in the presence or in the absence (control methylated) of the methyl-donor S-adenosylmethionine, following manufacturer instructions. Oligos were immediately purified (for gel-shift competition experiments) or labeled by employing a Klenow fill-in reaction and purified [for electrophoretic mobility-shift assay (EMSA)].
EMSAs.
pRc-CMV-DP1 together with either pRc-CMV-E2F1, pRc-CMV-E2F2, pRc-CMV-E2F3, pcDNA1-E2F4, or pcDNA3-E2F5 were in vitro transcribed and translated by using TNT-coupled reticulocyte lysate systems (Promega) following manufacturer recommendations. Preparation of nuclear extracts and binding reactions were carried out as described (24). Antibodies against E2F-1 (sc-193) and E2F-4 (sc-1082) were purchased from Santa Cruz Biotechnology. The anti-DP1 and preimmune rabbit polyclonal sera have been previously reported (24).
Construction of Luciferase Reporter Vectors.
The β-globin TATA box was cloned immediately upstream from the luciferase gene in the pGL2-basic plasmid, to generate pBG-LUC. Next, 300 ng of control irrelevant double-stranded oligonucleotide (control), 2xE2Fdhfr(wt), 2xE2Fdhfr(mut), 2xE2Fdhfr(Met), 2xE2Fc-myb(wt), 2xE2Fc-myb(mut), 2xE2Fc-myb(Met), 2xE2Fc-myc(wt), or 2xE2Fc-myc(Met) were ligated to ≈10 μg pBG-LUC, immediately upstream from the β-globin TATA box, for 3 h at 16°C with T4 DNA ligase (New England Biolabs). Ligations were phenol-extracted, were ethanol-precipitated, were resuspended in 20 μl of 1× TE (10 mM Tris/1 mM EDTA, pH 8.0), and were used for transfections without any further manipulation.
Transfections and Reporter Gene Assays.
Saos-2 cells were transfected by employing the calcium phosphate method (24). Transfections included ≈1 μg of appropriate luciferase reporter plasmid plus 0.5 μg of pCMV-βgal (25), and carrier plasmid [pBluescript KS(+) (Stratagene)], up to 30 μg. Where indicated, increasing amounts of pRc-CMV-E2F1, pRc-CMV-E2F3, and pRc-CMV-DP1 were also used. In control transfections, mock pRc-CMV was used. Luciferase assays and β-galactosidase assays (for normalization of luciferase values) were performed essentially as described (24). Further information is available at www.flemingtonlab.com.
Results
The Promoter Regions of the E2F-Regulated Genes dhfr, E2F1, cdc2, c-myb, and c-myc Are Associated with CpG Islands.
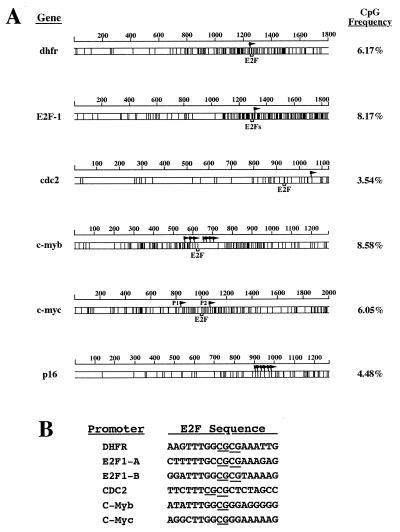
The CpG dinucleotide frequency in the human genome is ≈0.6% (approximately 10-fold lower than the statistically expected frequency). However, the frequency of CpG dinucleotides in the 5′ regulatory sequences of the dhfr, E2F1, cdc2, c-myb, and c-myc genes is significantly higher (Fig. 1A; the CpG density map of the well characterized p16 5′ regulatory region is also shown for comparison). The density of CpG sites is not uniform within these promoters, and, significantly, the E2F binding sites are located within subregions of high local CpG density (Fig. 1A). For example, the two E2F binding sites in the E2F1 promoter are located in the second half of the 5′ regulatory region that contains a CpG dinucleotide frequency of 14.64%. Further, the E2F elements in these promoters contain either one (c-myb and c-myc) or two (dhfr, E2F1-A, E2F1-B, and cdc2) CpG dinucleotides (see Fig. 1B), and are therefore likely candidates for in vivo regulation by DNA methylation.
Figure 1.
(A) CpG density maps of the E2F-regulated promoters dhfr, E2F1, cdc2, c-myb, and c-myc. Map plots are of the dhfr, E2F1, cdc2, c-myb, c-myc, and p16 (included as a reference) 5′ gene sequences, indicating the position of each CpG dinucleotide (vertical lines) and its frequency. The position of the E2F elements and transcription initiation sites (flags) are indicated. The two E2F elements within the E2F1 promoter are only 5 bp apart and are represented by only one bracket. P1 and P2 denote the two known c-myc promoters. The size of the analyzed gene sequences is indicated in base pairs above each map. (B) Alignment of E2F elements from the dhfr, E2F1, cdc2, c-myb, and c-myc promoters. The CpG dinucleotides within each element are underlined.
Differential Binding of E2F Family Members to Methylated DNA-Binding Sites.
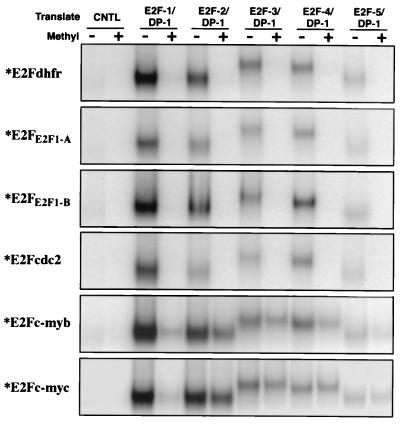
To begin to investigate how methylation of CpG residues within E2F binding sites influences the binding of different E2F family members, we tested the ability of in vitro translated E2F1, E2F2, E2F3, E2F4, and E2F5 (as co-translates with DP1) to bind methylated or unmethylated E2F elements. Oligonucleotides corresponding to the E2F elements derived from the dhfr (E2Fdhfr), cdc2 (E2Fcdc2), c-myb (E2Fc-myb), and c-myc (E2Fc-myc) genes, and the two E2F elements from the E2F1 gene (E2FE2F1-A and E2FE2F1-B) were either control methylated (the reaction was carried out in the absence of the methyl-donor, S-adenosyl-methionine), or methylated with the CpG methylase, Sss I. Methylated or unmethylated (control methylated) radiolabeled oligonucleotides were mixed with in vitro translated E2F heterodimers, and the interaction was analyzed by electrophoretic mobility-shift assays (EMSA). As shown in Fig. 2, E2F1, E2F2, E2F3, E2F4, and E2F5 interacted with each of the indicated unmethylated probes but failed to bind the methylated E2Fdhfr, E2FE2F1-A, E2FE2F1-B, and E2Fcdc2 probes. Notably, E2F2, E2F3, E2F4, and E2F5 bound nearly as well to the methylated vs. unmethylated E2Fc-myb and E2Fc-myc elements whereas E2F1 bound preferentially to the unmethylated E2Fc-myb and E2Fc-myc probes (Fig. 2). Therefore, methylation of a subset of E2F elements (those from the dhfr, E2F1, and cdc2) blocks the binding of all E2F factors tested whereas methylation of another subset of E2F elements (from the c-myb and c-myc promoters) does not significantly alter the binding of E2F2, E2F3, E2F4, and E2F5, but it blocks the binding of E2F1.
Figure 2.
Binding of in vitro translated DP1/E2Fs to different E2F elements is regulated by CpG methylation. DP1 and either E2F1, E2F2, E2F3, E2F4, or E2F5 were co-translated in vitro, and the interaction of the resulting heterodimers with the E2Fdhfr, E2FE2F1-A, E2FE2F1-B, E2Fcdc2, E2Fc-myb, and E2Fc-myc radiolabeled probes was analyzed by EMSA. Before labeling, the DNA probes were either methylated with Sss I (+) or control methylated (−). No cDNA was added to the control (CNTL) in vitro transcription-translation reaction. Only the retarded E2F/DNA complexes are shown.
Discrimination of Methylated E2F Elements by Endogenous E2F Factors.
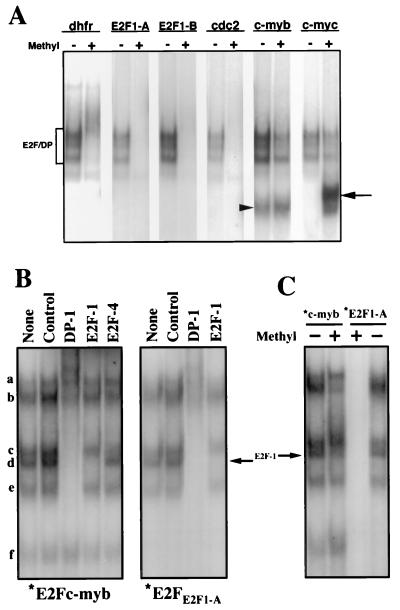
To investigate whether methylation of E2F elements affects the binding of endogenous E2F factors, nuclear extract from the nontransformed, EBV-immortalized B-lymphoblastoid cell line, X50-7, was analyzed by EMSA employing methylated and unmethylated E2F site probes. To specifically address the binding of “free” E2F proteins [i.e., not complexed with pRb family members—as was done by employing in vitro translated proteins (see above)], the free E2F fraction was enriched by treating the extracts with sodium deoxycholate, which disrupts E2F-pRb family member interactions (26). As shown in Fig. 3A, three major complexes are observed that are common to each of the unmethylated probes tested. Binding of these complexes is ablated by anti-DP1 Abs, but not anti-pRb, -p107, or -p130 Abs (data not shown), indicating that these are indeed free forms of E2F. Similar to the results obtained by employing in vitro translated E2Fs, these free forms of endogenous E2F bind to the methylated E2Fc-myb and E2Fc-myc E2F elements, but not the methylated E2Fdhfr, E2FE2F1-A, E2FE2F1-B, and E2Fcdc2 probes (Fig. 3A) (although methylation of the E2Fc-myb element resulted in a modest decrease in the binding of nuclear E2F factors).
Figure 3.
Binding of endogenous E2F factors to different E2F elements is regulated by CpG methylation. (A) Nuclear extracts from the B-lymphoblastoid cell line, X50-7, were incubated with the indicated methylated (+), or control methylated (−), radiolabeled E2F elements in the presence of sodium deoxycholate (to disrupt E2F/pRb-family member interactions). E2F-DNA complex formation was analyzed by EMSA. The position of free E2F/DP heterodimers is indicated by a bracket. The arrowhead and the arrow indicate specific complexes that bind only the E2Fc-myb probe and the methylated E2Fc-myc probe, respectively. (B) Nuclear extracts from S-phase synchronized NIH 3T3 cells were incubated with the indicated radiolabeled E2F elements. Extracts were preincubated in the absence (None) or in the presence of the indicated antibodies for 20 min before the addition of the labeled probes. Control, normal rabbit serum. (C) Nuclear extracts from S-phase synchronized NIH 3T3 cells were incubated with the indicated methylated (+) or control methylated (−) radiolabeled probes. The arrow indicates the position of the free DP1/E2F1 heterodimer.
Notably, E2F1 is not detectable as a major component of the free E2F forms present in X50-7 nuclear extracts (data not shown). To specifically address site discrimination by endogenous E2F1, extracts from S-phase synchronized NIH 3T3 cells were used to assess binding to methylated and unmethylated E2Fc-myb and E2FE2F1-A oligonucleotides (Fig. 3 B and C). Binding of the E2F1-containing complex (complex d) to the E2Fc-myb probe was significantly inhibited by methylation whereas formation of complex c (free DP1/E2F4) and complexes a and e was unaffected or increased. Formation of complex b was also partly inhibited by methylation of E2Fc-myb. [Note that, although complex b was not affected by the anti-E2F1 antibody, it was similarly not affected by antibodies against any other E2F family member (Fig. 3B; data not shown). This complex may contain a novel E2F family member, or, alternatively, the antibodies used in our experiments may not detect the epitope because of masking by interaction with pRb family members.] As expected, no E2F complexes bound to the methylated E2FE2F1-A probe (Fig. 3C).
In addition to binding E2F proteins, the E2Fc-myb element also forms a complex that does not contain E2F factors and migrates faster than free E2Fs in EMSA experiments (24). As shown in Figs. 3 and 4, this complex binds equally well to unmethylated and methylated E2Fc-myb. Interestingly, a new complex specifically interacts with the methylated but not with the unmethylated E2Fc-myc probe (Fig. 3A). Binding of this new factor(s) is specific because it is competed by an excess of a methylated self oligonucleotide but not other methylated or unmethylated E2F sites (data not shown). Further analysis of this complex is described elsewhere (M.R.C. and E.K.F., unpublished work).
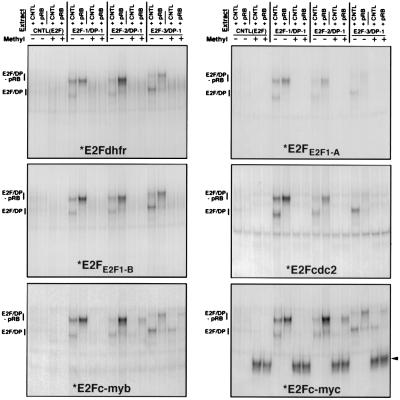
Figure 4.
Association of the retinoblastoma protein with E2F family members does not affect the specificity of binding to methylated E2F elements. U2OS cells were transfected with either E2F1/DP1, E2F2/DP1, E2F3/DP1, or the corresponding empty vector [CNTL(E2F)], together with either a retinoblastoma expression vector (+pRB) or the corresponding control vector (CNTL). Nuclear extracts from transfected cells were analyzed by EMSA employing the indicated methylated (+), or control methylated (−), radiolabeled E2F probes (*). The arrowhead points to a complex that binds uniquely to the methylated E2Fc-myc probe. Because unmethylated E2Fc-myc also binds a non-E2F factor that co-migrates with a free E2F form (M.R.C. and E.K.F., unpublished observations), the experiment shown in the E2Fc-myc panel was performed in the presence of a 100 molar excess of a cold competitor oligonucleotide that specifically binds this factor but not E2F proteins.
The Association of pRb Family Members with E2F Proteins Does Not Alter Methylated E2F Site Discrimination.
A previous report has suggested that the DNA binding specificity of E2F proteins can be modulated through their interaction with members of the pRb family (27). To address whether the binding of pRb family members alters the recognition specificity of E2F proteins for methylated E2F elements, EMSA analysis was performed by employing nuclear extracts generated from cells that were transfected with different DP1/E2F expression vectors plus either a control or a pRb expression vector. Two major shifted complexes are observed employing nuclear extracts from cells transfected with DP1/E2F1, DP1/E2F2, or DP1/E2F3 expression vectors plus the pRb control plasmid, and these complexes correspond to the respective overexpressed E2F factors in either a free form (E2F/DP), or an pRb-containing complex (because of the interaction with endogenous pRb) (Fig. 4). Co-transfection of these E2F expression vectors with a pRb expression plasmid results in predominantly the slower, pRb-containing complex. As shown in Fig. 4, both free and Rb-complexed E2F1, E2F2, and E2F3 bind to each of the unmethylated E2F elements but fail to interact with the methylated E2Fdhfr, E2FE2F1-A, E2FE2F1-B, and E2Fcdc2 sites. On the other hand, E2F2 and E2F3 bind to the methylated E2Fc-myb and E2Fc-myc probes, and their association with pRb only minimally effects their ability to interact with these methylated probes (Fig. 4). Similarly, E2F4 and E2F5 from corresponding transfected cells bind to unmethylated E2Fdhfr and E2Fc-myc and methylated E2Fc-myc but not to methylated E2Fdhfr (data not shown). Moreover, the association of p107 and p130 with E2F4 and E2F5, respectively, did not alter their capacity to discriminate between methylated or unmethylated E2Fdhfr and E2Fc-myc probes (data not shown). Therefore, the capacity of E2F family members to interact with methylated E2F-DNA binding sites is not overtly altered by interactions with pRb family members.
Differential Activation of Methylated E2F Elements by Different E2F Family Members in Vivo.
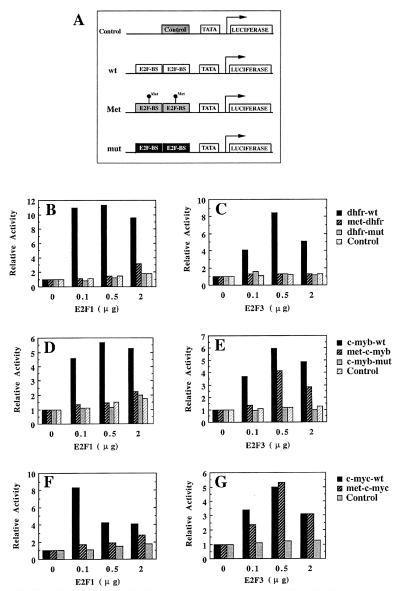
To address whether DNA methylation can regulate the ability of E2F family members to bind E2F recognition sequences in cells, we analyzed the ability of E2F1 and E2F3 to activate minimal reporter plasmids containing unmethylated or methylated E2F elements. Luciferase reporter plasmids containing a TATA box and two copies of the dhfr, c-myb, or c-myc E2F elements (see Fig. 5A) were used for these studies (see Materials and Methods for generation of methylated reporter plasmids). In addition to reporter plasmids containing unmethylated or methylated wild-type E2F elements, reporter plasmids containing E2F site point mutants that are unable to interact with E2F factors (DHFR-mut and C-MYB-mut), or an irrelevant sequence (Control) were also used. Luciferase reporter plasmids were co-transfected with different amounts of DP1 plus E2F1 or E2F3 expression vectors, and the luciferase activity of cell extracts was measured to assess the respective transactivation capacity of E2F1 and E2F3. As shown in Fig. 5 B, D, and F, E2F1 elicits transcriptional activation through wild-type, unmethylated E2F elements derived from the dhfr, c-myb, and c-myc genes but does not significantly influence reporters containing either mutant or methylated E2F elements, or the control sequence. E2F3 similarly activates a reporter containing the unmethylated, wild-type dhfr E2F element but does not activate methylated or mutated dhfr E2F elements (Fig. 5C). Unlike E2F1, however, E2F3 stimulates expression through methylated E2F binding sites derived from the c-myb and c-myc genes nearly as efficiently as through the corresponding unmethylated elements (Fig. 5 E and G). Both E2F1 and E2F3 fail to activate reporter plasmids containing mutant E2F binding sites or an irrelevant DNA sequence (Control) (Fig. 5). Therefore, methylation of CpG dinucleotides within E2F binding sites is a discriminative mechanism that allows E2F to control gene expression through specific E2F elements by regulating the interaction with selected E2F family members.
Figure 5.
E2F3, but not E2F1, efficiently activates transcription through a subset of methylated E2F-DNA binding sites. (A) Schematic representation of the reporter plasmids used. An irrelevant DNA sequence (Control) or two copies of wild-type and methylated E2F elements from the dhfr (dhfr-wt and met-dhfr), c-myb (c-myb-wt and met-c-myb), and c-myc (c-myc-wt and met-c-myc) genes were ligated immediately upstream to the β-globin TATA box. Mutant E2F binding sites from the dhfr (dhfr-mut) and c-myb (c-myb-mut) genes that do not interact with E2F proteins were used as additional controls. Note that, although the c-myb mutant E2F element does not bind E2F factors, it retains the ability to bind another factor that interacts with a region overlapping the E2F-binding site (Fig. 3; ref. 24). Because the unmethylated and methylated wild-type c-myb E2F site similarly bind this factor, this c-myb-mut is, therefore, a proper control. E2F-BS, E2F binding site. (B–G) Saos-2 cells were transfected with the indicated amounts of DP1/E2F1 (E2F1) and DP1/E2F3 (E2F3) expression vectors together with 0.5 μg of pRc-CMV-β-gal and ≈1 μg of the indicated luciferase reporter vectors described in A. Luciferase values from a representative experiment (normalized for β-galactosidase activity) are represented for each luciferase vector relative to cells transfected in the absence of E2F/DP expression vectors.
Discussion
Programmed alterations in the methylation status of genomic sequences regulates the progression of a variety of cellular responses. Soon after fertilization in mammals, the hypermethylated status of gametic genomes are reversed, resulting in global hypomethylation (28, 29). Previous studies have shown that both increases and decreases in promoter methylation regulate the expression of specific genes during terminal differentiation (22, 30–33). The development of tumors is frequently associated with both genome-wide hypomethylation and specific hypermethylation (generally localized to tumor suppressor genes) (4). These studies illustrate a number of systems through which cell cycle progression is controlled, in part, through specific programmed changes in genomic methylation. Moreover, E2F elements have been shown to be methylated in vivo (12), and known E2F responsive genes (pRb, p14ARF, and c-myc) have been previously shown to be methylated in either tumors or during terminal differentiation (10–12, 22, 23). As an initial step in understanding the role of E2F site methylation in regulating promoter activity, we have specifically analyzed the interaction between E2F family members and a panel of methylated E2F promoter elements.
Although several transcription factors, including AP-2, c-Myc, CREB, and NF-κB, do not bind DNA when their respective recognition sequences are methylated at cytosine residues, others, such as Sp1, CTF, and TCR-ATF, bind equally well to methylated and unmethylated binding elements (34). A previous study showed that methylation of the first cytosine residue within a GCGC motif (employing HhaI methylase) of the E2F element derived from the adenovirus E2 promoter inhibited the binding of unidentified E2F species from cell extracts (35). Here, we have shown that methylation of the E2F sites of several cellular promoters (dhfr, cdc2, and E2F-1) effectively blocks the binding of E2F-1, E2F-2, E2F-3, E2F-4, and E2F-5. We have also shown, however, that methylation of the E2F elements from the two early gene promoters, c-myc and c-myb, results in the selective abrogation of E2F-1 binding capacity but not that of E2F-2, E2F-3, E2F-4, or E2F-5. Together, these data indicate that the regulation of promoters through E2F site methylation exhibits the potential for a significantly greater level of complexity than that described for other transcription factors.
Although we have tested only a small sampling of E2F elements for their ability to bind E2F proteins when they are methylated, it may be significant that four sites that do not bind E2F proteins when they are methylated have two CpG dinucleotides within the core binding motif, but the two E2F elements that bind E2F proteins when methylated contain only one CpG dinucleotide. Although further testing is required to firmly establish this correlation, it is possible that this issue is predictive of whether the methylation of specific E2F elements will alter the binding of E2F family members.
A large number of studies carried out over the course of several years has provided a strong inverse correlation between promoter methylation and the level of gene expression (34). Several mechanisms that contribute to methylation mediated transcriptional suppression have been described. First, methylation of certain promoter elements blocks the ability of transcription factors to bind and activate transcription (34). Second, the binding of factors that specifically recognize methylated DNA, such as MeCP1 or MeCP2, can block the binding of transcription factors or transcription initiation complexes (30). In addition to steric hindrance, MeCP2 can actively elicit a condensed chromatin structure through the recruitment of mSin3A/HDAC complex that deacetylates local histone proteins (36–38).
Interestingly, E2F family members associate with factors that encode either acetylase (39) or deacetylase activity [through the interaction with pRb family members (40)], suggesting the possibility that E2F complexes play a role in chromatin remodeling. Our results indicate that the methylation of E2F sites in some promoters (such as the dhfr, cdc2, and E2F-1 promoters) blocks the binding of E2F factors and will therefore abrogate any possible functional influence of E2F family members, including histone deacetylation-mediated repression. The regulation of these promoters after methylation will instead be influenced by other factors that bind to methylated DNA sequences. In this regard, a recent study indicates that in vitro methylation of the E2F element derived from the pRb promoter elicits recruitment of MeCP2 (41). Because this element contains two CpG dinucleotides, it will be of interest to investigate whether other E2F elements that contain two methylated cytosines are also able to bind MeCP2 because recruitment of this factor may compensate for the lack of binding of repressor E2F complexes. On the other hand, binding of MeCP2 may not be required to inhibit transcription through E2F elements that contain only one methylated cytosine residue because they may still interact with E2F complexes with chromatin remodeling properties. This could be the case with the c-myc gene, whose E2F element contains only one CpG dinucleotide and which is hypermethylated and silenced in terminally differentiated K562 cells (22), but hypomethylated and overexpressed in hepatocellular carcinomas (5).
Most promoters that contain E2F elements also contain multiple SP1 sites. Because SP1 binds equally well to methylated and unmethylated DNA, it may still activate certain promoters, unless some other factor(s) counteract its activity. Importantly, however, E2F factors can have a dominant regulatory influence [either positive or negative (through suppression mediated by bound pRb family members)] over SP1 function in promoter activation. As a result, the loss of E2F binding to these promoters can have a significant impact on promoter activity. For example, the binding of E2F family members complexed with a pRb family member (and an associated histone deacetylase) to the E2Fc-myc element during methylation-associated promoter inactivation could play a role in inducing a more compacted, inactive chromatin structure that may counteract SP1-mediated transactivation. In contrast, the binding of free E2F species (in conjunction with a histone acetyl transferase enzyme, such as CBP) to methylated promoters before demethylation associated activation could help elicit a more open chromatin structure. Although little is known regarding the mechanisms through which demethylation is achieved in a compacted chromatin structure, a recent study has provided evidence for the active involvement of a sequence-specific DNA binding transcription factor in eliciting local promoter demethylation (42). In this regard, it is possible that E2F could facilitate the loading of a demethylase enzyme onto DNA by targeting chromatin acetylation.
Although there is a certain degree of functional redundancy between E2F family members, E2F-1 has several unique functions relative to other members of the family, including its role in causing exit from quiescence (43), certain apoptosis responses during mouse development (44, 45), and its involvement in tumor suppression (44, 46, 47). The characteristic(s) of E2F1 that endow it with these unique activities is not known, but slight differences in DNA binding specificity is a reasonable possibility. Our studies provide evidence that the binding properties of E2F1 are indeed distinct from that of other members of the E2F family because it is the only family member tested that cannot bind significantly to the methylated c-myc and c-myb E2F elements. This suggests that the interactions between E2F1 and DNA is in some way unique relative to other family members, supporting the idea that it may recognize a distinct set of promoter elements. On the other hand, activities that are specific to other E2F family members, relative to E2F1, may similarly be attributable to differences in recognition specificity. Interestingly, it is possible that such functions might also be the direct result of their ability to interact specifically with methylated DNA sequences that E2F1 does not recognize.
Acknowledgments
We thank Wilhelm Krek, Doron Gingsberg, and Claude Sardet for plasmids. We also thank Arthur Pardee and Antonio Rodriguez for critical reading of the manuscript. This work was supported by a postdoctoral fellowship from the Human Frontier Science Program Organization to M.R.C. and a research grant from the National Institutes of Health (R01 GM48045) to E.K.F.
Abbreviation
- EMSA
electrophoretic mobility-shift assay
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100340697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100340697
References
- 1.Bird A P. Nature (London) 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 2.Cross S H, Bird A P. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 3.Cho K R, Hedrick L. Curr Top Microbiol Immunol. 1997;221:149–176. doi: 10.1007/978-3-642-60505-5_7. [DOI] [PubMed] [Google Scholar]
- 4.Jones P A. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 5.Laird P W, Jaenisch R. Hum Mol Genet. 1994;3:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 6.Melvin A J, McGurn M E, Bort S J, Gibson C, Lewis D B. Eur J Immunol. 1995;25:426–430. doi: 10.1002/eji.1830250218. [DOI] [PubMed] [Google Scholar]
- 7.Mikovits J A, Young H A, Vertino P, Issa J-P J, Pitha P M, Turcoski-Corrales S, Taub D D, Petrow C L, Baylin S B, Ruscetti F W. Mol Cell Biol. 1998;18:5166–5177. doi: 10.1128/mcb.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang Y, Norihisa Y, Benjamin D, Kantor R R S, Young H A. Blood. 1992;80:724–732. [PubMed] [Google Scholar]
- 9.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D S, Gnarra J R, Linehan W M. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtani-Fujita N, Fujita T, Aoike A, Osifchin N E, Robbins P D, Sakai T. Oncogene. 1993;8:1063–1067. [PubMed] [Google Scholar]
- 11.Sakai T, Toguchida J, N, O, Yandell D W, Rapaport J M, Dryja T P. Am J Hum Genet. 1991;48:880–888. [PMC free article] [PubMed] [Google Scholar]
- 12.Stirzaker C, Millar D S, Paul C L, Warnecke P M, Harrison J, Vincent P C, Frommer M, Clark S J. Cancer Res. 1997. 2229–2237. [PubMed] [Google Scholar]
- 13.Mancini D N, Rodenhiser D I, Ainsworth P J, O'Malley F P, Singh S M, Xing W, Archer T K. Oncogene. 1998;16:1161–1169. doi: 10.1038/sj.onc.1201630. [DOI] [PubMed] [Google Scholar]
- 14.Rice J C, Massey-Brown K S, Futscher B W. Oncogene. 1998;17:1807–1812. doi: 10.1038/sj.onc.1202086. [DOI] [PubMed] [Google Scholar]
- 15.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J-P J, Davidson N E, Sidransky D, Baylin S B. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 16.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S, Sidransky D. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 17.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 18.Johnson D G, Schneider-Broussard R. Front Biosci. 1998;3:447–458. doi: 10.2741/a291. [DOI] [PubMed] [Google Scholar]
- 19.Bartek J, Bartkova J, Lukas J. Curr Opin Cell Biol. 1996;8:805–814. doi: 10.1016/s0955-0674(96)80081-0. [DOI] [PubMed] [Google Scholar]
- 20.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 21.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 22.Baker S J, Pawlita M, Leutz A, Hoelzer D. Leukemia. 1994;8:1309–1317. [PubMed] [Google Scholar]
- 23.Robertson K D, Jones P A. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campanero M R, Armstrong M, Flemington E K. Mol Cell Biol. 1999;19:8442–8450. doi: 10.1128/mcb.19.12.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacGregor G R, Caskey C T. Nucleic Acids Res. 1989;17:2365. doi: 10.1093/nar/17.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirodkar S, Ewen M, DeCaprio J A, Morgan J, Livingston D M, Chittenden T. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 27.Tao Y, Kassatly R F, Cress W D, Horowitz J M. Mol Cell Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 29.Razin A, Shemer R. Hum Mol Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- 30.Ammerpohl O, Schmitz A, Steinmuller L, Renkawitz R. Nucleic Acids Res. 1998;26:5256–5260. doi: 10.1093/nar/26.23.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benvenuto G, Carpentieri M L, Salvatore P, Cindolo L, Bruni C B, Chiariotti L. Mol Cell Biol. 1996;16:2736–2743. doi: 10.1128/mcb.16.6.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lubbert M, Miller C W, Koeffler H P. Blood. 1991;78:345–356. [PubMed] [Google Scholar]
- 33.Takei, S., Fernandez, D., Redford, A. & Toyoda, H. (1996) . [DOI] [PubMed]
- 34.Tate P H, Bird A P. Curr Opin Genet Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 35.Kovesdi I, Reichel R, Nevins J R. Proc Natl Acad Sci USA. 1987;84:2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 37.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Stronboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 38.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 39.Trouche D, Cook A, Kouzarides T. Nucleic Acids Res. 1996;24:4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nevins J R. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 41.Di Fiore B, Palena A, Felsani A, Palitti F, Caruso M, Lavia P. Nucleic Acids Res. 1999;27:2852–2859. doi: 10.1093/nar/27.14.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsieh C-L. Mol Cell Biol. 1999;19:45–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z M, Yang H, Livingston D M. Proc Natl Acad Sci USA. 1998;95:15583–15586. doi: 10.1073/pnas.95.26.15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Field S J, Tsai F-Y, Kuo F, Zubiaga A M, Kaelin W G, Livingston D M, Orkin S H, Greenberg M E. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 45.Tsai K Y, Hu Y, MacLeod K F, Crowley D, Yamasaki L, Jacks T. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 46.Pan H, Yin C, Dyson N J, Harlow E, Yamasaki L, Van Dyke T. Mol Cell. 1998;2:283–292. doi: 10.1016/s1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]
- 47.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]