Abstract
Dynamic repositioning of telomeres is a unique feature of meiotic prophase I that is highly conserved among eukaryotes. At least in fission yeast it was shown to be required for proper alignment and recombination of homologous chromosomes. On entry into meiosis telomeres attach to the nuclear envelope and transiently cluster at a limited area to form a chromosomal bouquet. Telomere clustering is thought to promote chromosome recognition and stable pairing of the homologs. However, the molecular basis of telomere attachment and movement is largely unknown. Here we report that mammalian SUN-domain protein Sun2 specifically localizes to the nuclear envelope attachment sites of meiotic telomeres. Sun2–telomere association is maintained throughout the dynamic movement of telomeres. This association does not require the assembly of chromosomal axial elements or the presence of A-type lamins. Detailed EM analysis revealed that Sun2 is part of a membrane-spanning fibrillar complex that interconnects attached telomeres with cytoplasmic structures. Together with recent findings in fission yeast, our study indicates that the molecular mechanisms required for tethering meiotic telomeres and their dynamic movements during bouquet formation are conserved among eukaryotes.
Keywords: meiosis, SUN-domain proteins, telomere attachment
Meiosis is a unique type of cell division that ensures the proper segregation of genetic material and the generation of viable haploid gametes. This is accomplished by a complex series of chromosomal interactions during first meiotic division that include alignment, pairing, synapsis, and recombination of the homologous chromosomes during prophase I, which in turn are prerequisites for the reductional segregation. Despite its incontestable significance for sexual reproduction and the generation of genetic variability, many questions regarding the most crucial events remain to be answered, among which is the understanding of the mechanisms that lead to specific alignment and stable pairing of the homologous chromosomes.
Induction of meiosis causes a unique redistribution of telomeres, which is highly conserved among eukaryotes. During early meiotic prophase telomeres migrate to the nuclear periphery, where they become physically attached to the nuclear envelope (NE). Once attached, they start to move along the inner NE and transiently cluster at a limited area to form a so-called chromosomal bouquet (for review see refs. 1 and 2). The bouquet configuration disappears at pachytene stage, and telomeres, while still remaining attached, redisperse around the inner surface of the NE. Because of the close temporal correlation between telomere clustering and homologue pairing as well as synapsis, it was suggested that the unique telomere dynamics during meiotic prophase are required for facilitating chromosome recognition and stable pairing of the homologs (1, 3–5). This hypothesis has been supported by previous studies in yeast. In mutants that are characterized by the absence of telomere clustering, homologue pairing and recombination are significantly delayed or disrupted (6–9). Formation of a bouquet is certainly not sufficient to ensure pairing and synapsis as evidenced by several mutants that are defective for early recombination events such as formation of double-strand breaks (for review see ref. 10). Nevertheless, the bouquet mutants demonstrated that telomere clustering is essential for proper meiotic progression, most likely by supporting recognition and alignment of the homologs. However, the molecular mechanisms that are required for attachment and movement of chromosomal ends are largely unknown.
In the past, several studies provided evidence that the NE for its part may play an important role in meiotic chromosome dynamics (for review see refs. 2, 11, and 12). Support for this came from observations that the NE of a meiotic cell differs remarkably from that of a somatic one. For instance, compared with somatic cells, in mammalian spermatocytes the nuclear lamina, i.e., the structural element of the NE that is critically involved in many fundamental cellular and developmental processes (for a recent review see ref. 13), shows significant differences regarding its composition and organization. Somatic lamins A, C, and B2 are absent. Instead, these cells express somatic lamin B1 together with lamin C2, a N-terminally shortened meiosis-specific variant of the lamin A gene (14–16). Contrary to somatic lamins, lamin C2 forms discontinuous domains at the NE of meiotic cells. Moreover, it is highly enriched at the attachment sites of the telomeres, indicating that in the immediate vicinity of these sites the NE becomes locally modulated. It was suggested that lamin C2 might lead to a local flexibility of the NE at the attachment sites that in turn enables the movement of telomeres and therefore supports pairing, recombination, and synapsis (17, 18). Beside the changes in its composition, at the attachment sites of meiotic chromosomes the NE exhibits some additional characteristic features as revealed by ultrastructural analyses (12, 19–21). The attachment of meiotic telomeres to the NE involves a terminal morphological specialization of the axial elements (AEs) of the synaptonemal complexes (SCs) called attachment plates. These structures appear as disk-shaped electron-dense plates that are intimately associated with the nuclear face of the inner nuclear membrane (INM) (21). Adjacent to the plates the nuclear membranes are more dense than in other regions. Thin fibrils arise from the plates, some of which traverse the perinuclear space and emanate into the cytoplasm, suggesting that chromosomal ends are directly interconnected with cytoplasmic structures. Such filaments bridging the nuclear membranes have been observed at zygotene and pachytene telomeres in a variety of different species, but up to the present their composition and function have remained elusive (12, 19, 20).
Recently, a new family of INM proteins has been identified that share a conserved SUN (Sad1/UNC-84 homology) domain. In somatic cells SUN-domain proteins were shown to be part of a protein complex spanning the nuclear membranes that provides a structural linkage between the cytoskeleton and nuclear components, which in turn is crucial for nuclear anchoring, migration, and positioning (22–27). In mammals two INM-localized SUN-domain proteins were identified, Sun1 and Sun2, that are derived from two different genes (28, 29). Both proteins contain a N-terminal nucleoplasmic domain that was found to bind nuclear lamins. The C terminus containing the conserved SUN domain extends into the perinuclear space, where it interacts with the KASH domain of both nesprin1 and nesprin2, which are outer nuclear membrane proteins that for their part are directly connected to the actin cytoskeleton (22–24). Thus, in somatic cells SUN-domain proteins appear to be at the very center of a nucleocytoplasmic bridge that is essential for nuclear motility. Since during meiotic prophase I a filamentous nucleocytoplasmic connection is established specifically at the attachment sites of meiotic telomeres, SUN-domain proteins may take part in forming these bridging filaments and therefore represent quite good candidates for being involved in tethering meiotic telomeres to the NE. In the present study we focused our interest on Sun2, and we report on its specific localization pattern and behavior in the context of meiotic chromosome dynamics. Our findings provide novel insights into meiotic chromosome dynamics, and we show further support for the existence of a nuclear–cytoplasmic linkage that is suggested to be required for homologue pairing and synapsis occurring in the nuclear interior.
Results
During Meiotic Prophase I Sun2 Is Selectively Located at the Attachment Sites of Meiotic Chromosomes.
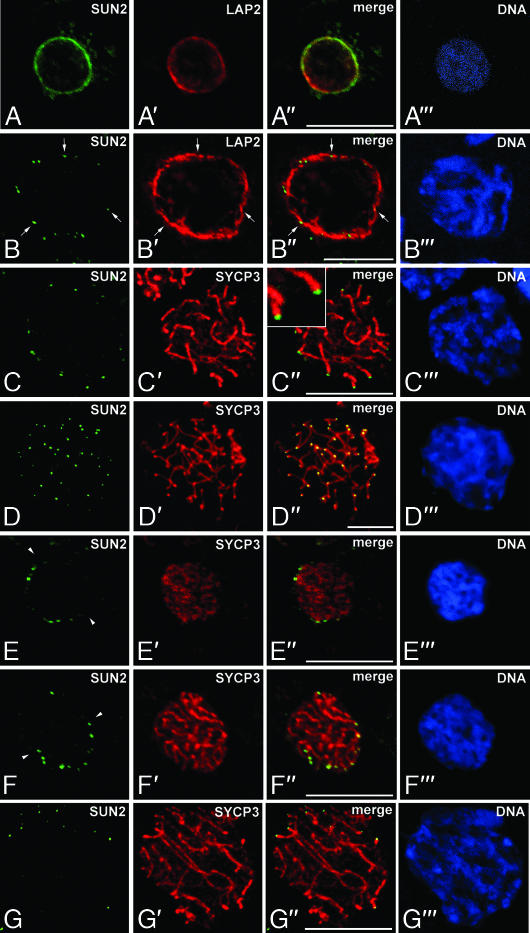
As described above, Sun2 is part of a membrane-spanning protein complex that was suggested to interconnect the nucleoskeleton with cytoplasmic actin filaments in somatic cells. Remarkably, in meiotic cells thin fibrils have been observed at the attachment sites of telomeres that arise from the attachment plates, traverse the perinuclear space, and apparently emanate into the cytoplasm (see below). Therefore, we hypothesized that Sun2 may be involved in forming these interconnecting filaments. To test this hypothesis we investigated the localization of Sun2 in the context of meiotic telomere anchoring to the NE. Thus, we made use of an affinity-purified anti-Sun2 antibody (29), which we applied to detailed immunofluorescence microscopy. Consistently with previous findings, in somatic cells of rat testes Sun2 showed a homogeneous rim-like pattern within the NE (29) (see Fig. 1A). However, in contrast to the situation in somatic interphase cells, this pattern is not maintained when cells enter meiosis. In rat as well as mouse pachytene spermatocytes we found a remarkable redistribution of Sun2. Instead of the homogeneous distribution throughout the NE, we observed a distinct dotted staining pattern with the signals nevertheless being located along the nuclear periphery (Figs. 1 B–D and 2A). As shown in Fig. 1B′ and in agreement with our previous findings (17), such a redistribution is not seen in the case of LAP2 isoforms, which represent another group of integral proteins of the INM (for review see ref. 30). In meiotic cells that are characterized by a punctured distribution of Sun2, integral LAP2 are evenly spread throughout the NE. This indicates that the punctured distribution is a specific property of Sun2 and not a general behavior of INM proteins during meiotic prophase I (Fig. 1 B–B″).
Fig. 1.
Sun2 localizes to the NE attachment sites of meiotic telomeres. (A) Sun2 (green) shows a homogeneous rim-like pattern within the NE of rat somatic testicular cells. (B–G) Localization of Sun2 in prophase I spermatocytes. Sun2 displays a punctate pattern in pachytene (B–D) as well as leptotene (E), zygotene (F), and diplotene (G) stages. (A′–G′) The same cells were costained for LAP2 or SYCP3 (red). Overlays are seen in A″–G″. All sections were labeled with the DNA-specific fluorochrome Hoechst 33258 (A‴–G‴). Some of the Sun2 spots are denoted by arrows; arrowheads indicate the midplane of nuclei. Images were taken from testis cryosections by confocal laser scanning microscopy, with the exception of D, which represents a whole-cell preparation recorded with wide-field epifluorescence microscopy. (Scale bars: 10 μm.)
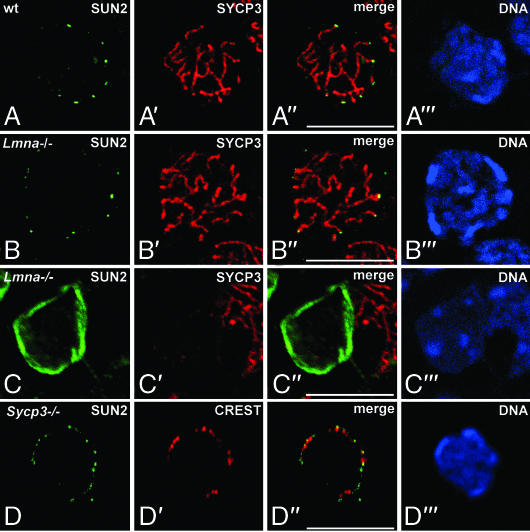
Fig. 2.
Sun2 localization is independent of A-type lamins and does not require AE/LE assembly. Shown is immunolocalization of Sun2 (green) in spermatocytes (A, B, and D) or somatic testicular cells (C) from wild-type (A), Lmna−/− (B and C), and Sycp3−/− (D) mice. Sections were costained with antibodies against SYCP3 (A–C) or alternatively with CREST serum, which labels centromeres that, in the mouse, are located at one end of each chromosome (D) (red). All sections were labeled with the DNA-specific fluorochrome Hoechst 33258 (A‴–F‴). Overlays are seen in A″–D″. (Scale bars: 10 μm.)
Due to the fact that the punctate pattern observed with the Sun2 antibody is reminiscent of the telomere distribution pattern during meiotic prophase I (see, e.g., ref. 31), we asked whether there is a spatial relationship between Sun2 signals and the attachment sites of meiotic telomeres. In double immunofluorescence analyses using Sun2 antibody together with an anti-SYCP3 antiserum, which labels the AEs/lateral elements (LEs) of the SCs (16), we observed that Sun2 signals were closely associated with the sites where AEs/LEs make intimate contact to the NE (Fig. 1 C and D). Evaluation of >600 Sun2 signals in whole-cell preparations of rat pachytene spermatocytes recorded with wide-field epifluorescence microscopy displayed a mean of 42.93 spots per nucleus (± 0.26 SD), whereby all spots were associated with ends of the bivalents (Fig. 1 D–D″). This clearly indicates that virtually every SC end is decorated with a Sun2 signal (in the rat there are 40 ends corresponding to the autosomal bivalents and three ends corresponding to the partially paired X and Y chromosomes). By closer inspection using confocal microscopy we could find Sun2 residing directly on top of the SC ends (Inset in Fig. 1C″), pointing to a close contact between Sun2 and the chromosomal ends at the NE. This observation could be corroborated by immunogold electron microscopy. In mouse pachytene spermatocytes we found strong Sun2 labeling within NE domains that are associated with the attachment sites of telomeres, whereas adjacent NE areas as well as the surrounding cytoplasm and nucleoplasm were virtually free of gold particles (Fig. 3 B–D). Remarkably, most gold particles decorated thin filaments that arise form the attachment plates (Fig. 3 B–D), suggesting that Sun2 is part of the filamentous structures that originate from the attachment plates, traverse the perinuclear space, and protrude into the cytoplasm (Fig. 3A) (19, 20).
Fig. 3.
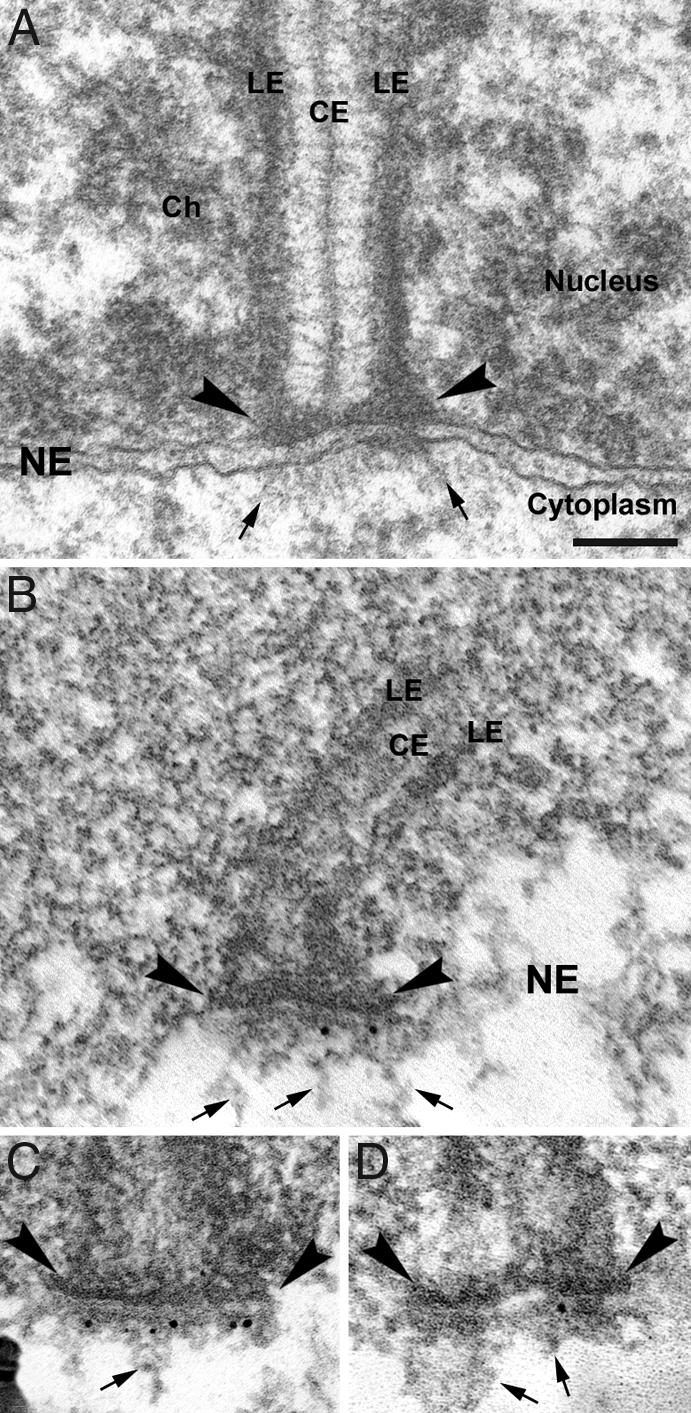
Immunogold EM examination of Sun2 in mouse spermatocytes. (A) Electron microscopic analysis of a telomere attachment site in pachytene. Ch, chromatin; CE, central element of the SC. (B–D) Electron microscopic immunolocalization of Sun2. Attachment plates are denoted by arrowheads; arrows indicate some of the cytoplasmic fibrils that are attached to the NE. (Scale bar: 100 nm.)
Sun2 Redistribution Within the NE Is Temporally Associated With Meiotic Telomere Dynamics.
One of the most striking observations during early meiotic prophase is that telomeres, once attached, start to move on the plane of the NE and congregate at one pole to form a so-called bouquet (1, 2). The observations described above clearly demonstrate that in pachytene stage of meiotic prophase I Sun2 localizes to the NE attachment sites of meiotic chromosomes. Given the role that Sun2 connects nuclear components to the actin cytoskeleton via its interaction with nesprin2 Giant (nesp2G) (22, 23), and because of the fact that SUN-domain proteins are critically involved in nuclear motions (25, 32), we propose that Sun2 might also be crucially involved in telomere movement during bouquet formation. If so, Sun2 localization has to be closely related to the attachment sites of the chromosomal ends during the whole process when telomeres move actively along the plane of the NE. To address this issue we started to investigate the specific behavior of Sun2 in the context of meiotic telomere dynamics. Remarkably, as early as leptotene stage when AEs become formed and telomeres, having attached to the NE, actively congregate at one pole (for review see ref. 1), Sun2 already displayed a discontinuous, rather punctate pattern along the nuclear periphery (Fig. 1E). This pattern reflects the above-described situation in pachytene cells in principle (Figs. 1 B–D and 2A). However, in contrast to the pattern in pachytene and concomitant with the specific congregation of telomeres during early meiotic prophase, Sun2-containing domains were not found evenly distributed throughout the NE, but they were clearly polarized to one half of the nucleus (Fig. 1E). A similar behavior of Sun2 could be observed in early zygotene stage as well, i.e., Sun2-containing spots were restricted to one side of the cell nucleus (Fig. 1F). Moreover, during the latter stage Sun2 could be found exclusively residing at the sites where the now-established AEs are in contact with the NE (Fig. 1F). Finally, after resolution of the bouquet configuration, from pachytene to diplotene stage Sun2 domains, although being directly associated with the chromosomal ends, are randomly distributed throughout the NE (Figs. 1 B–D and G, 2A, and 3). Taken together, these results provide strong evidence that the close contact between Sun2-containing structures and the attachment sites is established early in meiotic prophase and maintained throughout the process of dynamic movement of meiotic telomeres.
Sun2 Localization in Spermatocytes Is Independent of A-Type Lamins and Does Not Require AE/LE Assembly.
In a previous study we demonstrated that meiotic cells express a single A-type lamin isoform, namely the meiosis-specific lamin C2, which shows a remarkable behavior as it is discontinuously distributed within the NE, highly enriched at the attachment sites of meiotic chromosomes (16, 17). Because it was shown that somatic A-type lamins interact with Sun2 (23), Sun2 localization to the attachment plates might be a direct consequence of lamin C2 enrichment at these sites. To test this possibility we investigated Sun2 distribution in mice lacking A-type lamins (33). Previously it was shown that in mammalian somatic cells the absence of A-type lamins has only weak consequence for Sun2 distribution, if any (see refs. 22–24 and 34). Consistently, we found that Sun2 distribution in somatic cells of Lmna−/− testes is not affected, i.e., Sun2 showed a continuous rim-like pattern throughout the NE (Fig. 2C). Similar to the situation in somatic cells, within meiotic prophase cells the absence of the meiotic A-type lamin, lamin C2, has virtually no effect on Sun2 localization as well. We could find that Sun2 was still distributed in a punctured pattern whereby the close association with the attachment sites of meiotic chromosomes was maintained (Fig. 2B). Thus, our results clearly demonstrate that meiosis-specific redistribution of Sun2 and its association with chromosomal ends are independent of lamin C2 enrichment at these sites.
Due to the fact that AEs/LEs were shown to be in continuity with the filaments emanating into the cytoplasm (for review see ref. 1), we asked whether the formation of AEs might be responsible for the recruitment of Sun2 to the attachment sites of meiotic chromosomes. Therefore, we analyzed Sun2 distribution in spermatocytes of mice lacking SYCP3, a protein that represents the major structural component of the AEs/LEs (21, 35). In Sycp3−/− males cells enter meiosis, but meiotic progression is impaired, leading to apoptotic cell death in zygotene stage of meiotic prophase I (35). Furthermore, ultrastructural analysis of Sycp3−/− spermatocytes revealed that SC formation is disrupted with AE/LEs being absent (21). In analyzing Sun2 localization in male Sycp3−/− meiocytes, we could find that Sun2 distribution is indistinguishable between normal spermatocytes and Sycp3−/− spermatocytes lacking AE/LEs. Similar to the wild-type situation, Sun2 is concentrated in small-sized spots residing at the attachment sites of telomeres (Fig. 2 A and D). Thus, this result provides strong evidence that Sun2 does not require AE assembly for its localization to the attachment sites of chromosomal ends, which is consistent with our previous observation that the fibrillar material extending into the cytoplasm can still be found associated with the attachment plates in Sycp3−/− spermatocytes (21).
Discussion
In previous studies in yeast it has been demonstrated that meiotic telomere clustering is mandatory for meiotic progression (6–9). However, up to the present the molecular requirements for NE attachment and congregation of chromosomal ends during meiotic prophase I remained largely unknown. Progress in this issue came from recent observations in fission yeast. In meiosis of Schizosaccharomyces pombe, telomeres become clustered at the NE adjacent to the spindle pole body (SPB) (36). This clustering is mediated by the SPB-associated protein Sad1p, which is thought to function as a transmembrane linker involved in interconnecting meiotic telomeres with the SPB (37, 38). Sad1p belongs to a novel group of INM proteins, termed SUN-domain proteins, that are found across all eukaryotes. SUN-domain proteins were shown to be required for nuclear anchoring, migration, and positioning in somatic cells. Thus, they hold a key position at the nuclear–cytoplasmic interface, where they appear to be part of a transmembrane complex that links nuclear components to the cytoskeleton (for recent overview see refs. 27 and 39). Recently, Tomita and Cooper (40) hypothesized that, like Sad1p in S. pombe, SUN-domain proteins may be essentially involved in mammalian bouquet formation as well, i.e., in NE tethering and directed movement of telomeres. However, until now the expression and behavior of SUN-domain proteins in mammalian meiotic cells remained speculative.
In our present study we could demonstrate that with Sun2 at least one of the mammalian SUN-domain proteins is expressed during mammalian male meiosis. Sun2 was previously identified as an INM protein that in somatic cells is homogeneously distributed throughout the NE (29). However, within meiotic cells it shows a remarkably different behavior, as we could find Sun2 exclusively residing at the attachment sites of telomeres. This specific Sun2–telomere association appears as early as leptotene stage and is maintained throughout the dynamic movement of chromosomal ends. Based on these results we suggest that within meiotic cells Sun2 represents a constitutive component of the meiotic attachment complex that structurally links telomeres to the NE. Consistently, by EM immunolocalization we found that Sun2 is concentrated at membrane-spanning fibrillar complexes that connect to telomeres along the inner nuclear surface, traverse the perinuclear space, and radiate into the cytoplasm. Such telomere-associated filaments have been observed in a variety of different species, but up to the present their function and composition have remained elusive (12, 19, 20). Hence, our results now provide a hint for the molecular nature of these filaments, with Sun2 appearing to be a central component. Moreover, our findings suggest that the mammalian meiotic cell recruits NE components also expressed in somatic cells for tethering telomeres to the NE, and they provide clear support for the model described above whereby in mammals, with Sun2, a SUN-domain protein appears to be implicated in forming a meiotic telomere attachment complex that in turn might be functional in telomere clustering (ref. 40 and see below).
Recently, Chikashige et al. (38) presented compelling evidence that in fission yeast SUN-domain protein Sad1p holds a key position in meiotic telomere clustering. During meiotic prophase, the nucleoplasmic domain of Sad1p interacts with Bqt1p, which in turn binds telomere-associated Rap1p in the presence of Bqt2p, resulting in a structural linkage between telomeres and the NE (38, 41). On the other hand, the C-terminal conserved SUN domain of Sad1p, which most likely is located within the perinuclear space, was shown to interact with the KASH domain of Kms1p, an outer nuclear membrane protein that directly connects to the SPB (32, 42–44). Thus, Sad1p appears to hold a central position in a linker complex that interconnects meiotic telomeres with cytoskeletal structures. Our findings in the present study suggest a similar function for Sun2 in mammalian meiotic cells: (i) during meiotic prophase I, Sun2 is strictly associated with telomeres, which resembles Sad1p behavior in S. pombe; (ii) during bouquet formation telomeres that are linked to the Sun2-containing attachment complex congregate near the centrosome, the animal equivalent of the yeast SPB (2, 21); and (iii) Sun2 takes part in forming fibrillar structures that provide a nucleocytoplasmic bridge, hence interconnecting telomeres with cytoplasmic structures. Interestingly, it was recently found that Sun2 interacts with the KASH domain of outer nuclear membrane protein nesp2G to form a bridge across the NE (23), thus reflecting the Sad1p-Kms1p NE bridging complex formed in fission yeast. Taken together, the overt correlation between S. pombe Sad1p and mammalian Sun2 suggests that telomere anchoring and clustering follow a general mechanism that is conserved upon eukaryotes. In agreement with this assumption, in Caenorhabditis elegans SUN-1/matefin, a germ line-specific SUN-domain protein, was found to be essential for germ cell viability; however, up to the present it is completely unknown whether SUN-1/matefin is functionally involved in telomere attachment and movement (45). In mammals the INM of somatic cells contains a second SUN-domain protein, namely Sun1, which was suggested to have overlapping functions with Sun2 (23, 28). Up to now, it is not known whether Sun1 is expressed in the germ line as well. Thus, this point has to be clarified before a putative involvement of Sun1 in meiotic events can be postulated.
Because Sun2 was shown to interact with nesp2G and given that nesp2G connects to the actin cytoskeleton (23, 46) we propose a model describing NE attachment and movement of meiotic telomeres in mammals (Fig. 4). Upon entry into meiosis telomeres move toward the nuclear periphery to contact the NE. Then, Sun2 initially dispersed in the INM becomes recruited to telomeres by a yet-unknown mechanism, forming a mechanically stable telomere–INM connection. As we have shown here, this specific Sun2 redistribution is independent of AE formation as well as of lamin C2 enrichment at the attachment sites and thus requires other proteins, perhaps yet unidentified orthologues of S. pombe Bqt1p and Bqt2p that were shown to recruit Sad1p, hence tethering telomeres to the NE (38, 40, 41). Finally, through its interaction with nesp2G, Sun2 could link telomeres directly to the actin cytoskeleton. Thus, our model would favor a cytoplasmic actin-based mechanism that generates the forces needed for meiotic telomere dynamics, including their congregation during bouquet formation. Notably, in budding yeast inhibition of actin polymerization leads to disruption of telomere clustering, indicating the existence of such a mechanism; however, it is not known whether cytoplasmic or nuclear actin is involved in these processes (47). Also unknown is whether actin-dependent bouquet formation applies to other species like mammals. Beside a putative role in telomere clustering, linkage of telomeres to the cytoskeleton via the Sun2-containing attachment complex might contribute to more basic nuclear events occurring during early meiotic prophase I, such as the vigorous nuclear rotations and chromosomal oscillations observed during leptotene/zygotene in the rat that are thought to be required for alignment of the homologs (48). The movements of the entire nucleus could be induced by simply pulling at individual and/or groups of telomeres with respect to the centrosome, hence placing the affected telomeres near the centrosome (see also ref. 31). However, further analyses are required to clarify these important questions.
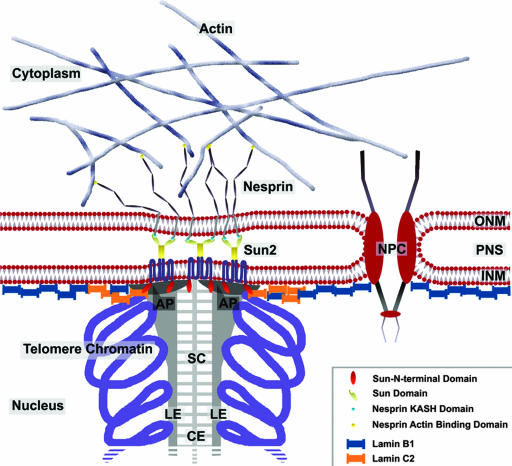
Fig. 4.
Model for the organization of the NE at the attachment sites of meiotic chromosomes. In meiotic cells Sun2 is exclusively located at the attachment plates, where it is involved in tethering meiotic telomeres to the NE. Within the perinuclear space its C-terminal SUN domain binds to the KASH domain of nesp2G, thus forming a stable fibrillar complex bridging both nuclear membranes. Because nesp2G interacts with cytoplasmic actin via its actin binding domain, the formation of such a complex would link telomeres to the actin cytoskeleton. This model is consistent with recent findings that identified an actin-dependent mechanism for bouquet formation. AP, attachment plate; CE, central element of the SC; NPC, nuclear pore complex; PNS, perinuclear space; ONM, outer nuclear membrane.
Materials and Methods
Animals and Tissue Preparation.
Male Wistar rats (39–42 days old) were obtained from Charles River (Sulzfeld, Germany). Wild-type, Lmna−/−, and Sycp3−/− C57BL/6 mice used in this study were described previously (33, 35). Animals were anesthetized and killed with CO2. Testes were immediately resected, shock-frozen for 5 min in 2-methylbutane (Carl Roth, Karlsruhe, Germany), cooled with liquid nitrogen to −140°C, and stored at −70°C in the presence of 2-methylbutane until further use. For immunofluorescence microscopy, 6-μm-thin cryosections were made with a Reichert-Jung Kryostat 2800 Frigocut-E (Leica, Bensheim, Germany).
For testis suspensions, fresh rat testes were transferred in a glass Petri dish containing ice-cold PBS (140 mM NaCl/2.6 mM KCl/6.4 mM Na2HPO4/1.4 mM KH2PO4, pH 7.4). After removing the tunica albuginea, seminiferous tubules were cut into small pieces and carefully resuspended. To obtain a fraction of singularized cells enriched with spermatogenic stages, the suspension was filtered through a nylon filter (mesh size 30 μm). The cells were washed once with ice-cold PBS, resuspended in a suitable volume of PBS, and transferred to Superfrost Plus coverslips (Menzel, Braunschweig, Germany). Attached cells were then fixed as described below.
Antibodies.
The following primary antibodies were used: affinity-purified rabbit anti-Sun2 antiserum (29), affinity-purified guinea pig anti-SYCP3 (16), mAb 13d4 against LAP2 (49), and human CREST serum (gift from Georg Krohne University of Würzburg, Germany). Secondary antibodies conjugated to Cy2 or Texas red were purchased from Dianova (Hamburg, Germany).
Immunofluorescence Microscopy.
Cryosections or cells from testes suspensions were fixed for 5 min in PBS containing 1% formaldehyde and permeabilized for 10 min in PBS containing 0.1% Triton X-100. After blocking for 1 h with PBT (PBS containing 1.5% BSA and 0.1% Tween 20), slides were incubated simultaneously with selected primary antibodies for 20 min. Then specimens were washed twice in PBS, and bound antibodies were detected with corresponding secondary antibodies. DNA was visualized with the fluorochrome Hoechst 33258 (Hoechst, Frankfurt, Germany). After a final washing in PBS, the cells were embedded in Mowiol.
Epifluorescence analyses were done with an Axiophot stereo fluorescence microscope (Zeiss, Jena, Germany) equipped with a Pixelfly digital system (PCO Computer Optics, Kelheim, Germany). For confocal microscopy, we used a Leica TCS-SP2 AOBS confocal laser scanning microscope. Settings were identical for all scanning procedures. Single optical sections were taken with a HCX PL APO lbd.Bl ×63/1.4 oil-immersion objective with zoom 4, a pinhole setting at 114.78 μm, and ×4 accumulations. All fluorochromes were scanned sequentially. Cy2 was excited with a wavelength of 488 nm, Texas red with 561 nm, and Hoechst 33258 with 405 nm. Series of three sections with a step size of 0.122 μm were taken. The maximum projection algorithm of the Leica software was then used to calculate information sectored in different ranks to a two-dimensional projection with a defined thickness of 0.244 μm. The obtained data were processed by using Photoshop (Adobe Systems, San Jose, CA).
EM.
Mouse testes were fixed in 2.5% cacodylate buffered glutaraldehyde (1 h, 4°C) and then postfixed with 1% osmium tetroxide (1 h). After overnight staining with 0.5% uranyl acetate, testes were dehydrated in ethanol series and embedded in Epon. Ultrathin sections of Epon-embedded testes were double-stained with uranyl acetate and lead citrate according to standard procedures.
For immunogold localization of Sun2, mouse testis cryosections were processed similar to the immunofluorescence protocol with the exception that Fluoro Nanogold− anti-rabbit (Nanoprobes, New York, NY) was used as secondary antibody. After washing in PBS, the cryosections were refixed for 10 min in 2% glutaraldehyde in PBS and washed several times in distilled water. For silver enhancement, the Aurion R-GENT SE-EM Kit (Aurion, Wageningen, The Netherlands) was used. Sections were dehydrated in ethanol series and embedded in Epon according to standard protocols. Ultrathin sections were stained as described above. Micrographs were obtained with a Zeiss EM-10 electron microscope.
Acknowledgments
We thank Elisabeth Meyer-Natus for excellent technical assistance. This work was supported by Deutsche Forschungsgemeinschaft Grants Al 1090/1–1 (to M.A.) and Be 1168/6–2 (to R.B.) and Graduate School 1048 of the University of Würzburg Grant A4 (to R.B.). D.H. was supported by a grant from the Muscular Dystrophy Association, and C.H. was supported by the Swedish Research Council.
Abbreviations
- SPB
spindle pole body
- NE
nuclear envelope
- SC
synaptonemal complex
- AE
axial element
- LE
lateral element
- INM
inner nuclear membrane.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Zickler D, Kleckner N. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 2.Scherthan H. Nat Rev Mol Cell Biol. 2001;2:621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- 3.Loidl J. Genome. 1990;33:759–778. doi: 10.1139/g90-115. [DOI] [PubMed] [Google Scholar]
- 4.Dernburg AF, Sedat JW, Cande WZ, Bass HW. In: Telomeres. Blackburn EH, Greider CW, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1995. pp. 295–338. [Google Scholar]
- 5.Niwa O, Shimanuki M, Miki F. EMBO J. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper JP, Watanabe Y, Nurse P. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- 7.Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- 8.Trelles-Sticken E, Dresser ME, Scherthan H. J Cell Biol. 2000;151:95–106. doi: 10.1083/jcb.151.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y. Dev Cell. 2004;6:329–341. doi: 10.1016/s1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 10.Lichten M. Curr Biol. 2001;11:R253–R256. doi: 10.1016/s0960-9822(01)00131-2. [DOI] [PubMed] [Google Scholar]
- 11.Wilson EB. The Cell in Development and Heredity. New York: Macmillan; 1925. [Google Scholar]
- 12.Holm PB. Carlsberg Res Commun. 1977;42:103–152. [Google Scholar]
- 13.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 14.Smith A, Benavente R. Differentiation. 1992;52:55–60. doi: 10.1111/j.1432-0436.1992.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 15.Furukawa K, Inagaki H, Hotta Y. Exp Cell Res. 1994;212:426–430. doi: 10.1006/excr.1994.1164. [DOI] [PubMed] [Google Scholar]
- 16.Alsheimer M, Benavente R. Exp Cell Res. 1996;228:181–188. doi: 10.1006/excr.1996.0315. [DOI] [PubMed] [Google Scholar]
- 17.Alsheimer M, von Glasenapp E, Hock R, Benavente R. Mol Biol Cell. 1999;10:1235–1245. doi: 10.1091/mbc.10.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alsheimer M, Liebe B, Sewell L, Stewart CL, Scherthan H, Benavente R. J Cell Sci. 2004;117:1173–1178. doi: 10.1242/jcs.00975. [DOI] [PubMed] [Google Scholar]
- 19.Esponda P, Gimenez-Martin G. Chromosoma. 1972;38:405–417. doi: 10.1007/BF00320159. [DOI] [PubMed] [Google Scholar]
- 20.Benavente R, Alsheimer M, von Glasenapp E. Chromosomes Today. 2004;14:119–126. [Google Scholar]
- 21.Liebe B, Alsheimer M, Höög C, Benavente R, Scherthan H. Mol Biol Cell. 2004;15:827–837. doi: 10.1091/mbc.E03-07-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 23.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee KK, Starr D, Cohen M, Liu J, Han M, Wilson KL, Gruenbaum Y. Mol Biol Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starr DA, Han M. J Cell Sci. 2003;116:211–216. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- 27.Tzur YB, Wilson KL, Gruenbaum Y. Nat Rev Mol Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- 28.Malone CJ, Fixsen WD, Horvitz HR, Han M. Development (Cambridge, UK) 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 29.Hodzic DM, Yeater DB, Bengtsson L, Otto H, Stahl PD. J Biol Chem. 2004;279:25805–25812. doi: 10.1074/jbc.M313157200. [DOI] [PubMed] [Google Scholar]
- 30.Dechat T, Vlcek S, Foisner R. J Struct Biol. 2000;129:335–345. doi: 10.1006/jsbi.2000.4212. [DOI] [PubMed] [Google Scholar]
- 31.Scherthan H, Weich S, Schwegler H, Heyting C, Harle M, Cremer T. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starr DA, Fischer JA. BioEssays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasan S, Güttinger S, Mühlhäusser P, Anderegg F, Bürgler S, Kutay U. FEBS Lett. 2006;580:1263–1268. doi: 10.1016/j.febslet.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 35.Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Höög C. Mol Cell. 2000;5:73–83. doi: 10.1016/s1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
- 36.Chikashige Y, Ding DQ, Imai Y, Yamamoto M, Haraguchi T, Hiraoka Y. EMBO J. 1997;16:193–202. doi: 10.1093/emboj/16.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagan I, Yanagida M. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 39.Worman HJ, Gundersen GG. Trends Cell Biol. 2006;16:67–69. doi: 10.1016/j.tcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Tomita K, Cooper JP. Cell. 2006;125:19–21. doi: 10.1016/j.cell.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 41.Tang X, Jin Y, Cande WZ. J Cell Biol. 2006;173:845–851. doi: 10.1083/jcb.200602152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, Niwa O. Mol Gen Genet. 1997;254:238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- 43.Miki F, Okazaki K, Shimanuki M, Yamamoto A, Hiraoka Y, Niwa O. Mol Biol Cell. 2002;13:930–946. doi: 10.1091/mbc.01-11-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miki F, Kurabayashi A, Tange Y, Okazaki K, Shimanuki M, Niwa O. Mol Genet Genomics. 2004;270:449–461. doi: 10.1007/s00438-003-0938-8. [DOI] [PubMed] [Google Scholar]
- 45.Fridkin A, Mills E, Margalit A, Neufeld E, Lee KK, Feinstein N, Cohen M, Wilson KL, Gruenbaum Y. Proc Natl Acad Sci USA. 2004;101:6987–6992. doi: 10.1073/pnas.0307880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, Karakesisoglou I, Korenbaum E. Exp Cell Res. 2004;295:330–339. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H. J Cell Biol. 2005;170:213–223. doi: 10.1083/jcb.200501042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parvinen M, Söderström KO. Nature. 1976;260:534–535. doi: 10.1038/260534a0. [DOI] [PubMed] [Google Scholar]
- 49.Alsheimer M, Fecher E, Benavente R. J Cell Sci. 1998;111:2227–2234. doi: 10.1242/jcs.111.15.2227. [DOI] [PubMed] [Google Scholar]