Abstract
Biochemically, the syntrophic bacteria constitute the missing link in our understanding of anaerobic flow of carbon in the biosphere. The completed genome sequence of Syntrophus aciditrophicus SB, a model fatty acid- and aromatic acid-degrading syntrophic bacterium, provides a glimpse of the composition and architecture of the electron transfer and energy-transducing systems needed to exist on marginal energy economies of a syntrophic lifestyle. The genome contains 3,179,300 base pairs and 3,169 genes where 1,618 genes were assigned putative functions. Metabolic reconstruction of the gene inventory revealed that most biosynthetic pathways of a typical Gram-negative microbe were present. A distinctive feature of syntrophic metabolism is the need for reverse electron transport; the presence of a unique Rnf-type ion-translocating electron transfer complex, menaquinone, and membrane-bound Fe-S proteins with associated heterodisulfide reductase domains suggests mechanisms to accomplish this task. Previously undescribed approaches to degrade fatty and aromatic acids, including multiple AMP-forming CoA ligases and acyl-CoA synthetases seem to be present as ways to form and dissipate ion gradients by using a sodium-based energy strategy. Thus, S. aciditrophicus, although nutritionally self-sufficient, seems to be a syntrophic specialist with limited fermentative and respiratory metabolism. Genomic analysis confirms the S. aciditrophicus metabolic and regulatory commitment to a nonconventional mode of life compared with our prevailing understanding of microbiology.
Keywords: anaerobic food chains, syntrophic metabolism, fatty acid and benzoate utilization
Syntrophus aciditrophicus is a rod-shaped, Gram-negative bacterium that degrades fatty acids, benzoate, cyclohexane carboxylate, cyclohex-1-ene carboxylate, and crotonate in coculture with hydrogen/formate-using methanogens or sulfate reducers (1, 2). Phylogenetically, S. aciditrophicus is a member of the delta subdivision of the Proteobacteria along with sulfate reducers and Geobacter species. It is representative of a broad class of syntrophic bacteria that have an essential role in the anaerobic recycling of organic matter to methane and carbon dioxide, along with the fermentative and methanogenic organisms. Whereas the carbon and energy yielding pathways of the latter two microbial groups are well understood, extremely little is known about the metabolism of syntrophs because so few strains are available in pure culture. They reproduce slowly (generation times of 24–50 h), with low yield, and require symbiotic partners to survive and express their syntrophic lifestyle. Biochemically, they constitute the missing link in our understanding of anaerobic flow of carbon in the biosphere.
A wide range of organic compounds including alcohols, fatty acids, aromatic acids, organic acids such as lactate and glycolate, many amino acids, sugars, and hydrocarbons including methane are degraded syntrophically under anaerobic conditions (3–9). The degradation of these compounds is thermodynamically unfavorable unless hydrogen and/or formate are maintained at low levels by hydrogen/formate-consuming microorganisms such as the methanogens (Table 1). This thermodynamically based interaction between microbial species is termed syntrophy. The ubiquity of syntrophic metabolism in many anoxic environments emphasizes that metabolic cooperation among microbial species is required for complete destruction of organic matter where the catalytic unit is a microbial consortium composed of two or more microbial species. Many syntrophic associations are highly organized, multicellular structures where the partners are in close physical proximity to each other (6, 10, 11). However, little is known about the molecular mechanisms involved in the formation and maintenance of these catalytic units. Even under optimal growth conditions, free energy changes are close to equilibrium (12–14) and the available free energy must be shared by the different organisms (4). Truly, syntrophy represents an extreme existence, that of a marginal energy economy.
Table 1.
Reactions involved in syntrophic metabolism
| Reactions | ΔG°′*, kJ/mol | ΔG′†, kJ/mol |
|---|---|---|
| Methanogenic H2 consumption | ||
| 4 H2 + HCO3− + H+ → CH4 + 3 H2O | −135.6 | −15.8 |
| Syntrophic metabolism | ||
| Propionate− + 3 H2O → Ace− + HCO3− + H+ + 3 H2 | +76.1 | −16.9 |
| Butyrate− + 2 H2O → 2 Ace− + H+ + 2 H2 | +48.6 | −39.2 |
| Benzoate− + 7 H2O → 3 Ace− + HCO3− + 3 H+ + 3 H2 | +70.1 | −68.5 |
*Calculated from the data in Thauer et al. (46) with the free energy of formation for benzoate given in ref. 47.
†Calculated on the basis of the following conditions observed in methanogenic ecosystems: partial pressures of H2 of 1 Pa and of CH4 of 50 kPa, 50 mM bicarbonate, and the concentrations of the substrates and acetate (Ace−) at 0.1 mM each.
A distinctive feature of syntrophic metabolism is the need for reverse electron transfer because critical oxidation-reduction reactions result in a negative ΔE′, i.e., a thermodynamically unfavorable direction. The production of hydrogen (E′ of −261 mV at 1 Pa H2) or formate (E′ of −258 mV at 1 μM formate) (4) from electrons generated in the oxidation of acyl-CoA intermediates to their respective enoyl-CoA intermediates [E′ of −10 mV; (15)] represents a considerable energy barrier (i.e., ΔE′ ∼ −250 mV). Thus, the membrane components involved in the generation and use of ion gradients are predicted to be key features of syntrophic metabolism.
The genome sequence of S. aciditrophicus provides the first glimpse of the genetic program directing energy conservation and reverse electron transport, which are essential features of the syntrophic lifestyle. S. aciditrophicus is a specialist with limited fermentative and respiratory metabolism, but the genome sequence suggests interesting mechanisms for the metabolism of aromatic and alicyclic compound and ATP synthesis from acetyl-CoA. The reliance of S. aciditrophicus on syntrophic fatty and aromatic acid metabolism delineates it from almost all organisms.
Results and Discussion
General Features.
The genome of S. aciditrophicus contains 3,179,300 base pairs in a single circular chromosome of an average G + C content of 51.46%. Of the 3,169 genes identified, annotated functions were assigned to 1,618 (51%) whereas 421 (13%) had no database match (Table 2). Although unusual for a bacterial species, the genome contains a single ribosomal RNA operon with one gene each for 5S, 16S, and 23S rRNA. With a normal complement of 48 tRNAs, this single rRNA cluster is compatible with the relatively long generation times of the organism (≈24–50 h). The chromosome has a well defined GC skew with inflection points at identifiable oriC- and termination sites [supporting information (SI) Fig. 3]. Two regions of foreign DNA were identified on the basis of deviation from the 51% average GC composition. One of these regions corresponds to a Mu-like prophage of ≈31 kb in size extending from 530,000 base pairs to ≈561,000 base pairs (SI Fig. 3). The second anomalous region includes a potential conjugative element of 19 kb at 1,742,000 base pairs to ≈1,761,000 base pairs. In addition, 37 IS-like structures were identified scattered throughout the genome (SI Table 3). These IS-like sequences fall into eleven groups based on sequence homology, and many seem to represent previously undescribed IS variants based on the absence of homologous sequences in the current IS database (ISFinder; http://www-is.biotoul.fr/).
Table 2.
| Category | Amount |
|---|---|
| DNA contigs | 1 |
| DNA sequenced | 3,179,300 bp |
| Coding sequence | 2,812,658 bp |
| G + C content | 51.46% |
| Transfer RNAs | 48 |
| Ribosomal RNA operons | 1 |
| Pseudogenes | 0 |
| ORFs‡, total | 3,169 (100%) |
| ORFs with assigned function | 1,618 (51%) |
| ORFs without assigned function | 1,551 (49%) |
| ORFs without function, no database match | 421 (13.3%) |
| ORFs in chromosomal clusters | 1,493 (47.1) |
| ORFs in paralog clusters | 786 (24.8%) |
| ORFs with Pfam matches | 1,852 (58.4%) |
| ORFs with COG matches | 1,986 (62.8%) |
*According to ERGO database (www.integratedgenomics.com).
†The protein similarity cutoff score used was 1.0e−05.
‡ORFs, open-reading frames, coding regions
Comparison with Other Genomes.
The S. aciditrophicus genome is the smallest of any of the δ-proteobacteria genomes sequenced thus far. When S. aciditrophicus ORFs were compared pair-wise to individual microbial genomes, best reciprocal BLAST hits revealed closest associations to the δ-proteobacteria: Geobacter metallireducens (1,118 reciprocal gene hits), Pelobacter carbinolicus (1,131), and Geobacter sulfurreducens (1,099) (SI Fig. 4). Approximately 1,100 genes are similar and are well conserved across the 25 Gram-negative species shown. The remaining genes (≈2,000) represent a recently discovered complement within the S. aciditrophicus genome.
In another comparison, the best BLAST hit to any microbial gene was determined (SI Fig. 5) and showed only 374, 250, and 224 closest hits to the genomes of G. metallireducens, G. sulfurreducens, and Pelobacter carbinolicus, respectively. Notably three archaeal genomes, Methanosarcina mazei, Methanospirillum hungatei, and Methanosarcina acetivorans gave 14–31 best BLAST hits each, suggesting the possibility of lateral gene transfer events from these potential syntrophic partners. A significant fraction of these genes have energy/Co-A related functions or are transposases.
Central Metabolism.
The 1,618 functionally assigned S. aciditrophicus genes were organized into pathways, revealing that most biosynthetic pathways considered essential for viability of a typical Gram-negative organism seem to be present (e.g., biosynthesis of amino acids, purines, pyrimidines, and many cofactors). The undetected steps in amino acid formation include genes encoding homoserine kinase (threonine biosynthesis), threonine deaminase (conversion of threonine to isoleucine), 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase or acetyldiaminopimelate aminotransferase (lysine biosynthesis), phosphoribosyl-ATP diphosphatase, and histidinol phosphatase (histidine biosynthesis). Also undetected were reactions for acetyl CoA carboxylase and enoyl-ACP reductase (NADH) (lipid biosynthesis), those needed to convert uroporphyrinogen to protoporphyrin IX (heme biosynthesis), and thymidylate synthase (DNA biosynthesis).
S. aciditrophicus has the gluconeogenic and pentose phosphate pathway genes to synthesize hexose- and pentose-phosphates from acetyl-CoA with pyruvate and/or phosphoenol pyruvate as intermediates. NADPH synthesis may occur by the combined activity of pyruvate carboxylase, malate dehydrogenase, and malic enzyme (Fig. 1) (16). The organism may also be able to synthesize and use glycogen and, although it has a complete Embden–Meyerhof–Parnas pathway for glycogen degradation, it lacks any apparent sugar uptake systems. However, S. aciditrophicus is unable to grow if either glucose or starch is provided in the medium. The genome contains four genes for alpha amylase (SYN0880, SYN2854, SYN0295, and SYN0687). S. aciditrophicus hydrolyzes starch but does not grow with it as a sole substrate (unpublished data).
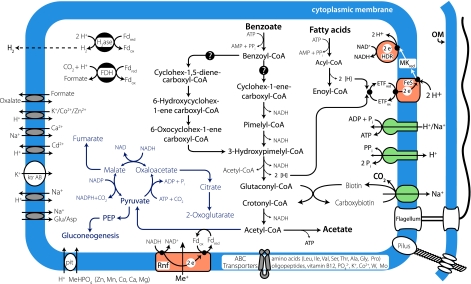
Fig. 1.
Overview of the metabolism of S. aciditrophicus. Pathways for aromatic and fatty acid degradation are shown in black, key biosynthetic steps are shown in blue, primary ion translocation pumps are in green, and electron transfer complexes are in orange. Membrane complexes involved in energy transduction and electron transport are arranged along the cellular membrane (CM). OM, outer membrane; pit, phosphate inorganic transport system; ETF, electron transfer flavoprotein; Me+, ion translocated by the RnF-like complex.
The S. aciditrophicus genome exhibits limited metabolic options to use any carbohydrates, amino acids, or purines/pyrimidines for energy conservation. Consistent with the limited metabolic capability, the genome does not seem to encode any recognizable carbohydrate transporters for mono-, di-, or larger oligosaccharides. Identifiable transporters for amino acids and small peptides are extremely limited: only genes encoding ABC transporters for the uptake of the branched-chain amino acids were identified along with transporters for inorganic nutrients (Fig. 1). This lack of general transport capability is mirrored in the paucity of degradation pathways for the above substrates. The reliance of S. aciditrophicus on syntrophic fatty and aromatic acid metabolism delineates it from other organisms.
S. aciditrophicus has an incomplete tricarboxylic acid cycle (Fig. 1): genes for malate dehydrogenase, isocitrate dehydrogenase, 2-oxyglutarate synthase, aconitase, and fumarase were identified. The genes for fumarate reductase, succinate dehydrogenase, citrate CoA transferase, citrate CoA lyase, and the si types of citrate synthase were not detected. 2-Oxoglutarate could be made by the oxidative arm of the TCA cycle if S. aciditrophicus contains a re type of citrate synthase as found in Desulfovibrio species (17, 18), which are also members of the δ-proteobacteria. The primary structure for re-citrate synthase is not known so it is not possible at this time to determine whether the gene is present in S. aciditrophicus.
Syntrophic Substrate Metabolism.
Enzymatic and metabolite tracking studies have revealed a number of the steps involved in the syntrophic benzoate and fatty acid oxidation (2, 19, 20). However, a number of questions still remain regarding the mechanism of aromatic ring reduction and cleavage.
The S. aciditrophicus genome contains multiple genes for AMP-forming ligases (Eq. 1 below) predicted to activate aromatic compounds (SYN1638, SYN2417, SYN2896, and SYN2898) and fatty acids (SYN1145, SYN2640, SYN2643, and SYN3128) (SI Table 4), consistent with its ability to use various fatty and aromatic acids as growth substrates (1, 2).
Analysis of the genome suggests a unique strategy for subsequent metabolism of the CoA-activated aromatic and alicyclic acids (Fig. 1). In denitrifiers and anaerobic phototrophs, benzoyl-CoA is reduced in an ATP-dependent reaction that requires four polypeptides (21–24). The S. aciditrophicus genome has four genes (SYN0368–SYN0371) with low homology (<35% identity) to two of the four types of genes needed to encode for the ATP-dependent, benzoyl-CoA reductase: genes for the other two subunits were not detected (BLAST e > −05). Based on proteomic and gene expression analyses in Geobacter metallireducens, Wischgoll et al. (25) proposed that benzoyl-CoA reduction in this organism occurs by a different enzymatic system. Two gene clusters in the S. aciditrophicus genome (Sa1 and Sa2) (SI Fig. 6 and SI Table 5) have related synteny and high similarity (>50% identity at the amino acid level) to the benzoate-induced gene cluster in G. metallireducens (25). Each S. aciditrophicus gene cluster (SI Fig. 6) contains a ligase gene, a gene for an aldehyde oxidoreductase whose protein product may function as the reductase (25), genes for selenium-containing heterodisulfide reductase subunits, and genes for NADH:quinone oxidoreductase-like components. The presence of the latter genes suggested to Wischgoll et al. that the membrane potential rather than ATP hydrolysis may drive the electron transfer needed for ring reduction by yet unknown membrane components (25).
The S. aciditrophicus genome contains three additional gene clusters (Sa3 to Sa5) with homologous heterodisulfide reductase subunit genes (SI Fig. 6 and SI Table 5). Two of these clusters (Sa3 and Sa4) also contain a gene for a tungsten-containing aldehyde oxidoreductase and genes for other redox proteins, but lack the genes for the NADH:quinone oxidoreductase-like components. The fifth cluster (Sa5) differs in having only the heterodisulfide reductase-type subunit, and an associated iron-sulfur protein. The linkage of S. aciditrophicus genes for oxidoreductases with those that encode proteins with different redox centers (iron-sulfur clusters and flavin- and pyridine nucleotide-binding domains) may provide a mechanism to control electron flow under the varying thermodynamic conditions as hydrogen and/or formate concentrations change in the environment. A molecular understanding of these complex multigene operons and their encoded protein complexes may provide considerable insight into syntrophy.
Although enzyme activities for the conversion of cyclohex-1-ene carboxyl-CoA to pimelyl-CoA (Fig. 1) were detected in cell-free extracts of S. aciditrophicus (2), genes homologous to those that encode these proteins in Rhodopseudomonas palustris (21, 22) were not detected in the S. aciditrophicus genome (<30% amino acid sequence identity). However, three genes (SYN1653–SYN1655) are present whose gene products have high sequence similarity to cyclohex-1,5-diene-1-carboxyl-CoA hydratase, BzdW (47% identity), 6-hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase, BzdY (66% identity), and the ring opening 6-oxocyclohex-1-ene-1-carboxyl-CoA hydrolase, BzdX (56% identity) of Azoarcus strain CIB (26). Thus, it is likely that once benzoyl-CoA is reduced, it is metabolized to 3-hydroxypimelyl-CoA by a pathway similar to that found in denitrifiers (Fig. 1) (21).
Interestingly, no genes homologous to the multifunctional isomerase-hydrolase-dehydrogenase (fadB) subunit, nor to the 3-oxoacyl-CoA thiolase (fadA) subunit of the common bacterial fatty acid degradation system, are recognizable within the S. aciditrophicus genomic sequences. The lack of these homologs is particularly striking because fatty acids and aromatic substrates are growth substrates for this organism. The genome contains six genes for acyl-CoA dehydrogenases, five genes for enoyl-CoA hydratases, and two genes for acetyl-CoA acetyltransferases (thiolase) (SI Table 4). However, only one gene is predicted to encode a 3-hydroxyacyl-CoA dehydrogenase (SI Table 4). Usually, microorganisms contain different gene systems for the metabolism of acyl-CoA intermediates derived from mono and dicarboxy fatty acids; dicarboxy-CoA intermediates are formed during alicyclic or aromatic acid metabolism (Fig. 1).
Substrate-Level Phosphorylation.
Surprisingly, S. aciditrophicus lacks a homolog for acetate kinase. The genome has two gene clusters, each with a gene for a butyrate kinase and two genes for phosphate acetyl/butyryl transferase (SI Table 4). Cell-free extracts of benzoate-grown cocultures and crotonate-grown pure cultures have low acetate kinase activity and phosphotransacetylase activity was not detected in cell-free extracts of crotonate-grown, pure cultures (2). The genome contains nine genes for ADP-forming, acetyl-CoA synthetases that would be able to synthesize ATP from acetyl-CoA (Fig. 1 and SI Table 4), thus indicating a major strategy for energy acquisition in syntrophs (reaction 2):
Acetyl-CoA synthetase (ADP-forming) is used by all acetate-forming Archaea, Entamoeba histolytica, and Giardia lamblia (27–30) to synthesize ATP during acetate formation. Future studies are needed to determine whether all nine genes are expressed, and to assign specificity/affinity to the gene products.
Electron Flow.
S. aciditrophicus exhibits severely limited genetic potential to use external electron acceptors (e.g., oxygen, nitrate, sulfate, iron, or commonly used organic molecules like fumarate). Two genes (SYN1975 and SYN1976) encode a cytochrome bd: ubiquinol-like oxidase. However, the genome lacks a proton-translocating, NADH:ubiquinone oxidoreductase (complex I) or succinate dehydrogenase (complex II) that would indicate an energy-yielding, oxygen metabolism. Two genes (SYN2615 and SYN2616) encode a soluble periplasmic nitrite-type reductase with a potential role in detoxification: however, nitrite metabolism has not been detected (1). Therefore, the S. aciditrophicus genome indicates the sole reliance on syntrophic metabolism (i.e., the reduction of protons to H2 and/or bicarbonate to formate) or the reduction of unsaturated fatty acids or benzoate to reoxidize reduced cofactors.
Pathway(s) for electron flow during syntrophic metabolism are unknown. Inspection of the genome reveals a limited number of respiratory options for transfer of electrons via FADH2- and NADH-linked reactions to hydrogen or formate. Potential membrane-bound, redox proteins are limited to six b-type cytochromes, three of which are associated with genes predicted to code for formate dehydrogenase or hydrogenase components, a putative, tetra-heme, c-type cytochrome, a flavoprotein, two Fe-S proteins, and three components of a predicted Rnf-type ion-translocating, electron transport complex (Fig. 1). A gene for a membrane-bound, iron-sulfur oxidoreductase (SYN2638) is clustered with a gene for acyl-CoA synthetase, a gene for a fatty acid transport, and the genes for the two subunits of the electron transfer flavoprotein (ETF). The linkage of these genes implicates the SYN2638 gene product in electron flow during the β–oxidation of acyl-CoA intermediates. Notably, the genome lacks the numerous soluble and membrane-bound, c-type cytochromes present in other δ-proteobacteria, Desulfovibrio spp. and Geobacter spp (31, 32).
The genome contains at least 46 genes encoding proteins with 4Fe-4S-binding domains and six genes for proteins with 2Fe-2S binding domains (SI Table 4). These genes, along with those discussed above for the heterodisulfide reductase gene clusters, indicate a soluble electron transfer machinery that couples substrate metabolism with redox carriers such as ferredoxin and/or NAD+ (Fig. 1, SI Fig. 6, and SI Table 4).
The presence of a menaquinone (Eo′ ∼ −80 mV) in S. aciditrophicus (R. Meganathan, personal communication) provides a potential membrane-diffusible carrier to link the oxidation of acyl-CoA intermediates (Eo′ ∼ −10 mV) with other redox complexes (15), possibly driven by the proton gradient or membrane potential (33) (Fig. 1).
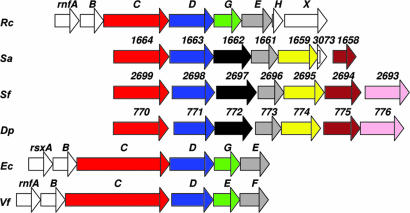
The genome contains a set of seven genes (SYN1664–1661, SYN1659, SYN1658, and SYN3073) with similarity to genes for the Rnf ion-translocating, electron transport complex found in Rhodobacter capsulatus, Clostridium tetani, and Methanosarcina acetivorans (34–36) (Figs. 1 and 2). In R. capsulatus, the RnfABCDGEH complex is involved in electron transport to nitrogenase, possibly functioning to drive the thermodynamically unfavorable reverse electron transfer from NADH to ferredoxin (35, 37, 38). Genes in Clostridium tetani and Clostridium perfringens (34) and Methanosarcina acetivorans (36) with similarity to the R. capsulatus rnf genes are believed to function in the generation of transmembrane ion gradients. The S. aciditrophicus proteins encoded by SYN1663, SYN1661, and SYN1659 are predicted to be integral membrane proteins homologous to RnfD, RnfE and RnfA, respectively. SYN1664 seems to encode RnfC-like protein with NAD+ and Fe-S-binding domains whereas two additional genes, SYN3073 and SYN1658, are predicted to encode periplasmic iron-sulfur proteins. Genes with the same synteny to SYN1658 to -1664 are found in the genomes of Desulfotalea psychrophila and the propionate-degrading, syntrophic bacterium, Syntrophobacter fumaroxidans (Fig. 2). The two predicted periplasmic components of the S. aciditrophicus, S. fumaroxidans, and D. psychrophila Rnf-like complexes seem to replace the RnfB and RnfG subunits of R. capsulatus. The amino acid sequence of the RnfG-like component (SYN1662 gene product) in S. aciditrophicus is 57% and 47% identical to the respective proteins in S. fumaroxidans and D. psychrophila, but shares <35% amino acid sequence identity to any other protein in the NCBI database. The RnfG-like component in S. acidi-trophicus is predicted to have a FMN-binding domain whereas the RnfB-like component is predicted to have a Fe-S center.
Fig. 2.
Comparison of the Rnf-type gene cluster in S. aciditrophicus. The rnfABCDGEH genes of R. capsulatus (Rc), the putative rnf genes of S. fumaroxidans (Sf), S. aciditrophicus (Sa), and D. psychrophila (Dp) are aligned 5′ to 3′. The rsxABCDEF genes of E. coli (Ec) and rnfABCDEF genes Vibrio fisheri (Vf) are shown below. Identically colored genes are homologous.
The unique features shared by the Rnf components in two syntrophic bacteria, S. aciditrophicus and S. fumaroxidans, argue for a pivotal function of this membrane complex. Because the production of hydrogen or formate from NADH is exergonic (ΔE′ ∼ +60 mV at 1 Pa H2 or 1 μM formate) under syntrophic growth conditions, the Rnf-like complex in S. aciditrophicus may function to catalyze the unfavorable reduction of ferredoxin needed for pyruvate synthesis (Fig. 2) unless thermodynamic conditions change.
S. aciditrophicus contains genes encoding a cytoplasmic Ni-Fe-type hydrogenase (SYN2219 to SYN2222) and a cytoplasmic Fe-only hydrogenase (SYN1370). The latter is adjacent to a gene predicted to code for a putative, sodium-translocating membrane protein, which would provide a unique mechanism for reverse electron transport. There are also two cytoplasmic and two periplasmic formate dehydrogenases, a formate channel, and an oxalate/formate antiporter (Fig. 1). Cytoplasmic- and periplasmic-oriented formate dehydrogenases suggest that energy conservation by a formate cycle could occur where proton consumption by the cytosolic formate synthesis is coupled to proton production in the periplasm by formate oxidation. The presence of two hydrogenases or four formate dehydrogenases would allow S. aciditrophicus to use either hydrogen or formate, thus providing a basis for understanding interspecies electron transfer depending on the metabolic capabilities of its partner.
Ion Movement and ATP Synthesis.
Besides the Rnf-like complex, other putative ion-translocation complexes include Na+-transporting, methylmalonyl-CoA/oxaloacetate decarboxylases (SYN0115 and SYN1434), which may interact with the glutaconyl-CoA decarboxylase (SYN0479 to -0481) to use the energy of decarboxylation to translocate sodium ions (20, 39, 40) and two ATP synthase complexes (SYN0543 to -0549 and SYN2101 to -2105) (Fig. 1), both of which may be Na+-translocating based on conservation of amino acids involved in ion coordination in the proteolipid c subunit (SI Fig. 7) (41). The presence of these systems suggests a critical role for sodium in the bioenergetics of S. aciditrophicus. Two proton-translocating pyrophosphatases (SYN2770 and SYN2772) are also present (42).
Stress.
The S. aciditrophicus genome contains genes for oxidative stress protection proteins including catalase, peroxiredoxin, thioredoxin, a Fe-Mn superoxide dismutase, and (S)-2-hydroxy-acid oxidases. Genes for the superoxide reductase system found in obligate anaerobes were not detected. There are several genes for efflux pumps that could confer resistance to certain metals or antimicrobial compounds.
Genes for heat shock include six chaperones, grpE, dnaK, and dnaJ, plus a small heat shock protein. Also present are five genes encoding cold shock proteins, two for the universal stress protein, and several solvent and acid stress response proteins. Several of the above genes are candidates for the 32-factor control (below).
Regulation and Signal Transduction.
The S. aciditrophicus genome contains a prototypical bacterial RNA core polymerase (RpoA, RpoB, and RpoC) that along with four sigma factors, confer promoter specificity (SI Table 4). The sigma factors include a general housekeeping 70 factor (RpoD), a heat shock 32 factor (RpoH), a flagella biogenesis 28 factor (FliA), and a 54 factor (RpoN) similar to that used for general nitrogen control in Escherichia coli. Strikingly, the genome contains seventeen sigma 54-interacting transcriptional regulators, suggesting a major role for the 54 factor in global control of S. aciditrophicus gene expression.
Numerous two-component regulatory systems (23 histidine kinase-type sensor transmitters, 18 response regulatory proteins, and 8 receiver-only domain proteins) are present in the genome. Roughly half are genetically linked to a cognate two-component protein member, thus suggesting an interacting partner in sensory transduction. Compared with other Gram-negative microbes, S. aciditrophicus has a relatively small number of primary transcription factors containing a helix-turn-helix motif (≈35 genes). Instead, S. aciditrophicus seems to have adopted a regulatory strategy highly reliant on 54 factor coupled signal transduction pathways.
Motility and Taxis.
S. aciditrophicus has a complete set of flagellar structural proteins (basal body, motor, hook, and filament) along with the associated flagellar biogenesis and regulatory genes including a σ 28 and an anti-28 factor (FlgM). Absent are the E. coli type master switch proteins, FlhCD. Four MCP sensory proteins act to detect as yet unknown attractants and/or repellants for signal transduction via two CheA, three CheW, and two CheY proteins. Although a full complement of flagellar and chemosensory genes are present, motility and chemotaxis have not been observed with laboratory cultures. Cell-density dependent gene systems (quorum signaling) are absent although genes for synthesis of Type IV pili are present that may conceivably facilitate cell-to-cell attachment of the syntrophic partners to facilitate interspecies hydrogen/formate transfer. S. aciditrophicus exhibits twitching motility (unpublished results).
Syntrophic Strategies.
Genomic analysis of S. aciditrophicus helps define a strategy for the syntrophic lifestyle. Previously, the syntrophic lifestyle was defined in a negative sense, i.e., as an organism that did not use common electron acceptors and did not have the ability to reoxidize reduced cofactors by making the typical fermentative end products (3). However, genomic analysis of S. aciditrophicus reveals previously undescribed specific systems that define the genetic potential for the syntrophic lifestyle. Distinctive features include unique approaches for carbon metabolism and reverse electron transport. The membrane components involved in the generation and use of ion gradients are critical. The genome of S. aciditrophicus contains a membrane-bound, ion-translocating complex (Rnf-like) that differs from all other sequenced representatives except those found in another syntrophic microorganism, S. fumaroxidans, and in the sulfate reducer, D. psychrophila. The latter has not been tested for syntrophic metabolism because its optimal growth (10°C) is well below that of culturally available hydrogen/formate-using partners (43). Further genomic sequencing and expression studies will be needed to determine whether the genes for the membrane-bound, iron-sulfur protein associated with β-oxidation genes and the RnfG-like complex are specific for syntrophic metabolism. If so, it can be used as a functional probe for this type of syntrophic metabolism.
Syntrophic metabolism proceeds close to thermodynamic equilibrium and the available free energy of the reactions depends on the terminal electron accepting reaction of the syntrophic partner (14). Multiple mechanisms are present to create and use ion gradients, which would help modulate the energy status of the cells in response to varying thermodynamic conditions. Examples of such membrane systems include an ion-translocating, electron transfer complex, proton- and sodium-translocating ATP synthases, proton-translocating pyrophosphatases, sodium-translocating glutaconyl-CoA decarboxylase, and hydrogenase linked with a putative sodium-translocating membrane protein. In addition, the separation of formate synthesis and hydrolysis across the cell membrane could form a proton/sodium gradient.
Genomic sequencing and expression studies of additional syntrophic microbes will be needed to determine whether the properties of S. aciditrophicus are hallmarks of syntrophic metabolism, or whether other biological solutions are evolved to explain this missing link in our understanding of the ecology and physiology of anaerobic carbon flow on Earth.
Methods
Media and Growth Conditions.
S. aciditrophicus was grown in basal medium (44) without rumen fluid with 20 mM sodium crotonate. Multiple 1-liter cultures were used to obtain cells for DNA extraction.
Genome Sequencing.
S. aciditrophicus genomic DNA was isolated by standard methods. Genomic DNA was mechanically sheared and 2- to 3-kb fragments were isolated. The ends were filled with Klenow enzyme and ligated into a modified pSMART vector to produce a high-copy number, small-insert library. Cosmid libraries were prepared from high molecular weight genomic DNA and partially digested with Sau3A, and 30- to 35-kb fragments were cloned into the BamHI site of Lorist 6. Undigested, unsheared DNA was used as template for PCR amplification of chromosomal regions not represented in the plasmid or cosmid libraries.
Whole-genome shotgun sequencing on ≈25,000 plasmids and 800 cosmids using Applied Biosystems (Foster City, CA) 3700 and 3730xls DNA sequencers resulted in an initial assembly containing ≈250 contigs larger than 1 kb. Gaps were closed by primer walking over cosmid clones, resulting in ≈90 contigs longer than 1 kb. The genome was completed by sequencing gap-spanning PCR products. The genome was finally assembled into a single contig representing a circular chromosome of 3,179 kb with average coverage of 11.4X at an average base quality (Phred) score of 93. The plasmid library was prepared and sequenced by Agencourt Biosciences (Beverly, MA) and the genome was finished and annotated by Integrated Genomics (Chicago, IL).
Genes were identified by a combination of Glimmer, Critica, and a coding sequence-calling program developed at Integrated Genomics. The finished genome of S. aciditrophicus underwent a round of automatic annotation followed by community-based manual curation organized within the ERGO bioinformatics suite. The results of this analysis, including annotations and partial metabolic reconstructions of S. aciditrophicus can be found in a public version of ERGO at www.ergo-light.com. A total of 3,169 potential CDS were identified, and functional assignments were made for 1,618. The prediction of transmembrane helices and signal peptides in gene products was done by using on-line bioinformatics tools (45, 46).
Supplementary Material
Acknowledgments
We thank Matthew McInerney for coding sequence analysis. This work was supported by National Science Foundation Award Grant NSF EF-0333294 (to R.P.G. and M.J.M.) and Department of Energy Awards DE-FG03-86ER13498 (to R.P.G.), and DE-FG03-96-ER-20212 (to M.J.M.).
Footnotes
Conflict of interest statement: J.W.C. and A.B. were employed by Integrated Genomics, Inc., which performed DNA sequencing and gene-calling analysis under contract to the University of California at Los Angeles. They may be viewed as having commercial interests in the product as a representation of their company's performance.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. CP000252).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610456104/DC1.
References
- 1.Jackson BE, Bhupathiraju VK, Tanner RS, Woese CR, McInerney MJ. Arch Microbiol. 1999;171:107–114. doi: 10.1007/s002030050685. [DOI] [PubMed] [Google Scholar]
- 2.Elshahed MS, Bhupathiraju VK, Wofford NQ, Nanny MA, McInerney MJ. Appl Environ Microbiol. 2001;67:1728–1738. doi: 10.1128/AEM.67.4.1728-1738.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McInerney MJ. In: The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications. Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH, editors. Vol 2. New York: Springer-Verlag; 1992. pp. 2048–2057. [Google Scholar]
- 4.Schink B. Microbiol Mol Biol Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stams AJ. Antonie Van Leeuwenhoek. 1994;66:271–294. doi: 10.1007/BF00871644. [DOI] [PubMed] [Google Scholar]
- 6.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jorgensen BB, Witte U, Pfannkuche O. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 7.Hinrichs KU, Hayes JM, Sylva SP, Brewer PG, DeLong EF. Nature. 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 8.Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. Science. 2001;293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- 9.Zengler K, Richnow HH, Rossello-Mora R, Michaelis W, Widdel F. Nature. 1999;401:266–269. doi: 10.1038/45777. [DOI] [PubMed] [Google Scholar]
- 10.Ishii S, Kosaka T, Hotta Y, Watanabe K. Appl Environ Microbiol. 2006;72:5093–5096. doi: 10.1128/AEM.00333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H. Appl Environ Microbiol. 1999;65:1280–1288. doi: 10.1128/aem.65.3.1280-1288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholten JC, Conrad R. Appl Environ Microbiol. 2000;66:2934–2942. doi: 10.1128/aem.66.7.2934-2942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams CJ, Redmond MC, Valentine DL. Appl Environ Microbiol. 2006;72:1079–1085. doi: 10.1128/AEM.72.2.1079-1085.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson BE, McInerney MJ. Nature. 2002;415:454–456. doi: 10.1038/415454a. [DOI] [PubMed] [Google Scholar]
- 15.Sato K, Nishina Y, Setoyama C, Miura R, Shiga K. J Biochem (Tokyo) 1999;126:668–675. doi: 10.1093/oxfordjournals.jbchem.a022501. [DOI] [PubMed] [Google Scholar]
- 16.Sauer U, Eikmanns BJ. FEMS Microbiol Rev. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y, Pingitore F, Mukhopadhyay A, Phan R, Hazen TC, Keasling JD. J Bacteriol. 2007;189:940–949. doi: 10.1128/JB.00948-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottschalk G. Eur J Biochem. 1968;5:346–351. doi: 10.1111/j.1432-1033.1968.tb00376.x. [DOI] [PubMed] [Google Scholar]
- 19.Auburger G, Winter J. Appl Microbiol Biotechnol. 1996;44:807–815. doi: 10.1007/BF00178623. [DOI] [PubMed] [Google Scholar]
- 20.Schöcke L, Schink B. Arch Microbiol. 1999;171:331–337. [Google Scholar]
- 21.Harwood CS, Burchhardt G, Herrmann H, Fuchs G. FEMS Microbiol Rev. 1998;22:439–458. [Google Scholar]
- 22.Egland PG, Pelletier DA, Dispensa M, Gibson J, Harwood CS. Proc Natl Acad Sci USA. 1997;94:6484–6489. doi: 10.1073/pnas.94.12.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boll M, Fuchs G. Eur J Biochem. 1995;234:921–933. doi: 10.1111/j.1432-1033.1995.921_a.x. [DOI] [PubMed] [Google Scholar]
- 24.Boll M, Albracht SS, Fuchs G. Eur J Biochem. 1997;244:840–851. doi: 10.1111/j.1432-1033.1997.00840.x. [DOI] [PubMed] [Google Scholar]
- 25.Wischgoll S, Heintz D, Peters F, Erxleben A, Sarnighausen E, Reski R, Van Dorsselaer A, Boll M. Mol Microbiol. 2005;58:1238–1252. doi: 10.1111/j.1365-2958.2005.04909.x. [DOI] [PubMed] [Google Scholar]
- 26.Barragan MJL, Carmona M, Zamarro MT, Thiele B, Boll M, Fuchs G, Garcia JL, Diaz E. J Bacteriol. 2004;186:5762–5774. doi: 10.1128/JB.186.17.5762-5774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mai X, Adams M. J Bacteriol. 1996;178:5897–5903. doi: 10.1128/jb.178.20.5897-5903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musfeldt M, Schonheit P. J Bacteriol. 2002;184:636–644. doi: 10.1128/JB.184.3.636-644.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez LB, Muller M. FEBS Lett. 1996;378:240–244. doi: 10.1016/0014-5793(95)01463-2. [DOI] [PubMed] [Google Scholar]
- 30.Reeves RE, Warren LG, Susskind B, Lo HS. J Biol Chem. 1977;252:726–731. [PubMed] [Google Scholar]
- 31.Heidelberg JF, Seshadri R, Haveman SA, Hemme CL, Paulsen IT, Kolonay JF, Eisen JA, Ward N, Methe B, Brinkac LM, et al. Nat Biotechnol. 2004;22:554–559. doi: 10.1038/nbt959. [DOI] [PubMed] [Google Scholar]
- 32.Methe BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, et al. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- 33.Schirawski J, Unden G. Eur J Biochem. 1998;257:210–215. doi: 10.1046/j.1432-1327.1998.2570210.x. [DOI] [PubMed] [Google Scholar]
- 34.Bruggemann H, Baumer S, Fricke WF, Wiezer A, Liesegang H, Decker I, Herzberg C, Martinez-Arias R, Merkl R, Henne A, Gottschalk G. Proc Natl Acad Sci USA. 2003;100:1316–1321. doi: 10.1073/pnas.0335853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumagai H, Fujiwara T, Matsubara H, Saeki K. Biochemistry. 1997;36:5509–5521. doi: 10.1021/bi970014q. [DOI] [PubMed] [Google Scholar]
- 36.Li Q, Li L, Rejtar T, Lessner DJ, Karger BL, Ferry JG. J Bacteriol. 2006;188:702–710. doi: 10.1128/JB.188.2.702-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. Mol Gen Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 38.Jouanneau Y, Jeong HS, Hugo N, Meyer C, Willison JC. Eur J Biochem. 1998;251:54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- 39.Buckel W. Biochim Biophys Acta. 2001;1505:15–27. doi: 10.1016/s0005-2728(00)00273-5. [DOI] [PubMed] [Google Scholar]
- 40.Beatrix B, Bendrat K, Rospert S, Buckel W. Arch Microbiol. 1990;154:362–369. doi: 10.1007/BF00276532. [DOI] [PubMed] [Google Scholar]
- 41.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 42.Schöcke L, Schink B. Eur J Biochem. 1998;256:589–594. doi: 10.1046/j.1432-1327.1998.2560589.x. [DOI] [PubMed] [Google Scholar]
- 43.Knoblauch C, Sahm K, Jorgensen BB. Int J Syst Bacteriol. 1999;49:1631–1643. doi: 10.1099/00207713-49-4-1631. [DOI] [PubMed] [Google Scholar]
- 44.McInerney MJ, Bryant MP, Pfennig N. Arch Microbiol. 1979;122:129–135. [Google Scholar]
- 45.Sonnhammer EL, von Heijne G, Krogh A. Proc Int Conf Intell Syst Mol Biol; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 46.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.