Abstract
Observations of numerous dramatic and presumably adaptive phenotypic modifications during human evolution prompt the common belief that more genes have undergone positive Darwinian selection in the human lineage than in the chimpanzee lineage since their evolutionary divergence 6–7 million years ago. Here, we test this hypothesis by analyzing nearly 14,000 genes of humans and chimps. To ensure an accurate and unbiased comparison, we select a proper outgroup, avoid sequencing errors, and verify statistical methods. Our results show that the number of positively selected genes is substantially smaller in humans than in chimps, despite a generally higher nonsynonymous substitution rate in humans. These observations are explainable by the reduced efficacy of natural selection in humans because of their smaller long-term effective population size but refute the anthropocentric view that a grand enhancement in Darwinian selection underlies human origins. Although human and chimp positively selected genes have different molecular functions and participate in different biological processes, the differences do not ostensibly correspond to the widely assumed adaptations of these species, suggesting how little is currently known about which traits have been under positive selection. Our analysis of the identified positively selected genes lends support to the association between human Mendelian diseases and past adaptations but provides no evidence for either the chromosomal speciation hypothesis or the widespread brain-gene acceleration hypothesis of human origins.
Keywords: molecular evolution, population size
Although humans and their closest living relatives, chimpanzees, are highly similar at the genomic level (1–6), they differ in many morphological, physiological, and behavioral traits (7). Phenotypically, modern humans appear to have changed considerably more than modern chimps from their common ancestors (7–10). Many of these evolutionary modifications in humans, such as the origins of bipedalism, speech and language, and other high-order cognitive functions, are widely thought to be adaptive (11–13). These observations led to a common belief that more genes underwent positive Darwinian selection in the human lineage than in the chimpanzee lineage. Indeed, there are more reports of positively selected genes (PSGs) in humans than in chimps (12, 13). Nonetheless, this difference may be largely due to a lack of study in chimps. To avoid such a bias, one could identify and compare all PSGs from the human and chimp genomes. Positive selection acting on a protein-coding gene may be detected by various population genetic and molecular evolutionary methods that use intraspecific polymorphism data, interspecific divergence data, or a combination of the two (14–16). However, because of the paucity of polymorphism data from chimps, a fair comparison between the two species would have to be limited to the divergence data. Such data can be used to estimate the ratio of nonsynonymous to synonymous substitution rates (ω). An ω value significantly >1 indicates the action of positive selection, whereas an ω significantly <1 indicates negative (or purifying) selection. Using this approach, two earlier studies (17, 18) pioneered the identification of human and chimp PSGs at the genomic scale, although no comparison was made between the numbers of human and chimp PSGs. In fact, the studies' results would be unsuitable for the comparison, owing to a number of deficiencies. First, both studies used the mouse as an outgroup, to distinguish between human-specific and chimp-specific nucleotide substitutions, because of the unavailability of genome sequences from any closer outgroups at that time. Because mouse is distantly related to human and chimp, this practice introduces errors. Second, one of the studies (17) was based on less reliable statistical methods and assumptions (19), whereas the other (18) used the draft chimp genome sequence (1) known to contain many more errors than the finished human genome sequence (20, 21). Because the majority of genes in a genome have ω < 1, and sequencing errors have an expected ω of 1, the errors inflate ω and the false detection of positive selection. In this work, we first design a protocol to rectify these problems and then use the protocol to identify and compare human and chimp PSGs. Our results show substantively more PSGs in chimpanzee evolution than in human evolution.
Results and Discussion
Study Design.
To compare human and chimp PSGs impartially, we made three improvements in the design of the analysis. First, to distinguish nucleotide substitutions that occurred in the human lineage from those that occurred in the chimp lineage, we used the macaque monkey as the outgroup. Because the divergence time between the macaque and human/chimp is approximately a quarter of that between the mouse and human/chimp (22–24), the reliability of our analysis was expected to increase significantly. Gene orthology determination and sequence alignment among the more closely related human–chimp–macaque gene trios is also more reliable than among human–chimp–mouse trios.
Second, we applied an improved branch-site likelihood method for identifying PSGs (25), which has been shown by computer simulation to produce good results even when some of the assumptions are violated (25). The method requires that the branches in a phylogenetic tree be separated into foreground and background branches a priori, where foreground branches are tested for the occurrence of positive selection. The method assumes that two classes of codons, either negatively selected (class 0) or neutral (class 1), exist in the background branches. This null model is compared with an alternative model in which a proportion of class 0 codons, and the same proportion of class 1 codons, become positively selected in the foreground branches. Positive selection in foreground branches is inferred for a gene if the likelihood of the observation of the gene sequences is significantly higher under the alternative model than under the null model. To further verify the suitability of the method in the present context, we conducted additional computer simulations specifically designed to mimic the evolution of human, chimp, and macaque genes [see supporting information (SI) Materials and Methods]. Our results showed that the false-positive rate is acceptable, except for extreme conditions when it slightly exceeds the nominal rate (see SI Tables 3 and 4).
Third, we used high-quality nucleotides from the 4× coverage chimp genome sequence to allow a fair comparison with the human sequence. Briefly, we assembled alignments of orthologous genes from human, chimp, and macaque, using publicly available genome sequences and annotations (see Materials and Methods). We then eliminated alignment gaps and those codons in which one or more chimp nucleotides did not meet our quality cutoff. Three different cutoffs, low (Q0), intermediate (Q10), and high (Q20), were used to generate three data sets. After removing alignments of <100 codons, we obtained our final data sets, containing 13,955, 13,924, and 13,888 genes for the Q0, Q10, and Q20 cutoffs, respectively (see SI Table 5). Even the smallest data set (Q20) has a total alignment length of 17,995,887 nucleotides, with a mean alignment length of 432 codons (standard deviation, 339 codons). All three data sets contain >50% of genes in a primate genome and cover >50% of all protein-coding regions in the genome. Using parsimony, we inferred the numbers of nucleotide substitutions in human and chimp lineages since their split. This inference is expected to be accurate because the three species studied here are closely related. We found that the ratio of the number of synonymous substitutions in the chimp lineage to that in the human lineage is r = 1.103 ± 0.009, 1.020 ± 0.008, and 0.985 ± 0.008 for the Q0, Q10, and Q20 data sets, respectively. Assuming identical mutation rates per year between human and chimp lineages, r is expected to be 1. If the mutation rate is 3% lower in humans than in chimps, as has been suggested (26), r is expected to be 1.03. Given these considerations, Q0 data, as used in an earlier study (18), are apparently unsuitable because the observed r is significantly higher than the expectation. To make our conclusion more conservative, we use Q20 rather than Q10 data. Two other independent assessments of the chimp genome sequence, one of which evaluated it against 172 kb of finished chimp sequence, also recommended the use of Q20 data for comparison with the human genome sequence (1, 20). Most importantly, the number of synonymous substitutions is already 1.5% lower in chimp than in human when the cutoff of Q20 is used, suggesting that the chimp sequencing errors become negligible at this quality level. The comparison between the 172 kb of draft and finished chimp sequences also showed that the use of cutoffs higher than Q20 is undesirable because many chimp-specific nucleotide changes tend to be lost (20). This is probably because polymorphic sites in the chimp individual that was sequenced, estimated to be 0.1% of all sites (1), tend to have lower qualities than homozygous sites. These polymorphic sites are excluded progressively as one increases the quality cutoff, which hampers a fair comparison with human because the human genome sequence contains polymorphic sites (1). Note that errors in the macaque genome sequence should not affect our analysis because the probability for a macaque error to occur at a nucleotide position where human and chimp differ is small. Even when such rare events occur, they should affect human and chimp equally and hence would not bias our results. Our human–chimp comparison should not be biased by indel errors because the detection of positive selection does not use indel information.
More PSGs in Chimp Evolution than in Human Evolution.
Applying the likelihood method and a P value of 5% for statistical significance (25), we identified 154 genes that were under positive selection in the human lineage (Table 1 and SI Table 6) and 233 in the chimp lineage (see SI Table 7). Thus, chimps have 51% more PSGs than humans have. As expected, the excess of chimp PSGs is even greater (157%) should the Q10 data be used (SI Table 5). The proportion of PSGs in the genome is 233/13,888 = 1.7% for the chimp lineage, significantly greater than that (154/13,888 = 1.1%) for the human lineage (P < 10−4, χ2 test). Because 13,888 statistical tests were conducted for each lineage, it is necessary to control for multiple testing. Under Bonferroni correction, two human genes and 21 chimp genes remain statistically significant (see SI Table 8). With use of a false discovery rate of 5%, the same two human genes and 59 chimp genes are significant (SI Table 8). The proportion of PSGs in the chimp genome remains significantly greater than that in the human genome (P < 10−4, χ2 test), even after the multiple-testing corrections (Table 1).
Table 1.
Genic positive selection in human and chimp lineages since their split
| Comparison | Chimp | Human | Chimp/human ratio | P, %* |
|---|---|---|---|---|
| No. of genes analyzed | 13,888 | 13,888 | 1 | > 5 |
| No. of PSGs | 233 | 154 | 1.51 | < 0.01 |
| No. of PSGs after Bonferroni correction | 21 | 2 | 10.5 | < 0.01 |
| No. of PSGs at 5% false discovery rate | 59 | 2 | 29.5 | < 0.01 |
| No. of synonymous changes in all genes | 29,644 | 30,083 | 0.985 | > 5 |
| No. of nonsynonymous changes in all genes | 17,701 | 19,000 | 0.932 | < 0.01 |
| Mean ω of all genes | 0.245 | 0.259 | 0.946 | < 0.01 |
| Mean ω of 13,508 non-PSGs | 0.238 | 0.252 | 0.944 | < 0.01 |
*Probability that the ratio = 1.
To further confirm our results, we analyzed the recently released 6× chimp genome assembly for the 233 chimp PSGs identified above. We found that 212 (or 91%) of them still show significant signals of positive selection (see SI Materials and Methods). Hence, when this new data set is used, chimps have 38% more PSGs than humans have (P = 0.002, χ2 test). Note that this is a conservative estimate because we did not consider non-PSGs from the 4× sequence that may become PSGs in the 6× sequence. Such incidences are possible because potentially more nucleotides per gene can be analyzed in the 6× sequence, leading to improved statistical power in identifying PSGs. Additionally, 4× and 6× sequences may differ at polymorphic sites, which can affect the outcome of PSG identification when the number of substitutions is small. Because the analyses of the 4× and 6× sequences both indicate substantially more PSGs in chimps than in humans, and because the 6× assembly is preliminary and unpublished, our subsequent analyses use the PSGs identified from the Q20 data of the 4× assembly. An additional reason for using the 4× assembly is the finding of a number of cases in which the 4× assembly is apparently more accurate than the 6× assembly (see SI Materials and Methods).
We found that the mean ω of all genes is 0.259 ± 0.002 in the human lineage, significantly larger than that (0.245 ± 0.002) in the chimp lineage (P < 10−4; Table 1). For the common set of 13,508 non-PSGs between humans and chimps, the mean ω is also significantly larger in human (0.252 ± 0.002) than in chimp (0.238 ± 0.002) (P < 10−4; Table 1). Because the majority of non-PSGs are under negative selection, as reflected in their low ω values, the above results indicate stronger negative selection in chimps than in humans. Multiple-population genetic data indicate that the long-term effective population size of humans (in the last 1–2 million years) is several-fold smaller than that of chimps and than that of the human–chimp common ancestor (2, 27–34). A recent analysis of 1 million base pairs of Neanderthal nuclear DNA also suggested that the common ancestor of modern humans and Neanderthals had a small effective population size (35). It is thus probable that the effective population size is greater in the chimp lineage than in the human lineage for a large portion of the divergence time between the two lineages. Population genetic theories (36) predict that both positive and negative selection are more effective in large populations than in small populations. Our observation that chimps have more PSGs but fewer nonsynonymous substitutions in non-PSGs than humans is consistent with these predictions.
Computer simulations showed that the branch-site likelihood method cannot detect all PSGs. Rather, the detection rate increases as the ω of background branches increases (see SI Table 9). If the overall strength of positive selection is weaker in humans than in chimps because of smaller populations of humans than chimps, a higher average background ω is required for PSGs to be detectable in humans than in chimps. We found that in the macaque branch of the human–chimp–macaque tree, the mean ω for all genes is 0.226 ± 0.001. For human PSGs, the mean ω in the macaque branch is 0.294 ± 0.007, significantly greater than the mean ω in the macaque branch (0.278 ± 0.005) for chimp PSGs (P < 0.05). Hence, these observations are consistent with the simulation result and further support the notion that positive selection was weaker in the human lineage than in the chimp lineage. Theories also predict that recombination can increase the efficacy of selection (37). Indeed, PSGs tend to be located in high-recombination regions, although this effect is significant in chimps (P = 0.041) but not in humans (P = 0.32) (see SI Fig. 4), probably as a result of a difference in statistical power caused by the difference in the number of PSGs in the two species.
Similarities and Differences Between Human and Chimp PSGs.
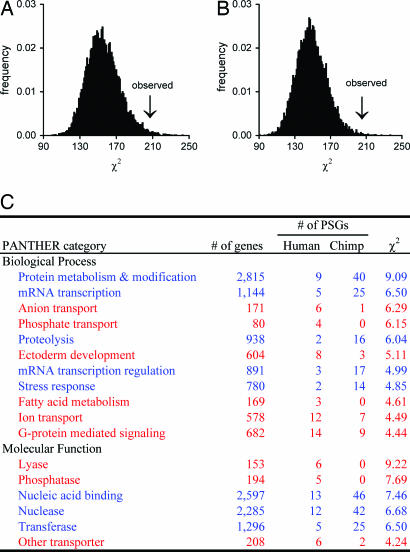
It has been claimed that genes of certain functional categories, such as olfaction and nuclear transport, were more frequently under positive selection in humans than in chimps, based on the ranking of all genes by their P values in the likelihood test of positive selection (17). Because genes with reduced negative selection also tend to have low P values (although unlikely to be as low as 0.05), such ranks potentially mix genes under positive selection with those under reduced negative selection. We took a more rigorous approach by limiting our analysis to the PSGs we detected. We found that seven genes are shared between the human and chimp PSGs (see SI Table 10), significantly greater than expected by chance (2.6; P < 0.02, binomial test), suggesting the presence of some common targets of positive selection in the two lineages. We classified all PSGs into biological process groups and molecular function groups, as defined in the PANTHER database (38). A randomization test indicated a significant difference in distribution of human and chimp nonoverlapping PSGs among biological process groups (Fig. 1A) and among molecular function groups (Fig. 1B). Those groups showing the greatest differences between the two species are listed in Fig. 1C. Interestingly, however, the majority of these groups (e.g., protein metabolism and modification, anion transport, phosphate transport, and lyase) do not correspond to the widely assumed adaptive phenotypic differences between humans and chimps (e.g., neurogenesis), suggesting the existence of yet-to-be-recognized adaptive phenotypic differences between the two species. We did not detect several previously reported PSGs that control brain size or cognitive functions (39–42) because previous identifications of these PSGs were based on a comparison of polymorphism and divergence data, whereas only divergence data are used here. As mentioned above, due to the paucity of chimp polymorphism data, any fair genome-wide comparison of human and chimp PSGs would have to be limited to divergence data at this time.
Fig. 1.
Functional differences between human and chimp unshared PSGs. (A and B) Human and chimp PSGs show a significantly larger difference in distribution across biological process groups (A) and molecular function groups (B) than by chance (P = 0.84% and 0.26%, respectively, one-tail randomization test). The bars show the frequency distribution of the χ2 values in 10,000 random divisions of the 373 unshared PSGs into 147 human PSGs and 226 chimp PSGs. The arrow indicates the observed χ2. Here, the randomization test is superior to the standard χ2 test because the functional groups are not independent of one another, and a single gene may belong to more than one group. Similar results are obtained when the seven shared PSGs are included. (C) Biological process and molecular function groups that show the greatest differences between human and chimp unshared PSGs, as ranked by individual χ2 values. Shown are the groups that each contribute at least 2% of the total χ2 of all groups. Groups with a higher frequency of human PSGs than chimp PSGs are shown in red; those with a higher frequency of chimp PSGs than human PSGs are shown in blue.
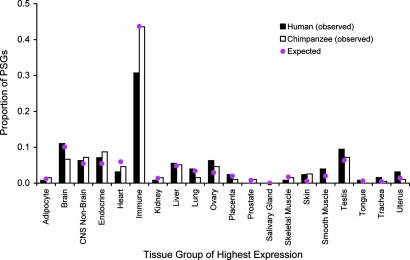
Using microarray data of human gene expression, we found that human and chimp PSGs are not significantly different in their distributions between the categories of tissue-specific genes and nonspecific genes (P > 0.5, χ2 test; and see SI Table 11). On examining the peak-expression tissue group for each gene (see SI Table 12), we again found no significant difference in the overall tissue distribution between human and chimp PSGs (Fig. 2). Notably, 14 (11%) human PSGs and 13 (6.7%) chimp PSGs have peak expressions in one or more parts of the brain, but the difference is not statistically significant (χ2 = 1.74, P = 0.19). On the contrary, for the central nervous system outside of the brain, human (8) has fewer PSGs than chimp (14) (χ2 = 0.09, P = 0.77). These findings are consistent with recent comparative genomic analyses (21, 43) and do not support more positive selection in humans than in chimps in regard to nervous system genes (44).
Fig. 2.
Frequency distribution of human and chimp PSGs across 20 peak-expression tissue groups. The overall difference between the distributions of the two species is not statistically significant (χ2 = 23.8, df = 19, P = 0.21). Only smooth muscle (χ2 = 7.7, P = 0.0056) shows a significant difference in proportion of PSGs between the two species, but the significance disappears when multiple testing is corrected for. Pink dots show the expected distribution of PSGs when there is no enrichment of PSGs in any tissue groups.
Genome-wide identification of human and chimp PSGs helps to test several evolutionary hypotheses. First, it has been argued that PSGs are more likely than non-PSGs to underlie known Mendelian disorders in humans because the current environment of humans is considerably different from that of earlier hominins and previous adaptive changes may become deleterious today (45, 46). Our data provide some support for this hypothesis. We found that 9.7% of human PSGs are disease-associated (see SI Table 13), significantly greater than that (6.1%) among the non-PSGs examined (P = 0.049; Table 2). Consistent with the prediction of the above hypothesis, the fraction of human PSGs underlying human diseases is greater than the fraction of chimp PSGs underlying human diseases (P = 0.044, Fisher's exact test). Furthermore, as expected, there is no significant difference in the proportion of genes underlying human diseases between chimp PSGs and non-PSGs (P = 0.23; Table 2).
Table 2.
Association of PSGs with human disease
| Gene type | No. of disease genes | No. of nondisease genes | Proportion of disease genes | P value* |
|---|---|---|---|---|
| Human | ||||
| PSGs | 15 | 139 | 0.097 | 0.049 |
| Non-PSGs | 832 | 12,902 | 0.061 | |
| Chimp | ||||
| PSGs | 11 | 222 | 0.047 | 0.230 |
| Non-PSGs | 838 | 12,817 | 0.061 | |
*Based on Fisher's exact test of no difference in proportion of disease genes among PSGs and non-PSGs.
Second, a recently proposed chromosomal speciation hypothesis asserts that chromosomal rearrangements facilitated the formation of reproductive isolation between populations that eventually led to modern humans and chimps (47). Several predictions of this hypothesis have been examined, with mixed results (47–53). One interesting prediction that has not been explicitly tested is that PSGs are preferentially located on rearranged chromosomes because such chromosomes are less likely to be introgressed after the initial separation of two lineages during speciation and thus are more likely to accumulate genes subject to local adaptations (47). Nine chromosomes (1, 4, 5, 9, 12, 15–18) contain pericentric inversions between humans and chimpanzees, and human chromosome 2 resulted from a fusion of two acrocentric chromosomes common to other great apes (54). These chromosomes are considered as rearranged chromosomes, whereas the other chromosomes are considered as colinear chromosomes. Our data, however, do not support the chromosomal speciation hypothesis for humans and chimps because the proportion of PSGs is even slightly lower on the rearranged chromosomes than on the colinear chromosomes in both the human and chimp lineages (Fig. 3).
Fig. 3.
Distributions of human and chimp PSGs among chromosomes. Contrary to the chromosomal speciation hypothesis, PSGs are slightly less abundant on rearranged chromosomes than on colinear chromosomes (P = 0.10 and 0.055 for the human and chimp lineages, respectively, χ2 test). The human chromosome numbers are used. The expected number of PSGs on each chromosome is calculated under the assumption that the probability of a gene being targeted by positive selection is not affected by the chromosome on which it is located.
Implications.
In summary, our genome-wide analysis showed that substantively more genes underwent positive selection in the chimp lineage than in the human lineage since their split. Although our study could not, and did not, detect all PSGs in human and chimp evolution, particularly those beneficial alleles that are yet to be fixed (12, 55, 56), it provides an unbiased comparison between the two lineages. Our results have several implications. First, in sharp contrast to common belief, there were more adaptive genetic changes during chimp evolution than during human evolution. Without doubt, we tend to notice and study human-specific phenotypes more than chimp-specific phenotypes, which may have resulted in the prevailing anthropocentric view on human origins. Our finding suggests more unidentified phenotypic adaptations in chimps than in humans. Although human and chimp PSGs show different distributions among molecular functions and biological processes, the differences do not ostensibly correspond to the widely assumed adaptive phenotypes in humans. Assuming that our statistical method is equally powerful in detecting PSGs of different biological processes, the finding shows how little is currently known about which traits are adaptive. Second, although the influence of population size on negative selection has been well documented (57, 58), the present study also demonstrates the impact of population size on positive selection at the genomic scale. Interestingly, even during human evolution when so many apparently dramatic phenotypic changes took place, the laws of population genetics prevailed. This being said, it is important to recognize that other factors also influence the frequency of positive selection. For example, it is possible that as a result of the relatively recent out-of-Africa migration of modern humans, many new advantageous alleles are yet to be fixed and thus are not identified by our method. Our results thus apply largely to completed selective sweeps in human and chimp lineages. Furthermore, a higher level of polymorphism in chimps than in humans could potentially lead to more predicted PSGs in chimps than in humans. But because some chimp polymorphic sites have been removed in the Q20 data, and because the number of synonymous changes is already 1.5% lower in chimp than in human for the Q20 data, we do not think this factor has affected our result. At any rate, it will be interesting to examine in other species whether the number of PSGs is strongly dependent on population size. Third, although we only studied positive selection on protein sequence changes and did not address positive selection on gene expression evolution (59, 60), a recent comparison between hominoids and murids in regard to regulatory sequence conservation showed that a reduction in population size also lowers the efficiency of natural selection on gene expression changes (61). Most interestingly, when conserved noncoding sequences, which often regulate gene expression, are examined, chimps show more incidences of accelerated evolution than humans do (62). Thus, it is likely that the total number of genes for which either the regulatory or coding regions underwent adaptive selection is also greater in chimp evolution than in human evolution.
Materials and Methods
Compilation of Human–Chimp–Macaque Gene Sequence Data.
Protein and corresponding nucleotide sequences of all predicted genes in the human, chimpanzee, and macaque genome sequences were downloaded from Ensembl (version 36, December 2005; www.ensembl.org). To identify orthologous genes, human protein sequences (n = 33,869) were used to conduct BLASTP searches (63) against the chimpanzee (n = 39,648) and macaque (n = 31,371) protein sequences. Reciprocal searches were performed using the chimpanzee and macaque proteins to query the human proteins. A total of 19,422 proteins with reciprocal best hits in both human/chimpanzee and human/macaque searches were retained for further analysis. Alignment of the human–chimpanzee–macaque orthologous proteins was performed using CLUSTALW version 1.83 (64). DNA sequence alignments were obtained by following the protein sequence alignments. Alignments containing <100 amino acids (n = 1,291) were discarded. Lineage-specific nucleotide substitutions were identified by parsimony as described in the next paragraph. Review of several alignments that had exceptionally high proportions of human- or chimpanzee-specific changes revealed that the apparent high level of lineage-specific changes resulted from incorrect alignment or nonorthology. Therefore, alignments containing >10% human- or chimpanzee-specific amino acid or nucleotide changes or >30% macaque-specific changes (n = 161) were discarded from analysis. Finally, each protein was assigned to a gene on the basis of the Ensembl annotation, and the protein sequence with the longest amino acid alignment was retained for each gene, resulting in the alignments of human, chimpanzee, and macaque sequences of 13,955 distinct genes (Q0 data set). Chimp genome sequence quality information was downloaded from the University of California, Santa Cruz, Bioinformatics web site (http://hgdownload.cse.ucsc.edu/goldenPath/panTro1/bigZips/chromQuals.zip). The average chimp quality score in the Q0 data set is 48.9526. The 13,955 alignments were scanned for codons in which one or more nucleotides had a chimp quality score <20 (i.e., an error rate of 1%) (65), and these codons were removed from the alignments. After this procedure, 67 alignments contained <100 amino acids and were removed from analysis. The remaining 13,888 alignments constituted the Q20 data set. The average chimp quality score in the Q20 data set is 49.3443. We similarly obtained the Q10 data set (i.e., a maximum error rate of 10% at any nucleotide site), comprising 13,925 genes. The average chimp quality score in the Q10 data set is 49.0695.
We applied the parsimony principle to identify human-specific and chimpanzee-specific substitutions, using the macaque as the outgroup. The numbers of synonymous (s) and nonsynonymous (n) nucleotide substitutions in the human and chimp lineages were counted. Using the modified Nei–Gojobori method (66) with a transition/transversion ratio of 2 (67), we estimated that the total number of nonsynonymous sites in the 13,888 genes of the Q20 data set was N = 12,783,034 and the total number of synonymous sites was S = 5,215,415, with their ratio being N/S = 2.45. Thus, for a set of genes, the mean nonsynonymous-to-synonymous rate ratio in a lineage can be computed by (n/s)/(N/S) = (n/s)/2.45 = 0.41n/s.
Identification of PSGs.
Using PAML (68), we applied the improved branch-site test of positive selection (test 2 in ref. 25) to identify putative cases of positive selection in the human lineage among the 13,888 genes (Q20 data). When we tested positive selection in the human lineage, the human branch was designated as the foreground branch and the chimp and macaque branches were designated as background branches. We tested positive selection in the chimp lineage similarly. Bonferroni correction (69) and a false discovery rate of 5% (70) were used to correct for multiple testing. We also analyzed the Q10 data set and identified 165 human and 424 chimp PSGs.
Use of the 6× Chimp Genome Assembly.
Our analysis of chimp PSGs using the 6× chimp genome assembly is described in SI Materials and Methods.
Comparison Between Human and Chimp PSGs.
Using the PANTHER database (38), we classified the 13,888 genes into different groups of biological processes and molecular functions. Note that these groups are not mutually exclusive and that a gene may belong to more than one group. To examine the distributional difference between human and chimp PSGs across PANTHER groups, we defined the statistic
 |
where xi and yi are the number of human and chimp PSGs, respectively, in PANTHER group i, and n is the total number of PANTHER groups. Because of the nonindependence of PANTHER groups, we used a randomization test to examine whether the observed χ2 was significantly different from the random expectation. Briefly, we randomly divided the 373 unshared human and chimp PSGs into 147 human PSGs and 226 chimp PSGs and computed χ2 by using the above formula. We repeated this procedure 10,000 times to obtain the null distribution of χ2, to which the observed χ2 is compared. Similar results were obtained when the seven shared PSGs were included.
The microarray gene expression data in 79 human tissues, and the nucleotide sequences for 27,215 probe sets on the array, were obtained from ref. 71. The probe set sequences were used to perform BLAST searches against the human coding sequences annotated by Ensembl. Probe sets that matched to multiple genes were considered ambiguous and were discarded. A total of 26,195 probe sets were unambiguously matched to 16,605 distinct genes. Among these 16,605 genes, 12,099 genes, including 127 human PSGs and 195 chimp PSGs, can be found in our Q20 data set. For genes that matched to more than one probe set, the expression levels measured by different probe sets were averaged for each tissue replicate. Two replicates were available for each tissue, and these were averaged to determine the expression level of a gene in each tissue. Identification of tissue specificity can be obscured if multiple tissues with very similar expression profiles are used (72). We therefore consolidated multiple tissues representing similar areas into tissue groups and took the highest expression level from any tissue in a group as the single representative expression level score for the tissue group (21) (SI Table 12). Expression levels in pathogenic tissues were not considered. A gene was considered to be tissue-specific if the expression level in the highest tissue group was greater than or equal to twice the expression level in the second highest tissue group. The 3,299 genes meeting this criterion are said to be tissue-specific in the highest tissue. We also considered the peak expression tissue for every gene.
Online Mendelian Inheritance in Man (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM) was used to identify all genes known to be involved in human Mendelian diseases. The chromosomal locations of all genes were obtained from Ensembl.
Recombination rate data for 1-megabase segments of human chromosomes were downloaded from University of California, Santa Cruz (http://genome.ucsc.edu/cgi-bin/hgTables). A recombination rate was assigned to each gene in the Q20 data set, based on the 1-megabase segment in which the midpoint of the gene lies. Of the 13,888 genes analyzed here, 13,714 are found in regions of known recombination rates. Among these 13,714 genes, 152 human and 228 chimp PSGs have available recombination rates. We then computed the mean recombination rate of the 152 human PSGs. To estimate the expected value of this mean, we randomly picked 152 genes from 13,714 genes and computed the mean. This procedure was repeated 10,000 times to estimate the probability that the observed mean is greater than the expected mean. The same procedure was applied to chimp PSGs, under the assumption that the recombination rate of a chimp gene is the same as for its human ortholog, which is probably correct for the majority of genes at the 1-megabase scale (73).
Performance of the Improved Branch-Site Likelihood Method.
The performance of the improved branch-site likelihood method is described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Soochin Cho, Wendy Grus, Ondrej Podlaha, Xiaoxia Wang, and especially Masatoshi Nei, for valuable suggestions; three anonymous reviewers for constructive comments; and the Washington University School of Medicine Genome Sequencing Center and Baylor College of Medicine Human Genome Sequencing Center for making available before publication the 6× chimp genome assembly and macaque genome assembly, respectively. This work was supported by the University of Michigan and by National Institutes of Health Grant GM67030 (to J.Z.).
Abbreviation
- PSG
positively selected gene.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701705104/DC1.
References
- 1.Chimpanzee Sequencing and Analysis Consortium. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 2.Chen FC, Li WH. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebersberger I, Metzler D, Schwarz C, Paabo S. Am J Hum Genet. 2002;70:1490–1497. doi: 10.1086/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britten RJ. Proc Natl Acad Sci USA. 2002;99:13633–13635. doi: 10.1073/pnas.172510699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wildman DE, Uddin M, Liu G, Grossman LI, Goodman M. Proc Natl Acad Sci USA. 2003;100:7181–7188. doi: 10.1073/pnas.1232172100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe H, Fujiyama A, Hattori M, Taylor TD, Toyoda A, Kuroki Y, Noguchi H, BenKahla A, Lehrach H, Sudbrak R, et al. Nature. 2004;429:382–388. doi: 10.1038/nature02564. [DOI] [PubMed] [Google Scholar]
- 7.Varki A, Altheide TK. Genome Res. 2005;15:1746–1758. doi: 10.1101/gr.3737405. [DOI] [PubMed] [Google Scholar]
- 8.Pilbeam D. Mol Phylogenet Evol. 1996;5:155–168. doi: 10.1006/mpev.1996.0010. [DOI] [PubMed] [Google Scholar]
- 9.Olson MV, Varki A. Nat Rev Genet. 2003;4:20–28. doi: 10.1038/nrg981. [DOI] [PubMed] [Google Scholar]
- 10.King MC, Wilson AC. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 11.Darwin C. The Descent of Man and Selection in Relation to Sex. New York: D. Appleton; 1871. [Google Scholar]
- 12.Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 13.Vallender EJ, Lahn BT. Hum Mol Genet. 2004;13(Spec No 2):R245–R254. doi: 10.1093/hmg/ddh253. [DOI] [PubMed] [Google Scholar]
- 14.Li W. Molecular Evolution. Sunderland, MA: Sinauer; 1997. pp. 237–267. [Google Scholar]
- 15.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ Press; 2000. pp. 51–71. [Google Scholar]
- 16.Nielsen R. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 17.Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, et al. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- 18.Arbiza L, Dopazo J, Dopazo H. PLoS Comput Biol. 2006;2:e38. doi: 10.1371/journal.pcbi.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J. Mol Biol Evol. 2004;21:1332–1339. doi: 10.1093/molbev/msh117. [DOI] [PubMed] [Google Scholar]
- 20.Taudien S, Ebersberger I, Glockner G, Platzer M. Trends Genet. 2006;22:122–125. doi: 10.1016/j.tig.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Shi P, Bakewell MA, Zhang J. Trends Genet. 2006;22:608–613. doi: 10.1016/j.tig.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Glazko GV, Nei M. Mol Biol Evol. 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- 23.Hedges SB. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 24.Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Nielsen R, Yang Z. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 26.Elango N, Thomas JW, Yi SV. Proc Natl Acad Sci USA. 2006;103:1370–1375. doi: 10.1073/pnas.0510716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stone AC, Griffiths RC, Zegura SL, Hammer MF. Proc Natl Acad Sci USA. 2002;99:43–48. doi: 10.1073/pnas.012364999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaessmann H, Wiebe V, Paabo S. Science. 1999;286:1159–1162. doi: 10.1126/science.286.5442.1159. [DOI] [PubMed] [Google Scholar]
- 29.Fischer A, Wiebe V, Paabo S, Przeworski M. Mol Biol Evol. 2004;21:799–808. doi: 10.1093/molbev/msh083. [DOI] [PubMed] [Google Scholar]
- 30.Ferris SD, Brown WM, Davidson WS, Wilson AC. Proc Natl Acad Sci USA. 1981;78:6319–6323. doi: 10.1073/pnas.78.10.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaessmann H, Wiebe V, Weiss G, Paabo S. Nat Genet. 2001;27:155–156. doi: 10.1038/84773. [DOI] [PubMed] [Google Scholar]
- 32.Ruvolo M. Mol Biol Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- 33.Wall JD. Genetics. 2003;163:395–404. doi: 10.1093/genetics/163.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahata N, Satta Y, Klein J. Theor Popul Biol. 1995;48:198–221. doi: 10.1006/tpbi.1995.1026. [DOI] [PubMed] [Google Scholar]
- 35.Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, Paunovic M, Paabo S. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 36.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge Univ Press; 1983. pp. 34–54. [Google Scholar]
- 37.Hill WG, Robertson A. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- 38.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Webb DM, Podlaha O. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J. Genetics. 2003;165:2063–2070. doi: 10.1093/genetics/165.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 42.Evans PD, Anderson JR, Vallender EJ, Gilbert SL, Malcom CM, Dorus S, Lahn BT. Hum Mol Genet. 2004;13:489–494. doi: 10.1093/hmg/ddh055. [DOI] [PubMed] [Google Scholar]
- 43.Wang HY, Chien HC, Osada N, Hashimoto K, Sugano S, Gojobori T, Chou CK, Tsai SF, Wu CI, Shen CK. PLoS Biol. 2006;5:e13. doi: 10.1371/journal.pbio.0050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 45.Young JH, Chang YP, Kim JD, Chretien JP, Klag MJ, Levine MA, Ruff CB, Wang NY, Chakravarti A. PLoS Genet. 2005;1:e82. doi: 10.1371/journal.pgen.0010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neel JV. Am J Hum Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 47.Navarro A, Barton NH. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Wang X, Podlaha O. Genome Res. 2004;14:845–851. doi: 10.1101/gr.1891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu J, Li WH, Wu CI. Science. 2003;302:988. doi: 10.1126/science.302.5647.988a. author reply 988. [DOI] [PubMed] [Google Scholar]
- 50.Osada N, Wu CI. Genetics. 2005;169:259–264. doi: 10.1534/genetics.104.029231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Innan H, Watanabe H. Mol Biol Evol. 2006;23:1040–1047. doi: 10.1093/molbev/msj109. [DOI] [PubMed] [Google Scholar]
- 52.Marques-Bonet T, Caceres M, Bertranpetit J, Preuss TM, Thomas JW, Navarro A. Trends Genet. 2004;20:524–529. doi: 10.1016/j.tig.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 53.Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- 54.Yunis JJ, Prakash O. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- 55.Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 56.Wang X, Grus WE, Zhang J. PLoS Biol. 2006;4:e52. doi: 10.1371/journal.pbio.0040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eyre-Walker A, Keightley PD. Nature. 1999;397:344–347. doi: 10.1038/16915. [DOI] [PubMed] [Google Scholar]
- 58.Ohta T. J Mol Evol. 1995;40:56–63. doi: 10.1007/BF00166595. [DOI] [PubMed] [Google Scholar]
- 59.Khaitovich P, Tang K, Franz H, Kelso J, Hellmann I, Enard W, Lachmann M, Paabo S. Curr Biol. 2006;16:R356–R358. doi: 10.1016/j.cub.2006.03.082. [DOI] [PubMed] [Google Scholar]
- 60.Rockman MV, Hahn MW, Soranzo N, Zimprich F, Goldstein DB, Wray GA. PLoS Biol. 2005;3:e387. doi: 10.1371/journal.pbio.0030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keightley PD, Lercher MJ, Eyre-Walker A. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prabhakar S, Noonan JP, Paabo S, Rubin EM. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 63.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ewing B, Hillier L, Wendl MC, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 66.Zhang J, Rosenberg HF, Nei M. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosenberg MS, Subramanian S, Kumar S. Mol Biol Evol. 2003;20:988–993. doi: 10.1093/molbev/msg113. [DOI] [PubMed] [Google Scholar]
- 68.Yang Z. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 69.Sokal RR, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Research. New York: Freeman; 1995. p. 240. [Google Scholar]
- 70.Storey JD, Tibshirani R. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winter EE, Goodstadt L, Ponting CP. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Serre D, Nadon R, Hudson TJ. Genome Res. 2005;15:1547–1552. doi: 10.1101/gr.4211905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.