Abstract
The proteolytic processing of bovine fibrinogen by MMP-2 (gelatinase A), which brings about the formation of a product unable to form fibrin clots, has been studied at 37 °C. Catalytic parameters, although showing a somewhat lower catalytic efficiency with respect to thrombin and plasmin, indeed display values indicating a pathophysiological significance of this process. A parallel molecular modelling study predicts preferential binding of MMP-2 to the β-chain of fibrinogen through its haemopexin-like domain, which has been directly demonstrated by the inhibitory effect in the presence of the exogenous haemopexin-like domain. However, the removal of this domain does not impair the interaction between MMP-2 and fibrinogen, but it dramatically alters the proteolytic mechanism, producing different fragmentation intermediates. The investigation at various pH values between 6.0 and 9.3 indicates a proton-linked behaviour, which is relevant for interpreting the influence on the process by environmental conditions occurring at the site of an injury. Furthermore, the action of MMP-2 on peroxynitrite-treated fibrinogen has been investigated, a situation possibly occurring under oxidative stress. The chemical alteration of fibrinogen, which has been shown to abolish its clotting activity, brings about only limited modifications of the catalytic parameters without altering the main enzymatic mechanism.
Keywords: fibrinogen, fragmentation, gelatinase A, kinetics, molecular modelling, pH-dependence
Abbreviations: DPA, N-3-(2,4-dinitrophenyl)-L-2,3-diaminopropionyl; MCA, (7-methoxycoumarin-4-yl)acetyl; MMP, matrix metalloproteinase; MMP-2, gelatinase A; MMP-9, gelatinase B
INTRODUCTION
MMPs (matrix metalloproteinases) are a class of Ca2+- and Zn2+-dependent endopeptidases, displaying an active site characterized by a Zn2+ atom co-ordinated to three histidine residues [1]. They show a multi-domain structural organization, usually made up of a propeptide domain (which is removed upon enzyme activation), a catalytic domain and a haemopexin-like domain, which are connected by a hinge domain. In addition, a MMP subclass composed of two MMPs, MMP-2 and MMP-9 (gelatinases A and B respectively), displays a unique collagen-binding domain, called a fibronectin-like domain, inserted on the catalytic domain and consisting of three 58-amino-acid fibronectin type II-like modules [2].
These enzymes, which are secreted as inactive zymogens, have the capacity of degrading several proteins of the extracellular matrix, participating in most of the tissue remodelling phenomena [3]. Furthermore, since they are released in the plasma from the polymorphonuclear leukocytes [4], it is very probable that they come into close contact with plasma proteins, which can then become physiological substrates for MMPs. As a matter of fact, a growing interest has been addressed toward the role of MMPs in the neovascularization events which play a key role in several physiological and pathological processes, from tumour dissemination to wound healing. In particular, MMP-2 is involved in a process termed the ‘angiogenic switch’ [5,6], which consists of the induction of a new vasculature by degrading the vascular basement membrane and the extracellular matrix, in order to allow endothelial cells to migrate into the perivascular space. This process can occur at different stages of tumour progression, depending on tumour type and environment. Premalignant lesions may also induce neovascularization and it has been shown that, during the neovascularization event, the role of several MMPs appears to be that of pericellular fibrinolysins [7], some of them being involved in the degradation of fibrinogen and cross-linked fibrin necessary for the migration of endothelial cells [8,9]. Therefore the enzymatic action by MMPs on components of the blood coagulation cascade is relevant both from the physiological and the pathological standpoint.
Fibrinogen is a 340 kDa dimeric glycoprotein, present in the blood, which is composed of six polypeptide chains (αβγ)2, joined by 29 disulfide bridges within the N-terminal E-domain, forming an elongated 45 nm structure consisting of two outer D-domains connected through a central E-domain by a coiled–coil segment. During blood coagulation, thrombin converts fibrinogen into fibrin monomers, which associate into staggered overlapping two-stranded fibrils and these domains contain binding sites which participate in the fibrinogen conversion into fibrin [10]. However, previous data clearly indicate a correlation between the activity of MMP-2 and MMP-9 and that of fibrinogen in the neovascularization process, since it has been shown that inhibition of the gelatinase action brings about an accumulation of unprocessed fibrinogen and fibrin, and this is accompanied by a blocking of the neovascularization process [11]. Further, during oxidative stresses connected to the blood vessels damage and their repair, a dramatic enhancement of the production of reactive oxygen species and reactive nitrogen species is observed, leading to vascular injury responsible for the development of atherosclerosis [12]. As a matter of fact, levels of nitrated proteins have been shown to be increased in atherosclerotic lesions [13–16], being predictors of a risk for coronary artery disease [17]. This observation provides additional support for the previous observation that post-translational modifications of fibrinogen by reactive nitrogen species, such as peroxynitrite [18], bring about dramatic alterations in the role of fibrinogen [19].
Therefore the elucidation of the mechanism by which the enzymatic activity of MMP-2 on native and oxidized fibrinogen is modulated can be of the utmost importance for a better comprehension of the different interplay between MMPs and coagulation properties, and of their physiopathological relevance for angiogenesis. In the present study we have analysed the catalytic efficiency of whole MMP-2 on native and oxidized fibrinogen, comparing it with that of the catalytic domain alone, in order to clarify the role of different domains of MMP-2 during fibrinogen degradation. It is interesting to observe that the efficiency of the enzymatic action of MMP-2 on fibrinogen turns out to be somewhat lower than that of thrombin [20] and similar to that displayed by plasmin [21]. Moreover, we accompany this investigation with a molecular modelling of the interaction between MMP-2 and fibrinogen in order to formulate a plausible mechanism based on structural and functional information. In addition, since pH may vary significantly from neutrality in injured areas and during inflammatory responses, we have performed an analysis of the pH-dependence of catalytic parameters for the processing of the α- and β-chains of fibrinogen by MMP-2 in order to provide more information on the modulation of this relevant process by environmental conditions.
EXPERIMENTAL PROCEDURES
Materials
Bovine fibrinogen (Sigma) was dissolved in water at room temperature (25 °C) to a final concentration of 1 mg/ml. The suspension was centrifuged for 1 h at 10000 g and the supernatant containing the dissolved fibrinogen was used. The amount of substrate was quantified using the method described by Bradford [22].
Peroxynitrite was synthesized according to the protocol reported by Uppu et al. [23] and stored at −80 °C. Bovine peroxynitrite-treated fibrinogen was prepared by treating native fibrinogen (5 mg/ml) with 40 mM of peroxynitrous acid, in 50 mM PBS (pH 7.2) and 0.1 mM DTPA (diethylenetriaminepenta-acetic acid).
MMP-2 proenzyme was either of commercial origin (R&D Systems) or a gift from Dr Chris Overall (Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, Canada), and no significant functional difference was detected between the two preparations after activation. The isolated purified MMP-2 was activated by incubating 0.1 ml of a 0.1 μg/ml progelatinase solution with APMA (p-aminophenyl mercuric acid; Sigma) at 37 °C for 30 min and immediately used for experiments. A control experiment using gelatin zymography showed that over the first 24 h no autocatalytic process was taking place under our experimental conditions (results not shown), as has already been described by Nagase et al. [24].
The haemopexin-like domain of MMP-2 was a gift from Dr Chris Overall (Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, Canada) and was prepared as previously reported [25].
Human recombinant MMP-2 catalytic domain (Biomol International) was dissolved in a solution of 50 mM Tris/HCl (pH 7.2), 0.1 M NaCl and 10 mM CaCl.
The quenched fluorogenic substrate MCA-Pro-Leu-Gly-Leu-DPA-Ala-Arg-NH2 [where MCA is (7-methoxycoumarin-4-yl)acetyl and DPA is N-3-(2,4-dinitrophenyl)-L-2,3-diaminopropionyl] was purchased from Calbiochem.
Plasmin enzyme and the fluorogenic peptide MUGB (4-methylumbelliferylguanidinobenzoate), an active site titrant of trypsin-like proteases, were of commercial origin (Sigma).
Activity assay
The active amount of MMP-2 was determined using gelatin zymography and using the fluorimetric assay, as previously reported [26], following the progressive decrease of hydrolysis of the quenched fluorogenic substrate MCA-Pro-Leu-Gly-Leu-DPA-Ala-Arg-NH2 upon addition of BB-94 (batimastat), a peptidomimetic inhibitor (provided by British Biotech Pharmaceuticals), which stoichiometrically inhibits MMPs.
Digestion of fibrinogen by MMP-2 and plasmin, and associated kinetics
For substrate fragmentation kinetics, activated whole MMP-2 or plasmin were added to fibrinogen solutions at a final concentration of 10 pM, whereas the catalytic domain of MMP-2 was added to fibrinogen solutions at a final concentration of 15–20 pM. The kinetics of MMP-2 were performed in 50 mM Tris/HCl, 0.1 M NaCl and 10 mM CaCl2 at 37 °C and at different values of pH, using different concentrations of bovine fibrinogen (spanning between 5.3 μM and 20 μM). The same range of substrate concentrations was used for peroxynitrite-treated fibrinogen, but in this case, as in the case of plasmin, kinetics were performed at only pH 7.1. In the case of the experiments in the presence of the haemopexin-like domain of MMP-2, 6 μM fibrinogen was equilibrated for 30 min in the presence of 200 μM haemopexin-like domain (a concentration required for the almost complete saturation of the binding site, see below), then whole MMP-2 (to a final concentration of 10 pM) was added to start the kinetic reaction.
Kinetics were performed keeping the mixtures at 37 °C and harvesting small aliquots at different time intervals. Reactions were stopped by the addition of SDS/PAGE loading buffer containing 20 mM EDTA and aliquots were frozen at −80 °C until use. The aliquots in the reducing sample buffer were separated on SDS/PAGE (10% gels), which were stained using 0.5% Coomassie Blue and destained in 10% acetic acid and 40% methanol until substrate bands were clearly visible. The broad spectrum protein markers (BioRad) were used as molecular mass standards.
Kinetic analysis
Electrophoretic spots corresponding to different aliquots at different time intervals were analysed by a laser densitometer (LKB 2202 UltraScan) and their intensity was calibrated (in order to obtain concentration values) using standard substrate solutions. It must be pointed out that different preparations of fibrinogen displayed different ratios of the relative amount of the three chains, as determined from the intensity of electrophoretic spots on SDS/PAGE under reducing conditions. For α- and β-chains the substrate disappearance rates were derived at each fibrinogen concentration employed.
The measurement of the initial velocity referred to a time period over which less than 10% of the substrate was degraded during the assay; this period never exceeded 10 h, which guaranteed the integrity of activated MMP-2, as reported previously by Nagase et al. [24]. Therefore even though we cannot rule out the possibility that at very long time intervals (i.e. >48 h) MMP-2 undergoes a partial autolysis, an investigation outside the purpose of the present paper, the catalytic parameters reported in Table 1 refer to the behaviour observed within the first 10 h of the reaction. In any case, the analysis was limited to the time interval over which linearity of the rate was observed and a steady-state condition for the first cleavage step was ensured, a prerequisite for the subsequent analysis step. This consisted of the verification for the applicability of the Michaelis–Menten approximation to the first cleavage step, which was based on the observation of an inverse linear correlation between velocity and substrate concentration according to the Lineweaver–Burk equation to obtain the catalytic parameters kcat and Km:
 |
(1) |
where E0 is the total enzyme concentration, v is the actual rate (expressed as mol/s), Km is the Michaelis–Menten equilibrium constant (expressed as mol), kcat is the rate-limiting step kinetic constant (expressed as s−1) and [S] is the substrate concentration.
Table 1. Catalytic parameters for the enzymatic processing at 37 °C and pH 7.1 of native fibrinogen by the whole MMP-2, the catalytic domain of MMP-2 and plasmin and of peroxynitrite-treated fibrinogen by whole MMP-2.
| kcat/Km (M−1·s−1) | kcat (s−1) | Km (M) | |
|---|---|---|---|
| α-Chain | |||
| Native fibrinogen | |||
| Whole MMP-2 | 2.29(±0.23)×105 | 36.4(±4.2) | 1.59(±0.18)×10−4 |
| Catalytic domain | 1.09(±0.12)×104 | 0.19(±0.02) | 1.72(±0.19)×10−5 |
| MMP-2 | |||
| Plasmin | 2.04(±0.21)×105 | 7.7(±0.9) | 3.77(±0.41)×10−5 |
| Oxidized fibrinogen | |||
| Whole MMP-2 | 1.05(±0.11)×105 | 3.1(±0.4) | 2.93(±0.35)×10−5 |
| β-Chain | |||
| Native fibrinogen | |||
| Whole MMP-2 | 6.61(±0.83)×104 | 12.5(±1.4) | 1.89(±0.20)×10−4 |
| Catalytic domain | 3.59(±0.42)×103 | 0.12(±0.02) | 3.27(±0.41)×10−5 |
| MMP-2 | |||
| Plasmin | 2.47(±0.34)×105 | 27.8(±2.9) | 1.12(±0.15)×10−4 |
| Oxidized fibrinogen | |||
| Whole MMP-2 | 1.34(±0.15)×105 | 1.06(±0.12) | 7.93(±0.88)×10−6 |
| γ-Chain | |||
| Native fibrinogen | |||
| Plasmin | 4.96(±0.6)×104 | 2.35(±0.36) | 4.74(±0.57)×10−5 |
Clotting experiments
The thrombin-induced clotting of fibrinogen after enzymatic processing by MMP-2 was measured spectrophotometrically, following the increase in absorbance at 350 nm as a function of time, in a Cary 1 dual-beam spectrophotometer, thermostatically controlled at 25 °C. Fibrinogen was incubated for 1 min in 50 mM Tris/HCl buffer (pH 8.2) and clot formation was triggered by addition of 1.2 units of human thrombin. Clotting curves were analysed as described previously [27].
Molecular modelling
Crystal structures isolated from the full-length proMMP-2 (PDB No. 1CK7) were used to model the interaction of fibrinogen with different truncated forms of MMP-2 (i.e. the autoinhibitory procatalytic domain, the catalytic domain, the catalytic domain lacking the fibronectin-like domain, the fibronectin-like domain alone and the haemopexin-like domain). The dimeric form of fibrinogen (PDB No. 1M1J) is symmetrical, so the structure used for the docking simulation was truncated to one monomer plus the N-terminal fragments of the second monomer to recreate the interface between the two identical monomers. The docking program employed for the molecular modelling was BiGGER (Biomolecular complex Generation with Global Evaluation and Ranking) [28].
RESULTS
Degradation of fibrinogen and peroxynitrite-treated fibrinogen by MMP-2
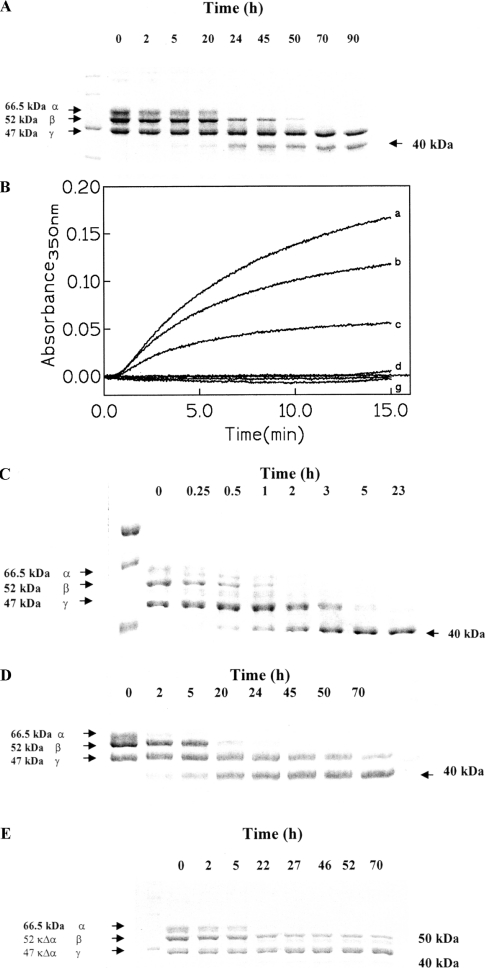
Figure 1(A) shows the three chains forming the fibrinogen molecule, and their enzymatic processing at 37 °C and pH 7.1 as a function of incubation time with whole MMP-2. It is interesting to observe that while α- and β-chains are progressively cleaved by MMP-2, the γ-chain turns out to be essentially immune from the cleavage action of MMP-2. This behaviour is significantly different from that reported for the proteolytic action on fibrinogen of MMP-8, MMP-12, MMP-13 and MMP-14 [29], since these MMPs seem to cleave the α-chains first and only at longer time intervals does the cleavage of the β- and γ-chains take place. However, it must be pointed out that the data in Figure 1(A) refer to whole MMP-2, whereas previous observations [29] were made using the catalytic domains of MMP-8, MMP-12, MMP-13 and MMP-14. An immediate comparison between the kinetics reported in the present paper and that reported by others [29] is not possible, since others have used a 1:10–1:50 enzyme/fibrinogen molar ratio whereas we have used a 1:104–1:105 enzyme/fibrinogen molar ratio (see the Experimental procedures section). Nonetheless, similar to that which has been reported for the other MMPs [29], we have observed the formation of lower molecular mass fragments of the fibrinogen chains, which grow as a function of time of the incubation (Figure 1A). It is important to emphasize that these fragments do not show any clotting activity upon incubation with thrombin (see Figure 1B), suggesting that the action of MMP-2 has relevant physiopathological consequences on the coagulation process connected to the impairment of fibrin formation and the promotion of angiogenetic processes. Such a result is not necessarily in contradiction with data reported by Bini et al. [8], who claimed that, unlike for MMP-3, fibrinogen cleaved by MMP-2 showed a slight clot formation after several hours of processing by thrombin. This statement underlies a gross alteration of the clotting properties for the fibrinogen processed by MMP-2, in line with that observed in the present study for the fibrinogen cleaved by whole MMP-2 (Figure 1B).
Figure 1. Kinetic proteolytic processing of bovine fibrinogen.
(A) SDS/PAGE electrophoresis of the processing of bovine fibrinogen by whole MMP-2 as a function of time (as indicated) at 37 °C and pH 7.1. α- (66.5 kDa), β- (52 kDa) and γ- (47 kDa) chains are indicated as well as the 40 kDa fragment. (B) Clotting induced by human thrombin on bovine fibrinogen (curves a–c) and on bovine fibrinogen after degradation by whole MMP-2 (curves d–g). Fibrinogen was incubated at 37 °C in 50 mM Tris/HCl buffer (pH 8.2) and the clotting was followed at 350 nm after addition of 1.2 units of human thrombin. Curves a–c: 0.24, 0.16 and 0.08 mg/ml of bovine fibrinogen. Curves d–g: 0.08, 0.16, 0.24 and 0.32 mg/ml of fibrinogen degraded by whole MMP-2. (C) SDS/PAGE electrophoresis of the processing of bovine fibrinogen by plasmin as a function of time (as indicated) at 37 °C and pH 7.1. α- (66.5 kDa), β- (52 kDa) and γ- (47 kDa) chains are indicated as well as the 40 kDa fragment. (D) SDS/PAGE electrophoresis of the processing of peroxynitritre-treated bovine fibrinogen by MMP-2 as a function of time (as indicated) at 37 °C and pH 7.1. α- (66.5 kDa), β- (52 kDa) and γ- (47 kDa) chains are indicated as well as the 40 kDa fragment. (E) SDS/PAGE electrophoresis of the processing of bovine fibrinogen by the catalytic domain of MMP-2 as a function of time (as indicated) at 37 °C and pH 7.1. α- (66.5 kDa), β- (52 kDa) and γ- (47 kDa) chains are indicated as well as the 50 kDa and the 40 kDa fragments. For further details, see text.
It is interesting to observe that under the same experimental conditions of pH and temperature, plasmin is able to cleave all three chains of fibrinogen (Figure 1C) at a different rate (see Table 1). In addition, we have observed the formation of fragments which show a similar molecular mass to that observed in the case of MMP-2, suggesting that the cleavage of the intact chains by plasmin occurs at a site topologically close to that of MMP-2. However, unlike the enzymatic action by MMP-2, in the case of plasmin we also observed at short intermediate time intervals the formation of larger fragments (probably derived from the cleavage of α- and β-chains), which then disappeared as the presence of the smaller fragments increased (see Figure 1C), indicating that plasmin could have an additional more efficient cleavage site.
Furthermore, peroxynitrite-treated fibrinogen (already incapable of any clotting activity even in the presence of thrombin [18]), displayed a similar proteolytic processing by whole MMP-2 (Figure 1D), producing a similar fragment to that observed for native fibrinogen, which suggested that the cleavage event(s) occur(s) at a similar site in native and in the peroxynitrite-treated fibrinogen. It is important to emphasize that from gradient electrophoresis observations no fragments of smaller molecular mass were observed (results not shown), ruling out the possibility of further fragmentation of the fibrinogen chains.
Degradation of fibrinogen by the catalytic domain of MMP-2
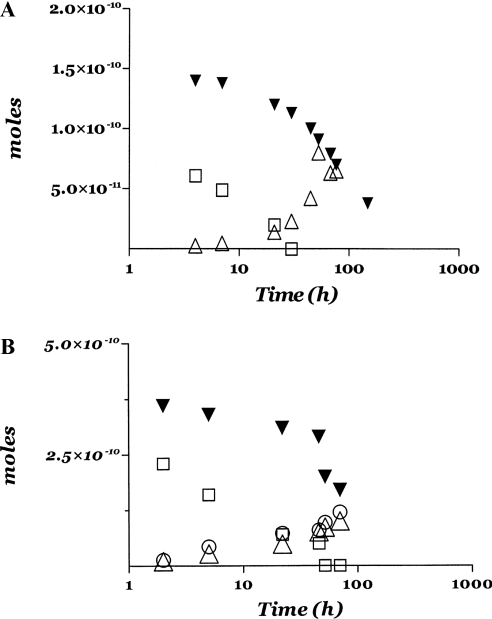
In order to clarify the role played by the haemopexin-like domain of MMP-2, we have also investigated the enzymatic processing of native fibrinogen by the catalytic domain of MMP-2. Figure 1(E) shows the time-dependent degradation of the three chains of fibrinogen by the catalytic domain of MMP-2 at pH 7.1 and 37 °C. It is immediately obvious that (i) the fragmentation occurs at a much lower rate with respect to the whole MMP-2 (Figures 1A and 2), and (ii) the proteolytic processing leads to the formation of a new fragment with a molecular mass intermediate between the β- and the γ-chain. This fragment looks similar to that observed during the early stages of the enzymatic processing of fibrinogen by plasmin (Figure 1C), but it is definitely not observed during the processing of native fibrinogen by whole MMP-2 (see Figures 1A and 1E). This difference becomes even more evident if we compare the time evolution of the increase in the different fragments with respect to the disappearance of the intact chains (Figure 2); thus, whereas in the case of whole MMP-2 the fragment(s) increase(s) concomitantly with the disappearance of the β-chain (Figure 2A), in the case of the fragment(s) formed by the catalytic domain of MMP-2 the time evolution of its appearance is correlated to the disappearance of both α- and β-chains (Figure 2B). Together, these data seem to indicate that cleavage of the α-chains by whole MMP-2 does not produce enough fragments of a large enough size to be detected, whereas cleavage of β-chains by whole MMP-2 occurs at a preferential class of sites, with the production of sizeable fragments. Furthermore, the lack of the haemopexin-like domain, when we have only the catalytic domain (Figure 1E), brings about cleavage events at additional sites with respect to those involved in the fragmentation by the whole MMP-2. Taken together, these data indicate an important role by the haemopexin-like domain in the correct recognition process, as already observed for MMP-8 toward collagen I [30]. It must be emphasized that the clotting activity was also abolished in the case of fibrinogen processed by the catalytic domain of MMP-2 (results not shown), in a similar fashion to that observed for the fibrinogen processed by whole MMP-2 (Figure 1B). This clearly indicates an important pathophysiological role for MMP-2 in modulating the clotting activity of fibrinogen, independently of its molecular state.
Figure 2. Temporal evolution of proteolytic processing of individual chains by MMP-2.
(A) Temporal evolution of the α- (□) and β- (▼) chain degradation and of the 40 kDa fragment formation (△) during the processing of bovine fibrinogen by whole MMP-2. The amounts of each species are expressed in terms of the number of moles. (B) Temporal evolution of the α- (□) and β- (▼) chain degradation and of the 50 kDa (○) and 40 kDa (△) fragment formation during the processing of bovine fibrinogen by the catalytic domain of MMP-2. Amounts of each species are expressed in terms of number of moles. T=37 °C and pH=7.1. For further details, see text.
In all cases (i.e. whole MMP-2 and catalytic domain with native and peroxynitrite-treated fibrinogen) the double reciprocal plot of the fibrinogen concentration and the enzymatic activity is linear for both chains of fibrinogen, clearly indicating that the first step occurs by a Michaelis–Menten mechanism (Figure 3). This consideration allows us to obtain the catalytic parameters for the cleavage of α- and β-chains of native fibrinogen by whole MMP-2 (Figure 3A) and its catalytic domain (Figure 3C), and by whole MMP-2 of fibrinogen treated with peroxynitrite (Figure 3B). These are reported in Table 1, showing that the lower enzymatic efficiency of the catalytic domain alone is due to a dramatically reduced kinetic constant for the rate-limiting cleavage step of the substrate.
Figure 3. Lineweaver–Burk plots of the proteolytic processing of bovine fibrinogen chains by MMP-2.
Lineweaver–Burk plots of the enzymatic processing by whole MMP-2 of α- (△) and β- (□) chains of native fibrinogen (A), peroxynitrite-treated fibrinogen (B) and of native fibrinogen by the catalytic domain of MMP-2 (C) at 37 °C and pH 7.1. Continuous lines have been obtained by non-linear least-squares fitting of data according to Eqn (1) and parameters are reported in Table 1. For further details, see text.
Molecular modelling
In order to verify whether the functional data described above have a structural explanation, we have performed molecular modelling of the interaction between MMP-2 and fibrinogen. Given that MMP-2 is likely to be flexible due to the long linker connecting the haemopexin domain to the catalytic domain, it would not be fruitful to attempt to dock the X-ray structure of the complete enzyme with fibrinogen as a rigid body. Therefore for the simulations of the docking between whole MMP-2 and fibrinogen we have regenerated a ‘flexible MMP-2’, deleting the 20 amino acid residues of the linker and forcing the distance between the N-terminus of the catalytic domain and the C-terminus of the haemopexin domain to be no longer than 20 Å (1 Å=0.1 nm) during the docking simulations. This provided us with the possibility of comparing the results for the two domains and, using constrained docking, we have attempted to simulate the mechanism by which the haemopexin-like domain may bring the catalytic domain into contact with fibrinogen at the correct region.
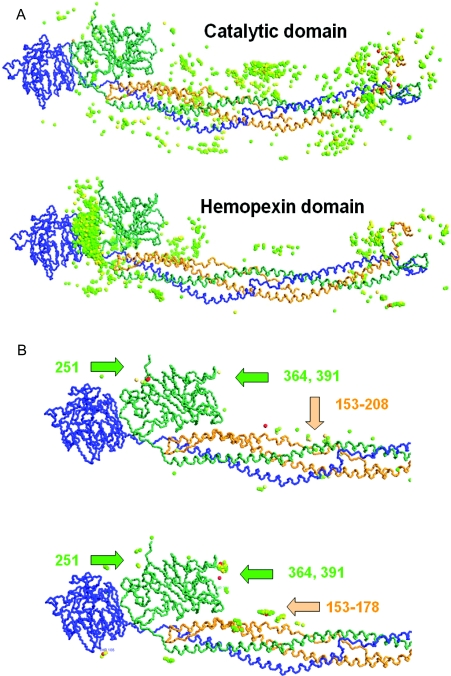
Figure 4(A) shows the results for the simulated docking of the catalytic domain (upper panel) and the haemopexin-like domain (lower panel) with fibrinogen. Both panels show 1000 models, with the domain of MMP-2 represented by a coloured sphere placed at its geometric centre in each model. The spheres are coloured according to the electrostatic interaction score, with red for the strongest (most negative) interactions, and green for the weakest interactions. We chose the electrostatic interaction score as the most representative of the interaction strength because it dominated strongly in the estimates given by BiGGER, with electrostatic energy being approx. 50 times stronger than the hydrophobic effects.
Figure 4. Molecular docking of MMP-2 on bovine fibrinogen.
(A) Comparison of 1000 models for the docking of the catalytic domain (top panel) and the haemopexin-like domain (bottom panel) with fibrinogen. The fibrinogen monomer is shown at the centre of each panel (with α-, β- and γ-chains identified by orange, green and blue respectively), and each MMP-2 domain is represented by coloured spheres placed at their geometric centres. The models were selected and coloured according to the electrostatic interaction score, with red indicating stronger interactions and green weaker interactions. For the results reported in (A) no constraints were used, as each domain was docked independently to compare the specificity of their interactions. (B) Comparison of the regions of fibrinogen found to be accessible to the catalytic domain. Spheres represent the position of the catalytic Zn2+. The top panel shows the models for the unconstrained docking of the catalytic domain; the bottom panel shows the models for the constrained docking of the catalytic domain with five complexes of the haemopexin-like domain and fibrinogen. Numbers indicate potential cleavage sites on the α- and β-chains of fibrinogen, as suggested by the docking of the complex between the catalytic domain and the haemopexin-like domain. For further details, see text.
These results (Figure 4A) suggest that the binding of the haemopexin-like domain is more specific, since a large fraction of the models place the haemopexin-like domain near the C-terminal region of fibrinogen (to the left on Figures 4A and 4B), whereas the catalytic domain models are more evenly spread throughout the structure of fibrinogen. Not apparent in the Figure, but also important, is that the top electrostatic interaction score given by BiGGER is 50% higher for the haemopexin-like domain than it is for the catalytic domain (−152 kCal/mol compared with −110 kCal/mol), suggesting that the interaction of the haemopexin-like domain is not only more specific but is also stronger.
The results shown in Figures 4(A) and 4(B) are for the docking of the active form of the catalytic domain, but we have also used the autoinhibitory procatalytic form of the catalytic domain, the catalytic domain lacking the fibronectin-like motif, and the fibronectin-like motif alone (results not shown). These simulations gave the results expected, producing neither a pattern of specific interactions nor interactions stronger than those for the active form of the catalytic domain of MMP-2. A closer examination of highest scoring models for the haemopexin-like domain docking indicated preferential binding sites which are grouped near the globular regions of the β-chains of the fibrinogen molecule. We selected the five highest ranking models from this simulation (according to the electrostatic score), and used the respective five complexes of fibrinogen with the haemopexin-like domain as targets for docking the catalytic domain of MMP-2. In addition, we imposed a constraint of 20 Å for the distance between the N-terminus of the haemopexin domain and the C-terminus of the catalytic domain, which corresponded to the length of the linker segment we deleted when separating these two domains from the X-ray structure of the complete enzyme [31] (see above). Figure 4(B) (lower panel) shows the models from these five docking simulations in which the catalytic Zn2+ atom of the catalytic domain is within 5 Å of the backbone of fibrinogen. The justification for this selection was the assumption that the haemopexin-like domain is responsible for the specific and stable binding to the fibrinogen, placing the catalytic domain near the regions where the backbone of fibrinogen is accessible to the catalytic centre. Figure 4(B) shows the comparison of these constrained docking results with a similar selection (catalytic Zn2+ within 5 Å of the fibrinogen backbone) of the models obtained by docking the catalytic domain alone to the fibrinogen with no constraints.
These docking simulation results are in agreement with the experimental results obtained with the digestion of fibrinogen by MMP-2, which show that the haemopexin-like domain greatly increases the catalytic rate of the catalytic domain, and suggest that the catalytic domain alone cleaves fibrinogen with less specificity than the complete enzyme does, leading to the formation of additional fragments (Figure 1E). Comparing the unconstrained with the constrained docking simulations for the catalytic domain (Figure 4B) we observed a similar pattern of models where the catalytic Zn2+ can interact with the fibrinogen backbone, but with a lower specificity in the case of the unconstrained docking (upper panel), which showed a more disperse distribution of the catalytic domain. More significantly, the docking simulations indicated cleavage sites on the α- and β-chains only, represented in yellow and green respectively (Figure 4B), and none on the γ-chain, again in agreement with the experimental data obtained in the digestion kinetic experiments.
An analogous analysis has not been possible for the α-chain, since most of this chain is not visible from available crystallographic data due to a widespread disordered structural arrangement of this chain. As a matter of fact, docking simulations focussed on the available portion of the α-chain did not show any preferential interaction with whole MMP-2.
Effect of the haemopexin-like domain on the digestion of fibrinogen by whole MMP-2
In order to test the reliability of predictions from molecular modelling on the crucial role played by the haemopexin-like domain in the interaction between whole MMP-2 and the β-chain (and probably the α-chain) of fibrinogen (Figure 4), we performed fragmentation kinetics by whole MMP-2 of fibrinogen incubated at 37 °C with 200 μM haemopexin-like domain. Thus if the hypothesis suggested by the molecular modelling is correct, at such a concentration the haemopexin-like domain should almost fully occupy the binding site, severely impairing the fragmentation of fibrinogen by whole MMP-2, as suggested by the Km value (Table 1). Figure 5 reports the outcome of this experiment performed in parallel with the fragmentation of fibrinogen alone, clearly showing that the presence of a 200 μM haemopexin-like domain greatly inhibits the fragmentation of the α- and β-chains of fibrinogen. This is a very important result since it (i) directly demonstrates the validity of the hypothesis formulated on the basis of molecular modelling, allowing us to propose the interaction of the haemopexin-like domain with the β-chain of fibrinogen as an actual mechanism and not simply as a working hypothesis; and (ii) rules out the possibility that other contaminating MMPs contribute significantly to the observed phenomenon, since the haemopexin-like domain can only interfere with intact MMP-2 and its reaction with fibrinogen is shown to essentially abolish the proteolytic cleavage.
Figure 5. Inhibitory effect of the haemopexin-like domain on the proteolytic processing of fibrinogen by MMP-2.
SDS/PAGE electrophoresis of fibrinogen degradation by whole MMP-2 after 20 h at 37 °C in the absence (lane 2) and in the presence of 200 μM haemopexin-like domain (lane 4). Lane 3 shows the intact fibrinogen. Lane 1 shows the the low molecular mass markers and lane 5 shows the broad molecular mass markers. Chains of fibrinogen and the haemopexin-like domain are indicated by arrows. For further details, see text.
The pH-dependence of catalytic parameters for MMP-2 on fibrinogen
The pH-dependence of catalytic parameters for MMP-2 on the α- and β-chains of native fibrinogen has been characterized and the values of pKaU (corresponding to values in the free enzyme) and pKaL (corresponding to values in the substrate-bound enzyme) have been determined on the basis of the pH-dependence of different catalytic parameters according to the following equations [26]
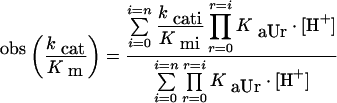 |
(2) |
 |
(3) |
 |
(4) |
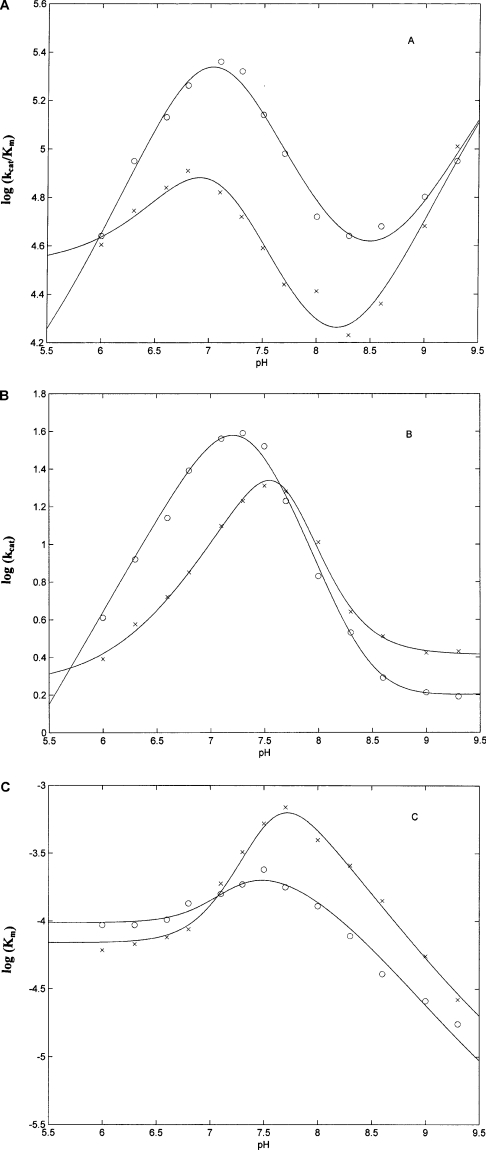
where obs(kcat/Km), obskcat and obsKm are the values of these catalytic parameters at a given pH, i (=0,1,…n) are the different protonation states [with (kcat/Km)0, kcat0 and Km0 the values of the catalytic parameters in the unprotonated form], KaUi (=10pKaUi) and KaLi (=10pKaLi) are the ith proton-binding constants for the free reactants and the enzyme–substrate complex respectively, and [H+] is the proton concentration. Experimental data have been fitted with Eqn 2–4, employing n=3 and this proton-linked modulation is described in Figure 6 with parameters reported in Table 2.
Figure 6. pH dependence of kcat/Km (A), kcat (B) and Km (C) for the processing of α- (○) and β- (x) chains of bovine fibrinogen by whole MMP-2 at 37 °C.
Continuous lines correspond to non-linear least-squares fitting of data according to Eqns 2, 3 and 4, and values of pKaU and pKaL for the two chains are reported in Table 2. For further details, see text.
Table 2. Values of pKaU and pKaL for the pH dependence of enzymatic processing of α- and β-chains of fibrinogen by whole MMP-2 at 37 °C.
| α-Chain | β-Chain | |
|---|---|---|
| pKaU1 | 10.04±0.15 | 10.04±0.17 |
| pKaU2 | 4.07±0.16 | 4.07±0.15 |
| pKaU3 | 9.97±0.17 | 9.97±0.18 |
| pKaL1 | 7.86±0.16 | 4.86±0.17 |
| pKaL2 | 4.52±0.16 | 7.72±0.15 |
| pKaL3 | 10.05±0.17 | 10.32±0.17 |
The data in Table 2 make it very clear that the pH-dependence profile of the overall enzymatic activity (i.e. kcat/Km) is similar for the two chains and quite complex (Figure 6A), requiring the involvement of (at least) three protonating groups. On the other hand, the pH-dependence of the rate-limiting step (i.e. kcat) shows a somewhat simpler bell-shaped profile and it appears to be slightly displaced to more alkaline pH values in the case of the β-chain (Figure 6B). As a consequence, a similar displacement is observed for Km (Figure 6C), which is affected only by those groups whose pKa values change upon substrate binding (see Table 2).
It is important to remark that resulting pKa values are different from those reported previously for MMP-2 against synthetic substrates [26]; this is quite obvious, since different residues are involved in the two cases, as indicated by the fact that MMP-2 interacts through the haemopexin-like domain with fibrinogen, whereas the binding of synthetic substrates only involves residues in the immediate vicinity of the catalytic site. Obviously, we cannot rule out the possibility that some of these pKa values also refer to residue(s) of fibrinogen, which change their value upon interaction with MMP-2.
From an overall view of the pH-dependence of these three catalytic parameters it emerges that the decrease of kcat/Km between pH 9.5 and pH 8.5 (Figure 6A) is due to a marked increase of Km (that is a decrease of substrate affinity) over the same pH range (Figure 6C), which is only modestly compensated by a small increase of kcat (Figure 6B). On the other hand, the enhancement of kcat/Km between pH 8.5 and pH 7.0 (Figure 6A) is mostly due to the increase of kcat, which attains the maximum value at pH 7.2 for the α-chain and at pH 7.5 for the β-chain (Figure 6B). Over the same range, Km is providing only a minor contribution, since it attains the minimum affinity value at pH 7.4 for the α-chain and at pH 7.8 for the β-chain (Figure 6C). The decrease of kcat/Km at pH<7.0 (Figure 6A) is again induced by the decrease of the rate-limiting step kcat (Figure 6B), which overwhelms the increasing affinity of MMP-2 for the substrate (as indicated by the decrease of Km; Figure 6C). Therefore we can conclude that the proteolytic activity of MMP-2 towards the two chains of fibrinogen is dominated at alkaline pH values by the substrate affinity, whereas as the pH is lowered the catalytic efficiency becomes dominated by the velocity of the rate-limiting step.
DISCUSSION
The role of MMP-2 in the neovascularization process is very well established and so is its capability to enzymatically process fibrinogen and cross-linked fibrin [8]. A very important aspect of the enzymatic action of MMP-2 on fibrinogen is the observation that the cleavage brings about an impairment in the formation of fibrin clots upon exposure to the action of thrombin (Figure 1B). It clearly means that this process induces a gross alteration in the coagulation process with important pathological consequences. Therefore since MMP-2 and fibrinogen are coming into close contact in the bloodstream, a kinetic analysis of the proteolytic processing by whole MMP-2 of native fibrinogen and of peroxynitrite-treated fibrinogen (a condition present under oxidative stress and which is known to impair the formation of clots [18]) is of the utmost importance for the comprehension of such a relevant event. This is clearly demonstrated by the fact that the catalytic efficiency of MMP-2 on fibrinogen (as indicated by kcat/Km) is somewhat lower than that of thrombin [20] and similar to that displayed by plasmin [21]. In addition, in order to uncover the determinants of the enzyme–substrate recognition process, the enzymatic inactivation of native fibrinogen by the catalytic domain of MMP-2 and by intact MMP-2 in the presence of a saturating amount of haemopexin-like domain has been investigated, allowing the role of the haemopexin-like domain in this relevant process to be unequivocally established. Furthermore, the pH-dependence of catalytic parameters for the processing of both fibrinogen chains by whole MMP-2 allows the substrate-binding mechanism and the modulation of enzymatic activity by environmental conditions to be clarified.
Table 1 reports the catalytic parameters for this process at pH 7.1 and 37 °C, which indicates that the overall enzymatic activity, as indicated by values of kcat/Km, of intact MMP-2 is faster on the α-chain as compared with the β-chain of native fibrinogen (mostly due to a higher kcat; see Table 1), while it is closely similar for the two chains in the peroxynitrite-treated fibrinogen. Furthermore, the removal of the haemopexin-like domain brings about a dramatic 20-fold reduction in the enzymatic activity with respect to the intact MMP-2, though keeping the approx. 3-fold preferential action for the α-chain (mostly due to a difference in Km; see Table 1), suggesting a difference for the substrate- recognition process according to whether MMP-2 contains the haemopexin-like domain or not. This is in accordance with a variation in the binding mode between the whole MMP-2 and the catalytic domain. Thus in the first case binding is driven by the haemopexin-like domain (which has a similar affinity for the two chains, as suggested by the similar Km of intact MMP-2; Table 1), whereas in the second case the process is regulated by the interaction of the catalytic domain (which interacts differently with the cleavage site of the two chains, as suggested by the different kcat in the whole MMP-2; Table 1). This result, which is supported by data on collagen reported by others [32,33], would suggest that MMP-2 interacts with fibrinogen by first binding through the haemopexin domain, which then acts as an anchor to force the catalytic domain into contact with the substrate. The occurrence of this interaction, which would also be in agreement with previous data indicating that binding of the haemopexin-like domain is driven by electrostatic forces [34], is demonstrated unequivocally by the experiment shown in Figure 5, which shows that in the presence of the haemopexin-like domain the degradation of fibrinogen by whole MMP-2 is severely impaired. As a whole, at pH 7.1 a picture emerges in which whole MMP-2 binds the α- and β-chains of native fibrinogen with equal probability (as seen from the closely similar values of Km; Table 1) by means of the haemopexin-like domain (Figure 4). Moreover, data reported in Table 1 indicate that MMP-2 cleaves the two chains at a different rate, mostly because the catalytic domain appears to preferentially process the α-chain, which is probably more exposed.
It is interesting to observe that comparing the behaviour of intact MMP-2 with plasmin shows a drastic difference in that plasmin is also able to enzymatically process the γ-chain of fibrinogen (Figure 1C), whereas intact MMP-2 does not. Furthermore, observing the catalytic parameters reported in Table 1, it emerges that the catalytic efficiency (as expressed by kcat/Km) of intact MMP-2 is closely similar to that of plasmin on the α-chain of fibrinogen, as a result of a counterbalancing between the velocity of the rate-limiting step (as expressed by kcat), which is higher for intact MMP-2 and the substrate affinity (as expressed by Km), which is higher for plasmin. On the other hand, plasmin shows a 4-fold better activity with respect to intact MMP-2 on the β-chain of fibrinogen, which is almost fully attributable to a faster kcat (Table 1). As a whole, it seems that the enzymatic action of plasmin and MMP-2 are fairly similar with the exception of the fact that plasmin is able to cleave the γ-chain, probably reflecting the smaller size of the enzyme and its higher flexibility.
Interpretation of data obtained for the catalytic domain of MMP-2 on native fibrinogen appears somewhat puzzling compared with that of intact MMP-2. Thus the removal of the haemopexin-like domain seems to bring about a dramatic decrease (by two orders of magnitude) for the rate constant of the rate-limiting step (i.e. kcat), partially compensated by an increased affinity by about one order of magnitude for both chains (as seen from the decrease of Km; Table 1). At first sight, this result seems to contradict the relevance of the role played by the haemopexin-like domain in the substrate recognition, since, this being the case, we would have expected a dramatic decrease of affinity, and thus an increase of Km, upon removal of the haemopexin-like domain. Actually, these data can be somehow reconciled with the picture given above for the whole MMP-2, suggesting a much more com-plex role of the haemopexin-like domain. Thus the drastically different value of kcat underlies a ‘correct’ substrate recognition (such that MMP-2 is able to cleave fibrinogen at the correct place) only in the presence of the haemopexin-like domain, suggesting an ‘unproductive’ enzyme–substrate complex in its absence, as also observed for collagen I [25,30,31,35]. Therefore the decrease of Km (Table 1) for the catalytic domain may be attributed to the appearance, consequent to the removal of the haemopexin-like domain, of a new interaction site between MMP-2 and fibrinogen, with the consequent formation of new fragments (Figures 1E and 2B) at a much less efficient rate.
A different effect must come into play in the cleavage mechanism of peroxynitrite-treated fibrinogen by whole MMP-2. In this case, it appears as if the peroxynitrite-induced oxidation of fibrinogen brings about a structural alteration of the molecule, which renders it more easily recognizable by MMP-2 (as seen by the lower value of Km; Table 1), but more resistant to the cleavage action (as seen by the lower value of kcat; Table 1). The net result is a similar overall enzymatic action by MMP-2 with respect to native fibrinogen (as seen by the similar value of kcat/Km, Table 1; and the same type of fragment produced by the cleavage action, Figure 1). This conformational change occurring in peroxynitrite-treated fibrinogen is also probably responsible for the disappearance of catalytic heterogeneity of the two chains. It makes the interaction of MMP-2 with both chains of fibrinogen much stronger (this feature being particularly evident for the β chain), but it also leads to a drastically reduced susceptibility of both chains to the proteolytic cleavage (the effect being especially pronounced for the β chain; Table 1).
An interesting aspect of catalytic parameters for the activity of intact MMP-2 on native fibrinogen is the close similarity of Km for both α- and β-chains (Table 1). The whole MMP-2 has the same affinity for α- and β-chains, suggesting the possibility that there is only one binding site for MMP-2 on fibrinogen. If this were true, the binding site could be located near the globular D-domain of the β-chain, as suggested above by the molecular modelling (Figure 4) and by the experiment in the presence of the haemopexin-like domain (Figure 5), and the same bound molecule could alternatively cleave either the α- and/or β-chains.
In order to discriminate between the possibility of a single binding site and that in which fibrinogen has two different binding sites for MMP-2 (one on the α-chain and one on the β-chain), we have investigated the pH-dependence of catalytic parameters for MMP-2 on the α- and β-chains of native fibrinogen. Thus following this approach the occurrence of a single binding site in fibrinogen for MMP-2 demands that the pH-dependence for Km is the same in the case of the α- and β-chain. This possibility is ruled out by the different pH-dependence of Km for the two chains (Figure 6C), unequivocally demonstrating that in fibrinogen there are two independent binding sites for MMP-2 (one at the α-chain and one at the β-chain).
A closer look at parameters reported in Table 2 shows that all three pKaU values are the same for both fibrinogen chains, indicating that the same groups of MMP-2 are involved in the proton-linked modulation of catalytic properties toward the two chains. This is also an indirect indication that two different MMP-2 molecules interact with the two chains, even though the interaction mode must differ for the two chains, as seen by the different pKa shifts (resulting in different pKaL values; Table 2).
The identification of groups involved in this proton-linked modulation of the enzymatic activity of MMP-2 towards fibrinogen is quite difficult, since an accurate identification would require an extensive study employing site-directed mutants. On the other hand, we cannot rule out the possibility that some of the pKa values refer to residue(s) of fibrinogen, which change their values upon interaction with MMP-2. However, some tentative attribution can be undertaken based on the pKa values and on their shift, taking into consideration the fact that not all three groups must necessarily be in close proximity to the active site.
In this respect, the residue responsible for pKa1, which displays a fairly high pKaU value (possibly referring to a tyrosine residue) before the enzyme–fibrinogen interaction, undergoes a drastic shift with a proton release upon substrate binding, much more pronounced for the interaction of MMP-2 with the β-chain (Table 2). This behaviour suggests that in the enzyme–substrate complex the environment is very polar and positively charged, with a consequent marked enhancement of its acidic character, which might render its protonation down to fairly low pH values very difficult.
On the other hand, the residue responsible for pKa2, which shows a quite low value for pKaU before the interaction (possibly referring to a histidine residue), changes very little upon the interaction of MMP-2 with the α-chain, but it is upshifted to a much larger extent when the enzyme interacts with the β-chain (Table 2). The different behaviour for the two chains suggests that this residue might be located in a region of MMP-2 which interacts with two drastically different regions of the two chains. Thus, unlike what is observed in the case of the residue responsible for pKa1 (see above), this residue weakens its acidic character taking up a proton upon interaction with the β-chain, while it is very marginally affected by the interaction with the α-chain.
In the case of the residue responsible for the pKa3, it displays a very marginal pKa increase upon substrate binding (by a similar extent for the two chains; Table 2), suggesting only a modest change in the environment upon substrate binding for both chains.
In conclusion, from the present study several novel aspects of this relevant process emerge, such that (i) different domains of MMP-2 contribute in a synergistic way to the proteolytic processing of fibrinogen; (ii) this proteolytic cleavage brings about the formation of an inactive form of fibrinogen, thus grossly affecting the coagulation process; (iii) MMP-2 binds with the haemopexin-like domain, enzymatically processing both α- and β-chains, whereas there is no evidence for the cleavage of the γ-chain; (iv) the peroxynitrite-treated fibrinogen undergoes a conformational change which affects the enzymatic mechanism by which MMP-2 processes fibrinogen; and (v) the pH-dependence of catalytic parameters for MMP-2 on fibrinogen required at least three protonating groups whose pKa values change upon substrate binding.
A final comment must emphasize that inactivation of fibrinogen by both whole MMP-2 and the catalytic domain of MMP-2 is of great pathophysiological relevance in view of the values of the catalytic parameters, not dramatically different from those observed for thrombin and plasmin [20,21] and for the important role played by MMP-2 in promoting inflammation and in favouring angiogenesis [7–9]. These data also give functional support to the crucial role played by MMP-2 in inducing vascular and circulatory alterations under chronic inflammatory conditions, such as those occurring in atherosclerosis and diabetes [36,37].
Acknowledgments
We acknowledge the financial support by the Italian Ministry of University and Research (MIUR COFIN 2003058409). We also express our gratitude to the COST (Co-operation for Science and Technology research) Action D21 for a short-term mission (to M.G.) at the Department of Chemistry of the Universidade Nova de Lisboa, and to Professor J.J. Moura (Departamento de Quimica, Facultade de Ciencias e Tecnologia, Universidade Nova de Lisboa, Portugal) for his hospitality and the use of the BiGGER software.
References
- 1.Sternlitch M. D., Werb Z. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell. Dev. Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan J. A., Docherty A. J., Barker P. J., Huskisson N. S., Reynolds J. J., Murphy G. Binding of gelatinases A and B to type-I collagen and other matrix components. Biochem. J. 1995;309:299–306. doi: 10.1042/bj3090299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borkakoti N. Matrix metalloproteases: variations on a theme. Prog. Biophys. Mol. Biol. 1998;70:73–94. doi: 10.1016/s0079-6107(98)00003-0. [DOI] [PubMed] [Google Scholar]
- 4.Schettler A., Thorn H., Jockusch B. M., Tschesche H. Release of proteinases from stimulated polymorphonuclear leukocytes. Evidence for subclasses of the main granule types and their association with cytoskeletal components. Eur. J. Biochem. 1991;197:197–202. doi: 10.1111/j.1432-1033.1991.tb15899.x. [DOI] [PubMed] [Google Scholar]
- 5.Van Hinsebergh V. W. M., Collen A., Koolwijk P. Role of fibrin matrix in angiogenesis. Ann. N.Y. Acad. Sci. 2001;936:426–437. doi: 10.1111/j.1749-6632.2001.tb03526.x. [DOI] [PubMed] [Google Scholar]
- 6.Fang J., Shing Y., Wiedereschain D., Yan L., Butterfield C., Jackson G., Harper J., Tamvakopoulos G., Moses M. A. Matrix metalloproteinase-2 is required for the switch to the angiogenic phenotype in a tumour model. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3884–3889. doi: 10.1073/pnas.97.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiraoka N., Allen E., Apel I. J., Gyetko M. R., Weiss S. J. Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell. 1998;95:365–377. doi: 10.1016/s0092-8674(00)81768-7. [DOI] [PubMed] [Google Scholar]
- 8.Bini A., Itoh Y., Kudryk B. J., Nagase H. Degradation of cross-linked fibrin by matrix metalloproteinase 3 (stromelysin 1): hydrolysis of the γ Gly404–Ala405 peptide bond. Biochemistry. 1996;35:13056–13063. doi: 10.1021/bi960730c. [DOI] [PubMed] [Google Scholar]
- 9.Bini A., Wu D., Schnuer J., Kudryk B. J. Characterization of stromelysin 1 (MMP-3), matrilysin (MMP-7), and membrane type 1 matrix metalloproteinase (MT1-MMP) derived fibrin(ogen) fragments D-dimer and D-like monomer: NH2-terminal sequences of late-stage digest fragments. Biochemistry. 1999;38:13928–13936. doi: 10.1021/bi991096g. [DOI] [PubMed] [Google Scholar]
- 10.Mosesson M. W., Siebenlist K. R., Meh D. A. The structure and biological features of fibrinogen and fibrin. Ann. N.Y. Acad. Sci. 2001;936:11–30. doi: 10.1111/j.1749-6632.2001.tb03491.x. [DOI] [PubMed] [Google Scholar]
- 11.Lambert V., Wielockx B., Munaut C., Galopin C., Jost M., Itoh T., Werb Z., Baker A., Libert C., Krell H.-W., Foidart J.-M., Rakic J.-M. MMP-2 and MMP-9 synergize in promoting choroidal neovascularization. FASEB J. 2003;17:2290–2292. doi: 10.1096/fj.03-0113fje. [DOI] [PubMed] [Google Scholar]
- 12.Diaz M. N., Frei B., Vita J. A., Keaney J. F., Jr Antioxidants and atherosclerotic heart disease. N. Engl. J. Med. 1997;337:408–416. doi: 10.1056/NEJM199708073370607. [DOI] [PubMed] [Google Scholar]
- 13.Buttery L. D. K., Springall D. R., Chester A. H., Evans T. J., Standfield N., Parums D. V., Yacoub M. H., Polak J. M. Inducible nitric oxide synthase is present within human atherosclerotic lesions and promotes the formation and activity of peroxynitrite. Lab. Invest. 1996;75:77–85. [PubMed] [Google Scholar]
- 14.Baker C. S. R., Hall R. J. C., Evans T. J., Pomerance A., Macflou J., Creminon C., Yacoub M. H., Polak J. M. Cyclooxygenase-2 is widely expressed in atherosclerotic lesions affecting native and transplanted human coronary arteries and colocalizes with inducible nitric oxide synthase and nitrotyrosine particularly in macrophages. Arterioscler., Thromb., Vasc. Biol. 1999;19:646–655. doi: 10.1161/01.atv.19.3.646. [DOI] [PubMed] [Google Scholar]
- 15.Leeuwenburgh C., Rasmussen J. E., Hsu F. F., Mueller D. M., Pennathur S., Heinecke J. W. Mass spectrometric quantification of markers for protein oxidation by tyrosyl radical, copper, and hydroxyl radical in low density lipoprotein isolated from human atherosclerotic plaques. J. Biol. Chem. 1997;272:3520–3526. doi: 10.1074/jbc.272.6.3520. [DOI] [PubMed] [Google Scholar]
- 16.Cromheeke K. M., Kockx M. M., De Meyer G. R. Y., Bosman J. M., Bult H., Vrints C. J., Herman A. G. Inducible nitric oxide synthase colocalizes with signs of lipid oxidation/peroxidation in human atherosclerotic plaques. Cardiovasc. Res. 1999;43:744–754. doi: 10.1016/s0008-6363(99)00148-0. [DOI] [PubMed] [Google Scholar]
- 17.Shishehbor M. H., Hazen S. L. Inflammatory and oxidative markers in atherosclerosis: relationship to outcome. Curr. Atheroscler. Rep. 2004;6:243–250. doi: 10.1007/s11883-004-0038-1. [DOI] [PubMed] [Google Scholar]
- 18.Lupidi G., Angeletti M., Eleuteri A. M., Tacconi L., Coletta M., Fioretti E. Peroxynitrite-mediated oxidation of fibrinogen inhibits clot formation. FEBS Lett. 1999;462:236–240. doi: 10.1016/s0014-5793(99)01500-8. [DOI] [PubMed] [Google Scholar]
- 19.Vadseth C., Souza J. M., Thomson L., Seagraves A., Nagaswami C., Scheiner M., Torbet J., Vilaire G., Bennett J. S., Marciano J.-C., Muzykantov V., Penn M. S., Hazen S. L., Weisel J. W., Ischiropoulos H. Pro-thrombotic state induced by post-translational modification of fibrinogen by reactive nitrogen species. J. Biol. Chem. 2004;279:8820–8826. doi: 10.1074/jbc.M306101200. [DOI] [PubMed] [Google Scholar]
- 20.Scheraga H. A. The thrombin–fibrinogen interaction. Biophys. Chem. 2004;112:117–130. doi: 10.1016/j.bpc.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Kastrikina T. F., Taran L. D., Kudinov S. A. Kinetic characteristics of fibrinogen and fibrin hydrolysis by plasmin 1 and 2 and miniplasmin. Thromb. Res. 1986;41:681–688. doi: 10.1016/0049-3848(86)90365-8. [DOI] [PubMed] [Google Scholar]
- 22.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Uppu R. M., Squadrito G. L., Cueto R., Pryor W. A. Synthesis of peroxynitrite by azide-ozone reaction. Methods Enzymol. 1996;269:311–321. doi: 10.1016/s0076-6879(96)69032-6. [DOI] [PubMed] [Google Scholar]
- 24.Nagase H., Suzuki K., Morodomi T., Enghild J. J., Salvesen G. Activation mechanisms of the precursors of matrix metalloproteinases 1, 2 and 3. Matrix Suppl. 1992;1:237–244. [PubMed] [Google Scholar]
- 25.Tam E. M., Moore T. R., Butler G. S., Overall C. M. Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): the differential roles of the MMP haemopexin c domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J. Biol. Chem. 2004;279:43336–43344. doi: 10.1074/jbc.M407186200. [DOI] [PubMed] [Google Scholar]
- 26.Fasciglione G. F., Marini S., D'Alessio S., Politi V., Coletta M. pH- and temperature-dependence of functional modulation in metalloproteinases. A comparison between neutrophil collagenase and gelatinases A and B. Biophys. J. 2000;79:2138–2149. doi: 10.1016/S0006-3495(00)76461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Cristofano R., Di Cera E. Phenomenological analysis of the clotting curve. J. Protein. Chem. 1991;10:455–468. doi: 10.1007/BF01025473. [DOI] [PubMed] [Google Scholar]
- 28.Palma P. N., Krippahl L., Wampler J. E., Moura J. J. G. BiGGER: a new (soft) docking algorithm for predicting protein interactions. Proteins. 2000;39:372–384. [PubMed] [Google Scholar]
- 29.Hiller O., Lichte A., Oberpichler A., Kocourek A., Tschesche H. Matrix metalloproteinases collagenase-2, macrophage elastase, collagenase-3, and membrane type 1-matrix metalloproteinase impair clotting by degradation of fibrinogen and factor XII. J. Biol. Chem. 2000;275:33008–33013. doi: 10.1074/jbc.M001836200. [DOI] [PubMed] [Google Scholar]
- 30.Gioia M., Fasciglione G. F., Marini S., D'Alessio S., De Sanctis G., Diekmann O., Pieper M., Politi V., Tschesche H., Coletta M. Modulation of the catalytic activity of neutrophil collagenase MMP-8 on bovine collagen I. Role of the activation cleavage and of the hemopexin-like domain. J. Biol. Chem. 2002;277:23123–23130. doi: 10.1074/jbc.M110873200. [DOI] [PubMed] [Google Scholar]
- 31.Morgunova E., Tuuttila A., Bergmann U., Isupov M., Lindqvist Y., Schneider G., Tryggvason K. Structure of human pro-matrix metalloproteinase-2: activation mechanism revealed. Science. 1999;284:1667–1670. doi: 10.1126/science.284.5420.1667. [DOI] [PubMed] [Google Scholar]
- 32.Patterson M. L., Atkinson S. J., Knäuper V., Murphy G. Specific collagenolysis by gelatinase A, MMP-2, is determined by the hemopexin domain and not the fibronectin-like domain. FEBS. Lett. 2001;503:158–162. doi: 10.1016/s0014-5793(01)02723-5. [DOI] [PubMed] [Google Scholar]
- 33.Tam E. M., Wu Y. I., Butler G. S., Stack M. S., Overall C. M. Collagen binding properties of the membrane type-1 matrix metalloproteinase (MT1-MMP) hemopexin C domain. The ectodomain of the 44-kDa autocatalytic product of MT1-MMP inhibits cell invasion by disrupting native type I collagen cleavage. J. Biol. Chem. 2002;277:39005–39014. doi: 10.1074/jbc.M206874200. [DOI] [PubMed] [Google Scholar]
- 34.Gohlke U., Gomis-Ruth F., Crabbe T., Murphy G., Docherty A. J., Bode W. The C-terminal (haemopexin-like) domain structure of human gelatinase A (MMP2): structural implications for its function. FEBS Lett. 1996;378:126–130. doi: 10.1016/0014-5793(95)01435-7. [DOI] [PubMed] [Google Scholar]
- 35.Overall C. M. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol. Biotechnol. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 36.Hayden M. R., Sowers J. R., Tyagi S. C. The central role of vascular extracellular matrix and basement membrane remodelling in metabolic syndrome and type 2 diabetes: the matrix preloaded. Cardiovasc. Diabetol. 2005;4:9–28. doi: 10.1186/1475-2840-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadoglou N. P., Daskalopoulou S. S., Perrea D., Liapis C. D. Matrix metalloproteinases and diabetic vascular complications. Angiology. 2005;56:173–189. doi: 10.1177/000331970505600208. [DOI] [PubMed] [Google Scholar]