Abstract
The amino-terminal histone tails are subject to covalent post-translational modifications such as acetylation, methylation, and phosphorylation. In the histone code hypothesis, these exposed and unstructured histone tails are accessible to a repertoire of regulatory factors that specifically recognize the various modified histones, thereby generating altered chromatin structures that mediate specific biological responses. Here, we report that lysine (Lys) 79 of histone H3, which resides in the globular domain, is methylated in eukaryotic organisms. In the yeast Saccharomyces cerevisiae, Lys 79 of histone H3 is methylated by Dot1, a protein shown previously to play a role in telomeric silencing. Mutations of Lys 79 of histone H3 and mutations that abolish the catalytic activity of Dot1 impair telomeric silencing, suggesting that Dot1 mediates telomeric silencing largely through methylation of Lys 79. This defect in telomeric silencing might reflect an interaction between Sir proteins and Lys 79, because dot1 and Lys 79 mutations weaken the interaction of Sir2 and Sir3 with the telomeric region in vivo. Our results indicate that histone modifications in the core globular domain have important biological functions.
Keywords: Chromatin, nucleosomes, histone modification, Sir proteins, gene silencing, histone methylation
Covalent post-translational modifications of amino-terminal histone tails by acetylation, methylation, and phosphorylation are well described (Strahl and Allis 2000; Jenuwein and Allis 2001). As histone tails are exposed and unstructured (Luger et al. 1997), they are likely to be accessible to a variety of nuclear factors in the context of the chromatin template. This includes enzymes that modulate the different states of modification as well as factors that specifically recognize the various modified forms. This forms the basis of the histone code hypothesis, which proposes that these histone modifications translate to biological signals through regulatory factors that recognize them.
The histone code hypothesis is strongly supported by the demonstration that protein modules bind specifically to the various modified forms of histone tails (Strahl and Allis 2000; Jenuwein and Allis 2001). For example, the bromodomains of Gcn5 and TAF1 have high affinity for acetylated histone H3 and/or H4 tails (Dhalluin et al. 1999; Hudson et al. 2000; Jacobson et al. 2000), whereas the chromodomain of HP1 binds specifically to methylated Lys 9 histone H3 tails (Bannister et al. 2001; Jacobs et al. 2001; Lachner et al. 2001). In fission yeast cells, localization of the HP1 homolog Swi6 depends on catalytically active Clr4, which methylates Lys 9 of histone H3 (Bannister et al. 2001; Nakayama et al. 2001). In mouse cells, heterochromatin localization of HP1 proteins also depends on the Lys 9 histone H3 methylases, Suv39h1 and Suv39h2 (Lachner et al. 2001). Therefore, modified histone tails are likely to provide a highly accessible platform for the assembly of factors that mediate nuclear processes such as transcriptional regulation and heterochromatin formation.
Histone methylation can occur at both arginine and lysine residues of the tail regions of histone H3 and H4 (Zhang and Reinberg 2001). CARM1 and PRMT1 are representatives of the histone arginine methylase family. These histone arginine methylases belong to a superfamily of methylases that display an AdoMet (S-adenosyl methionine) domain, which consists of a series of short motifs (Schluckebier et al. 1995). This methylase superfamily includes a wide variety of DNA, RNA, protein, and small-molecule methylases. Structural information from various methylases indicates that motifs I and post-I interact directly with the cofactor, AdoMet (Niewmierzycka and Clarke 1999). CARM1 preferentially methylates Arg 17 and Arg 26 of histone H3 (Ma et al. 2001; Schurter et al. 2001; Bauer et al. 2002), whereas PRMT1 methylates Arg 3 of histone H4 (Strahl et al. 2001; Wang et al. 2001b). However, it is also clear that histones are not the only substrates for CARM1, as they can also methylate other proteins like PABP1 and CBP/p300 (Xu et al. 2001; Lee and Bedford 2002). Thus far, the arginine histone methylases do not methylate other residues such as lysine.
Lysine methylation, on the other hand, is carried out by a novel class of enzymes that contains the SET domain. The first SET domain containing histone lysine methylase reported is the mammalian Suv39h1 and its fission yeast homolog Clr4, which methylate histone H3 at Lys 9 (Rea et al. 2000). Subsequently, other SET domain-containing proteins (Set1, Set2, Set7/Set9, G9a, ESET) were shown to methylate Lys 4, Lys 9, Lys 27, or Lys 36 of histone H3 tail (Briggs et al. 2001; Roguev et al. 2001; Tachibana et al. 2001; Wang et al. 2001a; Krogan et al. 2002; Nagy et al. 2002; Nishioka et al. 2002; Schultz et al. 2002; Strahl et al. 2002; Yang et al. 2002). In Saccharomyces cerevisiae, Lys 4 methylation is mediated by Set1 (Briggs et al. 2001; Roguev et al. 2001; Krogan et al. 2002; Nagy et al. 2002). Deletion of Set1 or mutation of Lys 4 leads to defects in ribosomal DNA silencing, suggesting that Lys 4 methylation is critical to maintain ribosomal DNA silencing (Briggs et al. 2001; Bryk et al. 2002). In contrast to the repressive role of Lys 4 methylation in S. cerevisiae, immunofluorescence and chromatin immunoprecipitation studies show that methylation of Lys 4 is strongly associated with certain transcriptional active regions of Schizosaccharomhyces pombe and mammalian genomes (Litt et al. 2001; Noma et al. 2001; Boggs et al. 2002). Four other SET domain-containing proteins (Set3, Set4, Set5, Set6) exist in S. cerevisiae, and these are strong candidates for methylases that modify histones or other proteins. However, it should be noted that not all SET domains are involved in catalysis, as the SET domain of trithorax binds histone tails (Katsani et al. 2001).
Although there is considerable knowledge about the roles and functions of histone tail modifications, very little is known about whether other solvent-accessible surfaces of the histones are similarly modified for mediating biological functions. Here, we identify a novel modification within the globular domain of histone H3, methylation of Lys 79, which is conserved in eukaryotic organisms. Surprisingly, methylation of Lys 79 is not mediated by a SET domain-containing protein, but rather by Dot1, a protein with methylase fold. Dot1 has been identified previously through its role in genomic silencing (Singer et al. 1998; San-Segundo and Roeder 2000), and we show this effect requires Dot1-mediated methylation of Lys 79. Furthermore, Lys 79 methylation of histone H3 by Dot1 is important for the association of the Sir proteins. These observations show that the modification of the histone H3 core domain has important biological consequences in vivo, and they suggest the possibility that the association of the Sir proteins with chromatin might involve an interaction with methylated Lys 79 of histone H3.
Results
Lys 79 of histone H3 is methylated in eukaryotic organisms
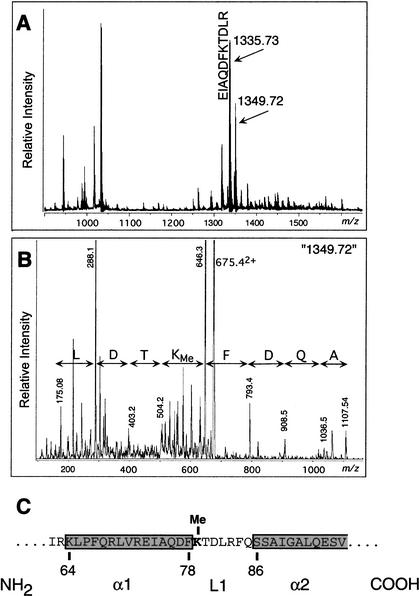
We initially sought to identify novel modifications on core histones by mass spectrometric fingerprinting of trypsin-generated peptides of histone H3 from calf thymus and human. The major ions match the calculated monoisotopic masses of predicted peptides (Fig. 1A). Interestingly, a pair of ions that differ by 13.99 amu between the monoisotopes (1335.73 and 1349.72) is observed in this spectrum. Such mass difference could arise as a result of methylation (DeltaMass database: http://www.abrf.org/index.cfm/dm.home). The lighter of the two ions corresponds to an H3 tryptic peptide (EIAQDFKTDLR; theoretical monoisotopic mass of the protonated form is 1335.69 Da; Δ = 0.04). This was confirmed by tandem (MS/MS) mass spectrometric sequencing using a triple quadrupole instrument with custom nano-electrospray ionization source (data not shown). The heavier, unaccounted ion (m/z = 1349.72) was also subjected to MS/MS analysis, as shown in Figure 1B. The results unequivocally show that it is derived from a peptide of identical sequence, but which is singly methylated on Lys 79.
Figure 1.
Identification of histone H3 Lys 79 methylation. (A) MALDI-TOF MS of histone H3. Calf thymus histone H3 was digested with trypsin and resulting peptides chromatographed on a Poros 50 R2 (Perseptive Biosystems) reversed-phase micro-tip and analyzed by matrix-assisted laser-desorption/ionization reflectron time-of-flight mass spectrometry (MALDI-reTOF MS) using a Reflex III instrument (Bruker Daltonics), as described (Erdjument-Bromage et al. 1998; Winkler et al. 2002). An ion pair differing by 13.99 amu is indicated; the lighter ion corresponds to an H3 tryptic peptide (sequence shown). (B) Nano-ES MS/MS. Tryptic digest mixture was analyzed by electropsray ionization (ESI) MS using an API 300 triple quadrupole instrument (Applied Biosystems/MDS SCIEX), modified with a continuous-flow NanoES source, as described (Geromanos et al. 2000; Winkler et al. 2002). The precursor ion (m/z = 675.4), corresponding to the doubly charged (z = 2) version of peptide ion marked 1349.72 in A was selected for collision-induced dissociation (CID)-based MS/MS analysis. The fragment ion spectrum was inspected for y” ions and the deduced sequence is indicated (note that y” ion series originate at the carboxyl terminus and, therefore, read backwards). (C) Location of Lys 79 of histone H3 relative to α-helices (α1 and α2) and loop 1 (L1) based on the crystal structure of nucleosome.
The X-ray crystal structure of nucleosome reveals that Lys 79 is positioned at the loop 1 region that lies between α1 and α2 helixes within the globular domain of histone H3 (Luger et al. 1997). Loop 1 of histone H3 does not contact DNA or other histones and is found on the solvent-accessible surface of the nucleosome.
Dot1 is required for Lys 79 methylation of histone H3 in yeast cells
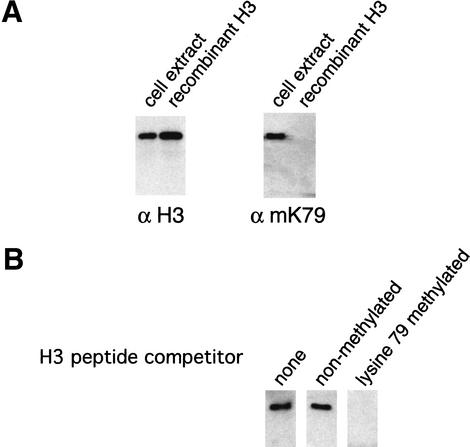
We next determined whether yeast histone H3 is methylated at Lys 79 using an antibody generated against a H3 peptide containing dimethylated Lys 79 (IAQDFmKTDLRF). This peptide is identical to the corresponding histone H3 sequence of many eukaryotic species (human, chicken, frog, and yeast), and the antibody binds to native histone H3 from these organisms (data not shown). As a control, we analyzed recombinant yeast histone H3 expressed and purified from bacteria, which should not contain eukaryotic post-translational modifications. This antibody specifically binds histone H3 isolated from a wild-type strain, but it does not react with recombinant histone H3 or histone H3 from strains with mutations at residue 79 (Fig. 2A; data not shown). Furthermore, competition with a peptide containing dimethylated Lys 79 specifically abolished reactivity of the antibody toward native yeast histone H3 (Fig. 2B).
Figure 2.
Loss of Dot1 abolishes histone H3 Lys 79 methylation in vivo. (A) Western blots of yeast whole-cell extracts and recombinant yeast histone H3 were probed with either antibodies against the histone H3 tail (residues 1–20) or against a peptide containing methylated Lys 79. (B) Specificity of the anti-methyl Lys 79 antibody. Antibodies (1/10,000 dilution) were preincubated with either no peptide, 25 μg nonmethylated peptide, or 25 μg of methylated Lys 79 peptide for 30 min at room temperature before probing yeast whole-cell extracts for histone H3. (C) Western blot screen for histone H3 Lys 79 methylase. Whole-cell extracts from the indicated deletion strains were probed with antibodies against methylated Lys 4, methylated Lys 79, and unmodified H3. (D) Restoration of Lys 79 methylation by constructs containing the DOT1 gene. Whole-cell extracts from the indicated strains were probed with either anti-methyl Lys 4, or anti-methyl Lys 79 antibodies.
Having established that yeast histone H3 is methylated at Lys 79, we performed a screen to identify potential enzymes responsible for this novel modification. We reasoned that deletion of the histone H3 Lys 79 methylase would eliminate or reduce Lys 79 methylation in vivo. To date, only SET domain-containing proteins (e.g., Suv39h1, Clr4, G9a, Set1, Set2, Set7/Set9, SETDB1, ESET) have been shown to methylate lysine residues within the histone tail regions (Rea et al. 2000; Briggs et al. 2001; Roguev et al. 2001; Tachibana et al. 2001; Wang et al. 2001a; Krogan et al. 2002; Nagy et al. 2002; Nishioka et al. 2002; Schultz et al. 2002; Strahl et al. 2002; Yang et al. 2002). Histone methylase activities of these enzymes are affected by mutations within the SET domain, suggesting that the SET domain is directly involved in catalysis. In S. cerevisiae, there are six proteins that contain a SET domain (Set1–Set6), and these were included for analysis. As S-adenosyl methionine (AdoMet) is a universal substrate for many methylation reactions involving DNA, RNA, and protein, we also examined proteins that show putative AdoMet-binding motif signatures. These include known protein methylases (Ctm1, Rmt1, Rmt2), , and proteins with a methylase fold (Dot1) or an AdoMet motif signature (Rms1, YHL039W, YDR198C, YBR030W).
Whole-cell extracts were prepared from these mutant strains and histone H3 Lys 79, and methylation was analyzed by Western blotting using anti-methylated Lys 79 antibody. Surprisingly, Lys 79 methylation is not affected in the set mutants, whereas the dot1 deletion strain completely lacks Lys 79 methylation (Fig. 2C). As a control, Lys 4 methylation is normal in all cases except for the set1 deletion strain, a result expected from the previous identification of Set1 as the Lys 4 methylase (Briggs et al. 2001; Roguev et al. 2001; Krogan et al. 2002; Nagy et al. 2002). The involvement of Dot1 in mediating Lys 79 methylation was also confirmed by independent deletion of Dot1 in other strain backgrounds (data not shown) and restoration of Lys 79 methylation upon introducing a plasmid expressing Dot1 into dot1 mutant cells (see below).
Dot1 methylates Lys 79 of nucleosomal histone H3 in vitro
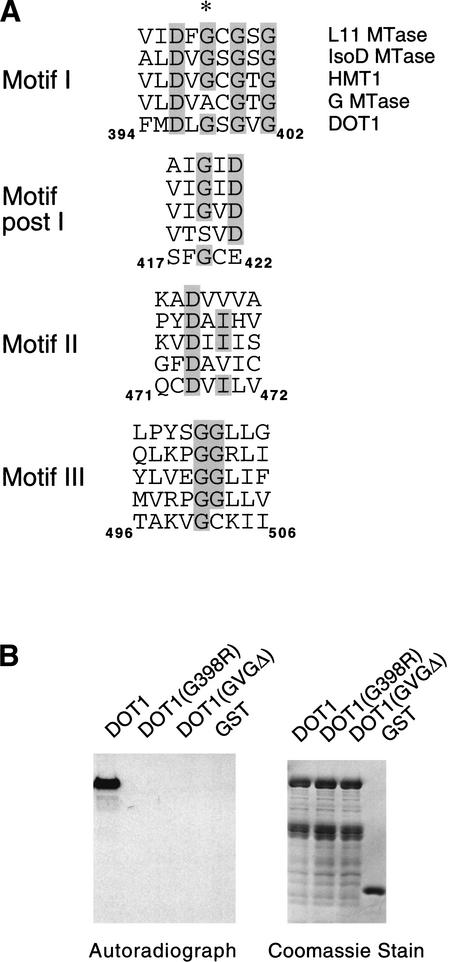
The homology between Dot1 and other methylases is weak, and it was not discovered in the initial computational screen for all putative yeast AdoMet-dependent methylases (Niewmierzycka and Clarke 1999). However, a subsequent analysis using a more powerful searching algorithm and structural modeling indicate that Dot1 has a similar fold pattern as a glycine N-methylase (Dlakic 2001). To characterize the enzymatic activity of Dot1, we expressed and purified GST–Dot1 and two mutant versions (G398R and ΔGVG400-402) from Escherichia coli cells (Fig. 3A). The point mutation and deletion are within motif I of the putative methylase-fold domain (Dlakic 2001). A UV cross-linking experiment reveals that wild-type recombinant Dot1 bound to Sadenosyl methionine in vitro, whereas the two mutant Dot1 derivatives do not (Fig. 3B).
Figure 3.
Dot1 methylates Lys 79 of histone H3 in vitro. (A) Alignment of S-adenosyl methionine (AdoMet) motifs (Dlakic 2001), with conserved residues shaded in gray and Gly 398 of DOT1 highlighted with an asterisk. G MTase is rat glycine N-methylase (accession no. P13255); HMT1 is yeast hnRNP methylase (accession no. NP_009590); L11 MTase is E. coli. L11 ribosomal protein methylase (accession no. P28637); IsoD MTase is human isoaspartate O-methylase (accession no. AAH07501). Amino acid positions of the indicated motifs are shown for Dot1. (B) Dot1 mutations abolish AdoMet-binding activity. GST derivatives of wild-type and mutant (G398R or ΔGVG400-402) Dot1 proteins were incubated with [3H] AdoMet and cross-linked by UV irradiation. The resultant proteins were resolved on SDS-PAGE (right) and tritium label was detected by fluorography (left). (C) Dot1 methylates nucleosomal histone H3. GST derivatives of Set2 and Dot1 were incubated with oligonucleosomes from HeLa cells, and histones were analyzed by autoradiography or Coomassie staining. (D) Dot1 methylation is specific to nucleosomal histone H3. GST–Dot1 was incubated with the indicated substrates and analyzed as described in C. (E) Dot1 mutant proteins are unable to methylate nucleosomal histone H3. Analysis was performed as described in C. (F) Dot1 methylates nucleosomal histone H3 at Lys 79 residue. GST–Dot1 was incubated with a crude nucleosomal preparation from dot1 cells, and the resulting material was analyzed by Western blotting using antibodies against metylated Lys 79 and unmodified H3.
Having established that Dot1 is an AdoMet-binding protein, we next assayed Dot1 methylase activity on free individual core histones, on a mixture of free core histones, and on oligonucleosomes. GST–Dot1 only methylates histone H3 in context of nucleosomes, and shows no activity toward free histone H3 under the conditions tested (Fig. 3C, D). As a positive control, we showed that recombinant Set2 protein specifically methylates H3. Dot1 can also methylate histone H3 of in vitro-assembled nucleosomes positioned on a 5S template, but not free HeLa histones (data not shown), thus confirming the requirement for a nucleosomal configuration for histone H3 methyl transfer. As expected, the Dot1 mutant proteins with alterations in motif I are unable to methylate histone H3 (Fig. 3E).
To show that Dot1 methylates histone H3 at Lys 79, we made use of micrococcal nuclease-released chromatin isolated from the yeast dot1 deletion strain as a substrate. Unlike histone H3 from HeLa cells, which contains a high level of Lys 79 methylation, the chromatin from dot1 cells is free of Lys 79 methylation, and, hence, permitted us to assay for methyl transfer by Dot1 in vitro. Upon incubation of recombinant Dot1 with the crude nucleosome preparation from dot1 cells, we detected methyl transfer to Lys 79 by Western blot analysis using antimethylated Lys 79 antibody (Fig. 3D). Therefore, Dot1 is a bona fide nucleosomal histone H3 Lys 79 methylase. We have not excluded the possibility that Dot1 might methylate other residues of histone H3.
Dot1 methylation of Lys 79 is important for telomeric silencing in vivo
Dot1 is a nuclear protein that was originally identified in a genetic screen for high-copy disruption of telomeric silencing (Singer et al. 1998; San-Segundo and Roeder 2000). In addition, a dot1 deletion strain is also defective for telomeric silencing. In diploid cells, Dot1 plays a role at the pachytene checkpoint, which ensures proper chromosome segregation by preventing meiotic progression when recombination and chromosome synapsis are defective (San-Segundo and Roeder 2000).
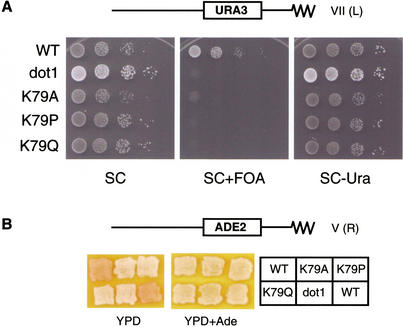
We used mutational analysis to address whether Dot1 affects telomeric silencing by methylating Lys 79. To this end, we first generated yeast strains whose sole source of histone H3 contained substitutions of Lys 79 mutations with other residues. As Lys 79 is found at a loop region connecting α1 and α2 helixes, we suspected that certain mutations might be deleterious to the general folding of histone H3. Therefore, we chose to mutate Lys 79 to three different nonmethylable residues (alanine, proline, and glutamine) in an attempt to identify a genuine property of Lys 79. These Lys 79 substitution strains, as well as the wild-type and dot1 deletion strains, also contain a URA3 reporter integrated at the telomere of the left arm of chromosome VII (Fig. 4A). As observed with the dot1 deletion strain, strains containing the various Lys 79 substitutions are viable and do not exhibit severe growth defect in rich (YPD) or synthetic complete (SC) medium containing glucose (Fig. 4; data not shown). In wild-type yeast cells, integration of the URA3 gene near the telomere results in gene silencing in a majority of the cell population, thereby permitting cells to grow on medium containing 5-fluoro-orotic acid (FOA) (Gottschling et al. 1990; Aparicio et al. 1991; Laurenson and Rine 1992). However, cells defective in telomeric silencing express URA3 and are therefore FOA sensitive. Strikingly, all three single amino acid substitution mutants of Lys 79 show the same degree of telomeric silencing defect as the dot1 strain.
Figure 4.
Dot1 methylation of Lys 79 of histone H3 is required for telomeric silencing. (A) URA3-based telomeric silencing assay in which equal numbers of wild-type (WT) and mutant (dot1, K79A, K79P, K79Q) strains were spotted at 10-fold dilution on synthetic complete (SC) medium in the presence or absence of 0.12% 5-fluoro-orotic acid (FOA) and/or Uracil (Ura), and incubated at 30°C for 3 d. (B) ADE2-based telomeric silencing assay in which the indicated wild-type and mutant strains were patched on YPD medium with or without 40 mg/L adenine for 2 d and transferred to 4°C for 2 d. These strains (WZY42 background) contain an ADE2 reporter integrated adjacent to the telomere of chromosome V.
As an independent measure of telomeric silencing, we used a strain with an ADE2 reporter integrated into another telomeric locus (right arm of chromosome V). Expression of ADE2 gives rise to white colonies, whereas repression of ADE2 results in pink colonies. Due to the reversible nature of telomeric silencing, wild-type colonies have a variegated appearance with pink and white sectors (Gottschling et al. 1990; Fig. 4B; data not shown). Both the dot1 deletion strain and the Lys 79 substitution strains are white (Fig. 4B), indicating that telomeric silencing is compromised by the Lys 79 mutations. Taken together, these observations provide strong evidence that the biological effect of Dot1 is mediated through Lys 79 of histone H3.
To address whether the histone methylase activity of Dot1 is important for telomeric silencing, we examined the phenotypes conferred by mutations in the AdoMet motif that presumably destroy the cofactor-binding site of the enzyme. Specifically, the dot1 deletion strain was transformed by plasmids expressing wild-type Dot1 or derivatives harboring the G398R and ΔGVG400-402 mutations. Importantly, these strains are defective for telomeric silencing (Fig. 5A), even though the level of Dot1 mutant proteins are comparable with the wild-type protein (Fig. 5B). Western blotting using anti-methylated Lys 79 antibody on crude cell extracts from these strains shows that the mutant Dot1 proteins do not restore Lys 79 methylation in vivo (Fig. 5B). Thus, telomeric silencing requires the histone methylase activity of Dot1.
Figure 5.
Catalytic activity of Dot1 is required for telomeric silencing. (A) Equal numbers of yeast cells of wild-type (WT) and dot1 mutant strains expressing the indicated Dot1 proteins were spotted at 10-fold dilution on synthetic complete (SC) medium in the absence of tryptophan (Trp) and/or Uracil (Ura) in the presence or absence of 0.12% FOA, and incubated at 30°C for 3 d. These strains (UCC1111 background) contain a URA3 gene integrated near the telomere of chromosome VII. (B) Analysis of Dot1 protein levels and histone H3 methylation at Lys 79 and Lys 4 in the above strains, as assayed by Western blotting with the indicated antibodies.
Methylation of Lys 79 by Dot1 is important for telomeric association of the Sir proteins
Telomeric silencing depends critically on the Sir proteins, which are recruited to telomeres by the DNA-binding protein Rap1 and associated proteins (Moretti et al. 1994; Grunstein 1997, 1998; Wotton and Shore 1997). We therefore used chromatin immunoprecipitation to analyze occupancies in vivo of Sir2 and Sir3 at positions 300 bp and 3.5 kb away from the right telomere of chromosome VI (Fig. 6A,B). Sir2 occupancy at a position 300 bp away from the telomere is significantly reduced in Lys 79 substitution and dot1 mutant strains; the effects are perhaps slightly less pronounced in the dot1 mutant strain. Sir2 occupancy in these mutant strains is much more reduced, and is nearly eliminated, at the position 3.5 kb away from the telomere. Similar results were observed at both positions for Sir3 occupancy.
Figure 6.
Dot1 methylation of Lys 79 of histone H3 is important for association of Sir proteins with telomeric regions. (A) Formaldehyde cross-linked chromatin from the indicated yeast strains was immunoprecipitated with affinity-purified Sir2 antibodies. Immunoprecipitated and input DNAs were quantified by PCR using primers that probe regions around 300 and 3500 bp away from chromosome end and the POL1-coding regions, which serves as a control for background signal. (B) Quantitation of relative Sir2 and Sir3 occupancy at TEL300 and TEL3500; the background signal from the POL1 region has been subtracted in all cases. Results presented are based on three independent experiments (standard deviation is shown). (C) Analysis of Sir2 and Sir3 protein levels in the indicated strains as assayed by Western blots of whole-cell extracts. (D) Lys 79 histone H3 methylation near the vicinity of telomere (right arm of chromosome VI) depends on Dot1 and Lys 79. Formaldehyde cross-linked chromatin from the indicated strains was immunoprecipitated with antibodies against methylated Lys 79 or Lys 4, and the resulting material quantitated by PCR.
These observations strongly suggest that methylation of Lys 79 by Dot1 is important for Sir protein association at telomeres. We cannot exclude the possibility that Lys 79 and dot1 mutations lead to a subtle reduction in the level of SIR or other silencing proteins that result in impaired silencing and reduced Sir2 and Sir3 occupancy, although the mutant strains appear to have comparable levels of Sir2 and Sir3 with that observed in the wild-type strain (Fig. 6C). Nevertheless, the reduction in Sir2 and Sir3 density and spreading along the telomeres provide a likely explanation for the underlying defect in telomeric silencing due to substitutions of Lys 79 and loss of Dot1. Our results are also consistent with the observation that Sir2 and Sir3 are delocalized in dot1 mutant strains as revealed by immunofluorescence (San-Segundo and Roeder 2000).
Using chromatin immunoprecipitation, we determined whether nucleosomes in the vicinity of telomeres are methylated at Lys 79 in vivo. The antibody against methylated Lys 79 clearly immunoprecipitates telomeric DNA, with only background signals being detected in immunoprecipitates from dot1 deletion or Lys 79 mutant strains (Fig. 6D). However, Dot1-dependent methylation of Lys 79 is also present at all other regions of the genome tested (data not shown), suggesting that Dot1 acts globally and is not specifically recruited to telomeres.
Discussion
Covalent modifications within the amino-terminal histone tails are recognized by specific regulatory proteins that affect gene regulation and heterochromatin structure. This forms the basis of the histone code hypothesis, in which specific histone modifications are translated into biological signals via direct interactions with particular regulatory proteins (Strahl and Allis 2000; Jenuwein and Allis 2001). Here, we identify a novel site of lysine methylation (Lys 79) within the globular domain of histone H3 that is observed in yeast, cow, human, and presumably other eukaryotic organisms. In S. cerevisiae, the silencing protein Dot1 mediates this modification, and we presume that homologous proteins perform the same function in other species. Dot1 represents a novel class of lysine histone methylases, as all previously described enzymes with this activity have SET domains. Our mutational analyses strongly suggest that Dot1 regulates telomeric silencing predominantly through methylation of Lys 79 of histone H3. Specifically, Dot1 catalytic activity and Lys 79 of histone H3 are important for telomeric silencing. We cannot understand why overexpression of Dot1 leads to a defect in telomeric silencing (Singer et al. 1998); perhaps hypermethylation of Lys 79 increases nonspecific binding of the Sir proteins throughout the genome, thereby lowering the concentration of Sir proteins at telomeres. More generally, our results show that cells can modify solvent-accessible surfaces on the histone core domains to regulate nuclear processes such as telomeric silencing.
Dot1-methylation of Lys 79 is important for telomeric association of the Sir proteins, a result that is likely to underlie the defect in telomeric silencing. By analogy with histone modifications in the amino-terminal tails, the simplest mechanism to account for telomeric silencing is a direct physical interaction between a Sir protein(s) and methylated Lys 79 of histone H3. This putative interaction is clearly not sufficient for Sir protein association, because Lys 79 methylation occurs over the entire genome, whereas Sir proteins are localized to telomeres and a few other selected genomic regions. At telomeres, it is presumed that the Sir proteins are initially recruited by Rap1 and associated proteins, whereupon they spread away from the telomere via protein–protein interactions between the Sir proteins themselves and between Sir proteins and histones (Grunstein 1997, 1998). As such, our results argue against the idea that Dot1 is targeted to telomeres (in this regard, we have not observed Dot1 association with telomeric regions by chromatin immunoprecipitation), but rather that the putative interaction between Sir proteins and Lys 79 stabilizes the association of Sir proteins with nucleosomes. Our observation that Dot1 methylation of Lys 79 is more important for Sir protein association at telomere-distal regions is consistent with this suggestion. In particular, Sir interactions with Rap1 and associated proteins might preferentially stabilize Sir proteins at telomere-proximal regions, thus making telomere-distal regions more dependent on the Sir-histone interactions.
Although our results are suggestive, they do not demonstrate that Sir proteins directly interact with Lys-79 of histone H3. An alternative model is that Lys-79 methylation affects a histone and/or nucleosome conformation that is important for the stability of Sir proteins on a chromatin template. In any event, our results extend the histone code hypothesis to residues within histone core domains, and perhaps to Sir proteins as effectors that recognize such modifications and contribute to transcriptional silencing and the heterochromatin-like structure at telomeres.
Materials and methods
Yeast strains and telomeric silencing assays
To screen for the gene encoding the histone H3 Lys 79 methylase, we used deletion derivatives of BY4741 (MATa, his3-Δ1, leu2-Δ0, met15-Δ0, ura3-Δ0) that were obtained from Research Genetics, except for the set1 mutant, which was generated by gene replacement (Longtine et al. 1998). Telomeric silencing assays were performed in derivatives of strains UCC1111 [MATα, ade2∷his3-Δ200, leu2-Δ0, lys2-Δ0, met15-Δ0, trp1-Δ63, ura3-Δ0, adh4∷URA3-TEL (VII-L), hhf2-hht2::MET15, hhf1-hht1::LEU2, pRS412(ADE2 CEN ARS)-HHF2-HHT2] (Kelly et al. 2000) and WZY42 [MATa, ura3-52, lys2-801, ade2-101, trp1-Δ63, his3-Δ200, leu2-Δ1, hht1-hhf1∷pWZ405-F2F9-LEU2, hht2-hhf2∷pWZ403-F4F10-HIS3, Ycp50-copyII HHT2-HHF2 (URA3 CEN ARS)] (Zhang et al. 1998). The ADE2-TEL(VR) derivative of WZY42 was constructed by integrating EcoRI-linearized pHR10-6 plasmid DNA and selecting clones on the basis of the colony color phenotype (Gottschling et al. 1990). This genotype of this strain was further confirmed by deleting Sir2 or Dot1, which resulted in loss of silencing and white appearance on medium deficient in adenine. Strains containing histone variants at residue Lys 79 were generated by plasmid shuffling. Wild-type and mutant histone alleles on TRP1-based centromeric plasmids were transformed into yeast strains harboring a copy of the histone H3 and H4 genes on a URA3 or ADE2 plasmid. For selecting cells that lack the URA3 plasmid, cells were first grown on medium supplemented with uracil and then transfered to medium containing FOA. For selecting cells that lack the ADE2 plasmid, cells were first grown on medium with 40 mg/L adenine and restreaked on medium containing 10 mg/L adenine (low adenine) to identify red colonies. Loss of wild-type plasmid and replacement with mutant histone alleles were confirmed by Western blotting using anti-methyl Lys 79 antibodies. The DOT1 gene (including 600 bp upstream of ATG, and 250 bp downstream of the termination codon) was cloned by PCR as a SphI–XmaI fragment into the corresponding sites pf YCplac22 (TRP, CEN, ARS). For 5myc–Dot1, 5 myc eptitopes were inserted before the ATG of DOT1. The G398R and ΔGVG400-402 mutations were introduced by PCR-mediated mutagenesis (Quik-Change site-directed mutagenesis kit; Stratagene) of plasmid pWZ414-F13, which contains a wild-type copy of histone H3 (Zhang et al. 1998). Telomeric silencing assays were carried as described (Gottschling et al. 1990; Kelly et al. 2000).
AdoMet binding and histone methylation assays
The Dot1 protein-coding regions of the wild-type and catalytically inactive derivatives were cloned by PCR into pGEX2T as a BamHI–EcoRI fragment. The Set2 protein-coding region was cloned by PCR into pGEX5T-1 as a XmaI–XhoI fragment. Nuclei and micrococcal nuclease-released chromatin preparations were performed as described previously (Pilon et al. 1997). Histone methylation assays were performed in 15-μL reactions containing 50 mM Tris.HCl (pH 8.0), 0.1 M NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol, 1 μL of [3H]AdoMet (Amersham Pharmacia Biotech; 82.0 Ci/mmole; 12 μM) for 2 h at 30°C with 0.5–1 μg of recombinant protein. Reactions were stopped by adding SDS loading buffer and boiled for 5 min. For nonradioactive methylation, reactions were performed with 0.25 mM AdoMet (New England Biolabs) for 12 h at 30°C. The UV cross-linking assay to detect AdoMet binding was carried out as described (Tang et al. 2000). Chicken core histones (isolated from erythrocytes; Upstate Biotechnology), calf thymus histone (Boerhinger Mannheim), recombinant Xenopus histones (Upstate Biotechnology), recombinant yeast histone H3 (kind gift from Danesh Moazed, Harvard Medical School, Boston, MA), and oligo-nucleosomes and acid-extracted free histones from HeLa nuclei (kind gift from Bob Kingston, Massachusetts General Hospital, Boston, MA) were used in these assays.
Western blotting
Yeast cells were lysed in RIPA buffer (50 mM HEPES at pH 7.9, 2 mM EDTA, 0.25 M NaCl, 0.1% SDS, 0.1% DOC, 1% Triton X-100) by the glass beads breakage method. Electrophoretically separated proteins were probed with affinity-purified anti-Sir2 and Sir3 antibodies (kind gifts from Danesh Moazed), anti-myc 9e10 monoclonal antibody (Santa Cruz), anti-dimethyl Lys 4 histone H3 antibody (AbCam), anti-histone H3 antibody (residues 1–20; Upstate Biotechnology), and anti-dimethylated Lys 79 histone H3 antibody (generated in this work and raised against the peptide IAQDFmKTDLRF).
Chromatin immunoprecipitation
Chromatin immunoprecipitation and quantitative PCR of input and immunoprecipitated samples were performed as described previously (Kuras and Struhl 1999). Sir2 and Sir3 occupancy values were calculated by measuring the apparent immunoprecipitation efficiency (obtained by dividing the signals from the immunoprecipitated samples by the corresponding input samples) and subtracting the values obtained in the POL1-coding region control.
Acknowledgments
We thank Danesh Moazed, David Sinclair, Dan Gottschling, Sharon Roth, Mark Parthun, Bob Kingston, Ned Sekinger, Jim Kadonoga, and Mark Levenstein for yeast strains, antibodies, polynucleosomes, and plasmids. We thank Lynne Lacomis for expert help with mass spectrometric analysis and preparation of the figure. This work was supported by postdoctoral fellowship from the Cancer Research Fund of the Damon Runyon Walter Winchell Foundation (H.H.N.) and research grants from the National Institutes of Health to P.T. (P30CA08748), Y.Z (GM63067), and K.S. (GM53720). Y.Z. is a Kimmel Scholar and is also supported by the American Cancer Society (RSG-00-351-01-GMC). H.H.N is on leave from the Department of Biological Sciences, National University of Singapore.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Corresponding authors.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1001502.
References
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bauer UM, Daujat S, Neilsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 Lys 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M, Briggs SD, Strahl BD, Curcio MJ, Allis CD, Winston F. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiaeby a Sir2-independent mechanism. Curr Biol. 2002;12:165–170. doi: 10.1016/s0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dlakic M. Chromatin silencing protein and pachytene checkpoint regulator Dot1p has a methyltransferase fold. Trends Biochem Sci. 2001;26:405–407. doi: 10.1016/s0968-0004(01)01856-4. [DOI] [PubMed] [Google Scholar]
- Erdjument-Bromage H, Lui L, Grewal A, Annan RS, MacNulty DE, Carr SA, Tempst P. Micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- Geromanos S, Freckleton G, Tempst P. Tuning of an electrospray ionization source for maximum peptide-ion transmission into a mass spectrometer. Anal Chem. 2000;72:777–790. doi: 10.1021/ac991071n. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiaetelomeres: Reversible repression of pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- ————— Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J Mol Biol. 2000;304:355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Katsani KR, Arredondo JJ, Kal AJ, Verrijzer CP. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes & Dev. 2001;15:2197–2202. doi: 10.1101/gad.201901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TJ, Qin S, Gottschling DE, Parthun MR. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol Cell Biol. 2000;20:7051–7058. doi: 10.1128/mcb.20.19.7051-7058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS: A histone H3 (Lysine4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivois stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Laurenson P, Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Bedford MT. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002;3:268–273. doi: 10.1093/embo-reports/kvf052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. Hormone-dependent CARM1-directed,arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes & Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Niewmierzycka A, Clarke S. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes & Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Pilon J, Terrell A, Laybourn PJ. Yeast chromatin reconstitution system using purified yeast core histones and yeast nucleosome assembly protein-1. Protein Expr Purif. 1997;10:132–140. doi: 10.1006/prep.1996.0716. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZQ, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiaeSet1 complex includes an Ash2 homologue and methylates histone 3 Lys 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol Biol Cell. 2000;11:3601–3615. doi: 10.1091/mbc.11.10.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluckebier G, O'Gara M, Saenger W, Cheng X. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J Mol Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJI. SETDB1: A novel KAP-1 associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccaromyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, et al. Methylation of histone H4 at arginine 3 occurs in vivoand is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukishima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275:7723–7730. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchres C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001a;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001b;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Winkler, G.S., Lacomis, L., Philip, J., Erdjument-Bromage, H., Svejstrup, J.Q., and Tempst, P. 2002. Isolation and mass spectrometry of transcription factor complexes. Methods (in press). [DOI] [PubMed]
- Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes & Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, Hickstein DD, Zhang Y. Molecular cloning of ESET, a novel histone H3-specific methyltransferse that interacts with ERG transcription factor. Oncogene. 2002;3:148–152. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]