Abstract
MicroRNAs (miRNAs) are an extensive class of ∼22-nucleotide noncoding RNAs thought to regulate gene expression in metazoans. We find that miRNAs are also present in plants, indicating that this class of noncoding RNA arose early in eukaryotic evolution. In this paper 16 Arabidopsis miRNAs are described, many of which have differential expression patterns in development. Eight are absolutely conserved in the rice genome. The plant miRNA loci potentially encode stem–loop precursors similar to those processed by Dicer (a ribonuclease III) in animals. Mutation of an Arabidopsis Dicer homolog, CARPEL FACTORY, prevents the accumulation of miRNAs, showing that similar mechanisms direct miRNA processing in plants and animals. The previously described roles of CARPEL FACTORY in the development of Arabidopsis embryos, leaves, and floral meristems suggest that the miRNAs could play regulatory roles in the development of plants as well as animals.
Keywords: miRNA, siRNA, ncRNA, Dicer, CARPEL FACTORY
A growing body of evidence suggests that ∼22-nucleotide (nt) noncoding RNA molecules play crucial roles as regulators of gene expression in eukaryotes. The first endogenous ∼22-nt RNAs to be identified were lin-4 RNA and let-7 RNA, both of which are key regulatory molecules in the pathway controlling the timing of larval development in the nematode Caenorhabditis elegans (Lee et al. 1993; Reinhart et al. 2000). When these RNAs are expressed, they pair to sites within the 3′ untranslated region (UTR) of target mRNAs, triggering the translational repression of the mRNA targets (Lee et al. 1993; Wightman et al. 1993; Reinhart et al. 2000; Slack et al. 2000). The mature lin-4 and let-7 RNAs are processed from the double-stranded region of RNA precursor transcripts by Dicer, a molecule with an N-terminal helicase and tandem C-terminal ribonuclease III domains (Bernstein et al. 2001; Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001). Argonaute homologs also influence the accumulation of the lin-4 and let-7 RNAs, but their biochemical roles are unclear (Grishok et al. 2001). Argonaute family members have a PAZ domain, which may allow protein–protein interaction with Dicer, as well as a Piwi domain, whose function is unknown (Cerutti et al. 2000).
The lin-4 and let-7 regulatory RNAs are now recognized as the founding members of a large class of ∼22-nt noncoding RNAs termed microRNAs (miRNAs), several of which are conserved from worms to humans (Pasquinelli et al. 2000; Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). RNAs are classified as miRNAs if they share the following features with lin-4 and let-7 RNAs: (1) The mature form of the RNA is a 20-nt to 24-nt species that is usually detectable on Northern blots. (2) The RNA has the potential to pair to flanking genomic sequences, placing the mature miRNA within an imperfect RNA duplex thought to be needed for its processing from a longer precursor transcript. In addition, miRNAs are typically derived from a segment of the genome that is distinct from predicted protein-coding regions. Thus far, >150 tiny RNAs that satisfy these criteria have been identified in animals (Lagos-Quintana et al. 2001, 2002; Lau et al. 2001; Lee and Ambros 2001; Mourelatos et al. 2002). The abundance of the miRNA genes, their intriguing expression patterns in different tissues or in different stages of development, and their evolutionary conservation imply that, as a class, miRNAs have broad regulatory functions in addition to the known roles of lin-4 and let-7 RNAs in the temporal control of developmental events. In support of this idea, six of the recently identified Drosophila miRNAs are complementary to 3′-UTR elements known to confer posttranscriptional regulation in this species (Lai 2002).
MicroRNAs are not the only small RNAs processed by Dicer. Dicer was originally identified as a nuclease involved in the RNA interference (RNAi) pathway of animals (Bernstein et al. 2001). This method of RNA silencing is triggered by long double-stranded RNA (dsRNA), typically introduced by injection or expression from a transgene (Fire et al. 1998). The dsRNA trigger is cleaved by Dicer into ∼22-nt RNAs (Bernstein et al. 2001). These ∼22-nt RNAs, known as small interfering RNAs (siRNAs), act as guide RNAs to target homologous mRNA sequences for destruction (Hammond et al. 2000; Zamore et al. 2000; Elbashir et al. 2001). RNAs ∼25 nt in length are also associated with posttranscriptional gene silencing (PTGS) in plants, and it has been suggested that a Dicer-like activity also produces these small RNAs (Hamilton and Baulcombe 1999; Matzke et al. 2001; Vance and Vaucheret 2001). RNAi, PTGS, and quelling of Neurospora are related pathways that require a conserved set of proteins (Hutvágner and Zamore 2002). For example, PTGS requires ARGONAUTE (Fagard et al. 2000), the RNA-directed RNA polymerase SDE1/SGS2, which may amplify dsRNA used as a trigger for silencing (Dalmay et al. 2000; Mourrain et al. 2000), and the RNA helicase SDE3 (Dalmay et al. 2001). Some aspects of RNA silencing may be species-specific, such as the RNA-directed DNA methylation required to maintain transgene silencing in plants (Morel et al. 2000; Bender 2001). Although RNA silencing has been proposed to have evolved as a viral defense mechanism (Vance and Vaucheret 2001), it can clearly be used by organisms for the regulation of endogenous genes. The Drosophila Argonaute family member aubergine is involved in the endogenous RNAi-like silencing of Stellate by dsRNA produced from both DNA strands of the Suppressor of Stellate locus (Aravin et al. 2001). It is possible that other animals or plants also generate endogenous siRNAs for gene regulation in development.
To further examine the roles of small RNAs in the regulation of plant gene expression, we cloned endogenous RNAs from Arabidopsis. Here we describe 16 plant RNAs that have the defining features of miRNAs. The presence of miRNAs in plants greatly expands the known phylogenetic distribution of this class of tiny noncoding RNAs and indicates that miRNAs arose early in eukaryotic evolution, before the last common ancestor of plants and animals. The presence of miRNAs in plants also suggests that the developmental defects of carpel factory (caf), a mutation in a Dicer homolog (Jacobsen et al. 1999), and mutations in ARGONAUTE family proteins (Bohmert et al. 1998; Moussian et al. 1998) could result from miRNA processing defects. In fact, we find that the accumulation of plant miRNAs is substantially reduced in the caf mutant. The ancient origin of miRNAs, together with the potential link between miRNAs and development, implies that miRNAs might have played roles during the origins and evolution of both plant and animal multicellular life.
Results
Identification of Arabidopsis miRNAs
Using methods designed to clone Dicer cleavage products, which are 20-nt to 24-nt RNAs with 5′-phosphate and 3′-hydroxyl groups (Bernstein et al. 2001; Elbashir et al. 2001; Hutvágner et al. 2001; Lau et al. 2001), ∼200 tiny RNAs were cloned from Arabidopsis seedlings and ∼100 were cloned from flowers. Of these, 18 sequences were represented by more than one clone and were the subject of further analysis. Of these 18 RNAs, 16 had striking similarities to the miRNAs of animals and have therefore been named miR156 through miR171, with genes designated MIR156 through MIR171 (Table 1). Six of the miRNAs represent three pairs of closely related RNA sequences differing only by one or two nucleotides. Interestingly, most of the plant miRNAs begin with a U, a trend previously observed in animal miRNAs (Lagos-Quintana et al. 2001; Lau et al. 2001).
Table 1.
MicroRNAs cloned from Arabidopsis
| miRNA gene
|
No.
of clones
|
miRNA sequence
|
miRNA length
(nt)
|
Oryza matches
|
Fold-back arm
|
Fold-back length
|
Chr.
|
Distance
to nearest
gene
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIR156a | 16 | U | G | A | C | A | G | A | A | G | A | G | A | G | U | G | A | G | C | A | C | 20–21 | 10 | 5′ | 82 | 2 | 3.2 kb downstream of At2g25100 (s) | |
| MIR156b | 5′ | 80 | 4 | 0.36 kb upstream of At4g30970 (a) | ||||||||||||||||||||||||
| MIR156c | 5′ | 83 | 4 | 3.2 kb downstream of At4g31875 (s) | ||||||||||||||||||||||||
| MIR156d | 5′ | 86 | 5 | 2.6 kb upstream of At5g10940 (s) | ||||||||||||||||||||||||
| MIR156e | 5′ | 96 | 5 | 1.6 kb downstream of At5g11980 (s) | ||||||||||||||||||||||||
| MIR156f | 5′ | 90 | 5 | 1.3 kb downstream of At5g26150 (a) | ||||||||||||||||||||||||
| MIR157a | 9 | U | U | G | A | C | A | G | A | A | G | A | U | A | G | A | G | A | G | C | A | C | 20–21 | — | 5′ | 91 | 1 | 1.8 kb downstream of At1g66780 (a) |
| MIR157b | 5′ | 91 | 1 | 2.7 kb downstream of At1g66790 (a) | ||||||||||||||||||||||||
| MIR157c | 5′ | 165 | 3 | 2.3 kb downstream of At3g18215 (a) | ||||||||||||||||||||||||
| MIR157d | 5′ | 173 | 1 | 1.0 kb upstream of At1g48470 (s) | ||||||||||||||||||||||||
| MIR158 | 8 | U | C | C | C | A | A | A | U | G | U | A | G | A | C | A | A | A | G | C | A | 20 | — | 3′ | 64 | 3 | 0.6kb upstream of At3g10750 (s) | |
| MIR159 | 8 | U | U | U | G | G | A | U | U | G | A | A | G | G | G | A | G | C | U | C | U | A | 21 | — | 3′ | 182 | 1 | 1.9 kb upstream of At1g73690 (s) |
| MIR160a | 4 | U | G | C | C | U | G | G | C | U | C | C | C | U | G | U | A | U | G | C | C | A | 21 | 4 | 5′ | 78 | 2 | 4.0 kb downstream of At2g39180 (a) |
| MIR160b | 5′ | 80 | 4 | 2.4 kb upstream of At4g17790 (a) | ||||||||||||||||||||||||
| MIR160c | 5′ | 81 | 5 | 1.5 kb upstream of At5g46850 (a) | ||||||||||||||||||||||||
| MIR161 | 16 | U | U | G | A | A | A | G | U | G | A | C | U | A | C | A | U | C | G | G | G | G | 20–21 | — | 5′ | 90 | 1 | 2.6 kb downstream of At1g48270 (a) |
| MIR162a | 3 | U | C | G | A | U | A | A | A | C | C | U | C | U | G | C | A | U | C | C | A | G | 21 | 1 | 3′ | 85 | 5 | 1.2 kb upstream of At5g08190 (s) |
| MIR162b | 3′ | 88 | 5 | 1.4 kb upstream of At5g23070 (s) | ||||||||||||||||||||||||
| MIR163 | 24 | U | U | G | A | A | G | A | G | G | A | C | U | U | G | G | A | A | C | U | U | CGAU | 24 | — | 3′ | 303 | 1 | 0.6 kb upstream of At1g66730 (s) |
| MIR164a | 21 | U | G | G | A | G | A | A | G | C | A | G | G | G | C | A | C | G | U | G | C | A | 21 | 2 | 5′ | 78 | 2 | 1.1kb upstream of At2g47590 (s) |
| MIR164b | 5′ | 149 | 5 | 2.4 kb upstream of At5g01750 (s) | ||||||||||||||||||||||||
| MIR165a | 2 | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | C | C | C | C | C | 20–21 | — | 3′ | 101 | 1 | 1.5 kb downstream of At1g01180 (a) |
| MIR165b | 3′ | 136 | 4 | 2.8 kb upstream of At4g00880 (s) | ||||||||||||||||||||||||
| MIR166a | 5 | U | C | G | G | A | C | C | A | G | G | C | U | U | C | A | U | U | C | C | C | C | 21 | 6 | 3′ | 136 | 2 | 4.7 kb upstream of At2g46690 (a) |
| MIR166b | 3′ | 112 | 3 | 3.5 kb upstream of At3g61900 (a) | ||||||||||||||||||||||||
| MIR166c | 3′ | 108 | 5 | 10 kb downstream of At5g08690 (s) | ||||||||||||||||||||||||
| MIR166d | 3′ | 101 | 5 | 22 kb downstream of At5g08740 (a) | ||||||||||||||||||||||||
| MIR166e | 3′ | 135 | 5 | 2.6 kb downstream of At5g41910 (a) | ||||||||||||||||||||||||
| MIR166f | 3′ | 91 | 5 | 1.1 kb downstream of At5g43600 (s) | ||||||||||||||||||||||||
| MIR166g | 3′ | 90 | 5 | 1.5 kb upstream of At5g63720 (s) | ||||||||||||||||||||||||
| MIR167a | 19 | U | G | A | A | G | C | U | G | C | C | A | G | C | A | U | G | A | U | C | U | A | 21 | 3 | 5′ | 101 | 3 | 4.7 kb upstream of At3g22890 (a) |
| MIR167b | 5′ | 90 | 3 | 0.19 kb downstream of At3g63370 (s) | ||||||||||||||||||||||||
| MIR168a | 3 | U | C | G | C | U | U | G | G | U | G | C | A | G | G | U | C | G | G | G | G | A | 21 | — | 5′ | 104 | 4 | 2.3 kb upstream of At4g19390 (a) |
| MIR168b | 5′ | 89 | 5 | 0.5 kb downstream of At5g45310 (s) | ||||||||||||||||||||||||
| MIR169 | 3 | C | A | G | C | C | A | A | G | G | A | U | G | A | C | U | U | G | C | C | G | A | 21 | 2a | 5′ | 190 | 3 | 1.9 kb downstream of At3g13400 (a) |
| MIR170 | 3 | U | G | A | U | U | G | A | G | C | C | G | U | G | U | C | A | A | U | A | U | C | 21 | — | 3′ | 64 | 5 | 0.5 kb downstream of At5g66040 (s) |
| MIR171 | 10 | U | G | A | U | U | G | A | G | C | C | G | C | G | C | C | A | A | U | A | U | C | 21 | 5b | 3′ | 92 | 3 | 0.5 kb downstream of At3g51380 (a) |
| — | — | 2 | in At2g45160 SCARECROW-like (a) | |||||||||||||||||||||||||
| — | — | 3 | in At3g60630 SCARECROW-like (a) | |||||||||||||||||||||||||
| — | — | 4 | in At4g00150 SCARECROW-like 6 (a) | |||||||||||||||||||||||||
Some miRNAs are represented by clones of different lengths due to heterogeneity of the RNA ends. The sequence of the most abundant clone is shown. Both miR156 and miR161 clones were found with 5′ or 3′ heterogeneity. MIR160b and MIR161 each had one clone of the same size but in a register shifted 5′ of the sequence shown by 2 and 8 nucleotides, respectively. The number of perfect matches to the available rice genomic sequence (Oryza matches) are indicated, as is the arm of the predicted stem-loop precursor that contains the miRNA (Fold-back arm) and the minimum number of nt that would be required to from a fold-back structure bounded by the miRNA and the segment of the predicted precursor that pairs to the miRNA (Fold-back length). Oryza fold-backs have the miRNA in the same arm as their Arabidopsis homologs (Supplemental data available online at http://www.genesdev.org). Chromosomal (Chr) positions, distance to the nearest annotated gene, and the position of the miRNA, sense (s) and antisense (a), relative to the nearest gene are noted for all matches in the Arabidopsis genome.
One of the miR169 Oryza matches is at the end of a contig, precluding prediction of a fold-back precursor structure.
As with Arabidopsis, only one of the miR171 Orzya matches has a predicted fold-back characteristic of miRNAs.
Five of the plant miRNA sequences have a single copy in the Arabidopsis genome, whereas each of the other 11 sequences correspond to multiple (2–7) loci (Table 1), most likely because of duplications in the Arabidopsis genome (The Arabidopsis Genome Initiative 2000). As expected for miRNA loci, nearly all (37 of 40) of the genomic loci lie outside of annotated segments of the genome, and thus do not correspond to previously identified genes. The three exceptions are for a single miRNA, miR171. Furthermore, each of these 37 loci place the cloned RNA sequence in a context where it can pair with a nearby genomic segment to form a dsRNA hairpin structure resembling those thought to be required for Dicer processing of miRNAs (Fig. 1; Supplemental data available online at http://www.genesdev.org). As with metazoans, the mature miRNA can be processed from either the 5′ or the 3′ arm of the fold-back precursor. Nevertheless, each miRNA with multiple matches to the genome is always present on the same arm of its potential precursors, suggesting that these loci share a common ancestry (see Supplemental data available online at http://www.genesdev.org). We do not know whether all of these loci are transcriptionally active or whether some might be pseudogenes.
Figure 1.
Fold-back secondary structures of Arabidopsis miRNA predicted precursors as determined by the RNAfold program. The miRNA sequences are in red. For miR156 and miR169, RNAs from the other side of the fold-back (boxed in blue) were each cloned once. The duplexes that could form between these RNAs and the miRNA from the other strand have ∼2-nt 3′ overhangs characteristic of Dicer cleavage (Elbashir et al. 2001).
The sizes of the predicted Arabidopsis hairpins are more variable than those of animals. For example, Caenorhabditis elegans miRNAs tend to be cleaved from precursors ∼70 nt in length, with the mature miRNA located only ∼2–10 bp from the terminal loop of the stem–loop (Lau et al. 2001). Although some of the Arabidopsis precursor predictions resemble those of C. elegans (Fig. 1), others are larger, as seen for the ∼190-nt predicted precursor of miR169 (Fig. 1).
In other systems, only one of the RNA strands accumulates following Dicer processing of miRNAs from the double-stranded region of the precursor, while the remainder of the precursor quickly degrades (Hutvágner et al. 2001). As a result, RNA from only one side of the miRNA precursor is typically cloned or detected on Northern blots, although on rare occasions RNA from the other side of the precursor is identified (Lau et al. 2001; Mourelatos et al. 2002), particularly if many clones are sequenced (E.G. Weinstein and D.P. Bartel, unpubl.). In contrast, Dicer processing of perfectly complementary dsRNA molecules in the RNAi pathway is thought to produce two stable overlapping ∼21-nt RNA molecules that pair to each other with ∼2-nt 3′ overhangs (Elbashir et al. 2001; Nykäken et al. 2001). As expected, for most (14/16) of the plant miRNAs, we cloned sequences from only one arm of the fold-back precursor. For two loci, we also cloned a single 21-nt sequence from the other arm of the fold-back (Fig. 1). The disparity in cloning frequency between the two sides, 16:1 in the case of MIR156, was similar to that seen for metazoan miRNAs (E.G. Weinstein and D.P. Bartel, unpubl.). The isolation of these two sequences generated from the opposite arm of the predicted fold-back supports the existence of these stem–loops as miRNA precursors. Furthermore, the duplexes that could be formed between the sequences isolated from both sides of the stems have 2-nt 3′ overhangs (Fig. 1), suggesting that they are products of a Dicer-like activity similar to that which processes the metazoan miRNAs (E.G. Weinstein and D.P. Bartel, unpubl.).
The Arabidopsis miRNAs display developmental expression differences
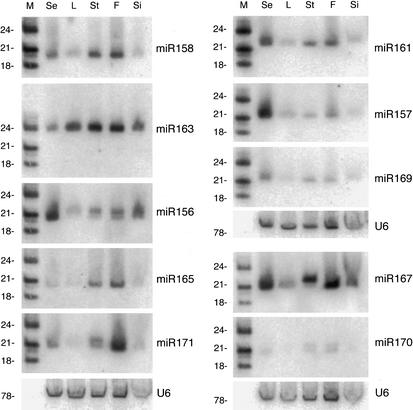
Northern analysis confirmed that the 16 miRNAs were stably expressed as ∼21-nt RNAs (Fig. 2). All are expressed at some level in seedlings, leaves, stems, flowers, and siliques (seed pods). Whereas miR163 accumulates in all tissues, with only slightly lower levels in seedlings and siliques, other miRNAs have quite variable levels among the tissues tested. For example, miR157 is most highly expressed in seedlings, and miR171 is most highly expressed in flowers, suggesting that they might play roles in the development of these stages/organs. The size of the RNAs detected approximately matches those that were cloned. In some cases, RNAs of two sizes can be detected, reflecting the heterogeneity of the cloned sequences (Table 1). For example, a probe to miR156 detects both 20-nt and 21-nt RNAs, and the miR156 clones were of both sizes. In another case, miR167, a 21-nt RNA accumulates in all tissues except stem, where a 22-nt RNA accumulates instead. This might reflect either differential transcription of the two MIR167 genes that have differently processed precursors or tissue-specific differences in the Arabidopsis miRNA processing machinery. We have not been able to reliably detect expression of RNAs in the size range of 60–200 nt that might correspond to the stem–loop precursors cleaved by Dicer.
Figure 2.
Developmental expression of Arabidopsis miRNAs. Total RNA from Columbia seedlings (Se), leaves (L), stems (St), flowers (F), and siliques (Si) was analyzed on Northern blots by hybridization to end-labeled DNA oligonucleotide probes complementary to the miRNA. The lengths of end-labeled RNA oligonucleotides run as a size marker (M) are noted to the left of each panel. Although miR165 and miR166 sequences and miR170 and miR171 sequences are too closely related to be reliably distinguished by hybridization probes, miR156 and miR157 should be specifically recognized (Lau et al. 2001), as reflected in their different levels of expression in seedlings and siliques. miR159 and miR164 show a similar expression profile to miR165, whereas miR160, miR162, and miR168 have similar profiles to miR158 (data not shown). The low expression level of most miRNAs in leaves and siliques might reflect a difference in the efficiency of small RNA recovery with the RNA isolation method used for these two tissues (see Materials and Methods). Blots were stripped and reprobed with an oligonucleotide probe complementary to U6 as a loading control.
Arabidopsis miRNAs are produced by CARPEL FACTORY
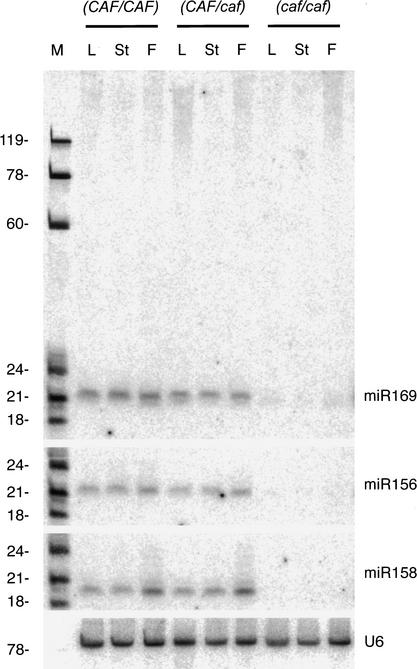
Although the presence of precursors in Arabidopsis was not detected on Northern blots, the potential for their production prompted us to investigate whether the ∼21-nt miRNAs might be processed from a longer dsRNA by proteins homologous to those that generate metazoan miRNAs. Dicer is thought to cleave the double-stranded region of the miRNA precursors in Drosophila, C. elegans, and humans (Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001; Lee and Ambros 2001). Mutations have been isolated in only one of the four Dicer homologs in Arabidopsis, CARPEL FACTORY (CAF; also named SHORT INTEGUMENT [SIN1]; GenBank accession no. AAG38019). The pleiotropic phenotypes associated with loss of CAF/SIN1 function, such as floral meristem proliferation defects, floral organ morphogenesis defects, and altered ovule development, emphasize the critical developmental role of RNAs processed by CAF (Robinson-Beers et al. 1992; Ray et al. 1996a,b; Jacobsen et al. 1999). Northern analysis showed that the expression level of the three miRNAs tested is significantly reduced in carpel factory homozygotes (Fig. 3). Although the level of miRNA precursors is increased when Dicer function is reduced in metazoans (Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001; Lee and Ambros 2001), we have not detected precursor accumulation in caf mutants (Fig. 3; data not shown).
Figure 3.
Expression of miR169 is dependent on CARPEL FACTORY. Total RNA from wild-type Landsberg erecta (CAF/CAF), heterozygous (CAF/caf), and homozygous (caf/caf) carpel factory leaves (L), stems (St), and flowers (F) was analyzed on a Northern blot. RNA size markers (M) are noted to the left. The blot probed for miR158 was stripped and reprobed with a U6 end-labeled DNA probe as a loading control.
Evolutionary conservation of Arabidopsis miRNAs in Oryza
The evolutionary conservation of miRNA sequences in different species indicates that they have important biological functions (Pasquinelli et al. 2000; Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). Eight Arabidopsis miRNAs have sets of identical matches in the genome of the rice Oryza sativa L. ssp. indica (Table 1), which was estimated to have 92% functional coverage at the time of our analysis (Yu et al. 2002). With rare exceptions (noted in Table 1), these sets of Oryzahomologs have adjacent sequences that could form stem–loop precursors analogous to those of Arabidopsis, with the miRNA sequence invariably on the same arm of the precursor in both species (see Supplemental data available online at http://www.genesdev.org). The Arabidopsis and Oryza sequences have drifted considerably in regions outside the miRNA sequence, but selective pressure can be seen in the segments predicted to base-pair with the miRNAs, resulting in only a few base changes in these segments and a conserved overall propensity for dsRNA formation (Fig. 4). For each set of related loci, the precursor duplexes extend beyond the length of the miRNA, but the sequence of the flanking duplex RNA is variable (see Supplemental data available online at http://www.genesdev.org). This conservation in secondary structure accompanied by variability in sequence provides added evidence that the secondary structural context of these RNAs is important, presumably for their processing from stem–loop precursors.
Figure 4.
Conservation between the Arabidopsis and Oryza predicted stem–loop precursors. (A) miR162 homologs. (B) miR164 homologs. Sequence homology is seen within the miRNA (in red), its paired sequences, and a few base pairs adjacent to the miRNA. The remainder of the sequence has drifted considerably, with the main constraint being the formation of a stem–loop structure.
An miRNA complementary to three related mRNAs
In nematodes, lin-4 and let-7 RNA recognize their target mRNAs through limited base-pairing to complementary sites within the 3′ UTR of their targets. The largest regions of uninterrupted complementarity are only ∼8 nt (Lee et al. 1993; Wightman et al. 1993; Reinhart et al. 2000; Slack et al. 2000). Consistent with this precedent, the plant miRNA sequences do not perfectly match coding regions, with the exception of miR171, which has four matches to the genome. One locus is 0.5 kb from the nearest predicted coding region and adjacent to genomic sequence that can form a classical miRNA precursor, consistent with the idea that it is a true miRNA. Further supporting this idea is the observation that a closely related sequence, miR170, was also cloned multiple times and has all the characteristics of the other plant miRNAs. However, the other three MIR171 loci differ from those of the other miRNAs (Table 1). They are anti-sense to the coding region of three SCARECROW-like genes of the GRAS family of putative transcription factors (DiLaurenzio et al. 1996; Pysh et al. 1999). This is the first example of a convincing miRNA candidate that is also the perfect anti-sense match to a coding region. Although this miR171 sequence identity might be a coincidence, the targets of this 21-nt RNA could include these three SCARECROW-like genes. miR171 (and perhaps the related miRNA, miR170) might act like a translational regulator similar to the lin-4 and let-7 RNAs, or it might pair with these three genes for a very different type of regulatory interaction. miR171 could direct cleavage of the messages as if it were an siRNA of the RNAi pathway, or it could direct a nucleic acid modification such as the methylation of genomic DNA seen in PTGS and transcriptional gene silencing of plants. Interestingly, the five perfect matches to miR171 in Oryza also include one miRNA homolog and four anti-sense matches to SCARECROW family members. This observation raises the possibility that these SCARECROW segments might be conserved based on their function as miRNA targets in addition to their function in coding proteins.
Other endogenous small RNAs
The other two RNAs cloned multiple times, Seq C and Seq F in Figure 5, are not likely to be miRNAs. Expression of Seq F but not Seq C can be detected on Northern blots (data not shown). Nonetheless, neither appears to have the potential to form extended pairing with the adjoining sequence like that seen for the other 16 sequences. Interestingly, both of these sequences match single loci in the same 2.3-kb region of Chromosome 2 that is also the source of four other ∼22-nt RNAs that we cloned once (Fig. 5). These RNAs are unlikely to be simply degradation products of mRNAs. Only two of these six sequences correspond to the same DNA strand as the two predicted protein-coding genes in this 2.3-kb region. Moreover, one of the single-clone RNAs (Fig. 5, Seq B) is a 2-nt-offset reverse-complement of Seq C. A duplex formed between them would have 1-nt and 2-nt 3′ overhangs, reminiscent of Dicer cleavage products during RNAi (Elbashir et al. 2001). The high density of 21-nt to 22-nt RNAs cloned from this region implicates either endogenous RNAi or some other, unknown Dicer-mediated event.
Figure 5.
A cluster of small RNAs derived from Chromosome 2. Arrows represent the two predicted genes in this region, and vertical lines represent the genomic positions of the six cloned RNAs. Sequences of the RNAs are listed, with cloning frequencies in parentheses.
Discussion
We have described 16 plant miRNAs that have the characteristic features of metazoan miRNAs. Like the miRNAs of animals, the plant miRNAs are 20-nt to 24-nt endogenous RNAs detectable on Northern blots and are derived from one arm of an apparent stem–loop precursor through the action of Dicer. As with most of the metazoan miRNAs, most plant miRNAs begin with a U, are transcribed from independent genes, and are evolutionarily conserved. The discovery that the phylogenetic distribution of miRNAs extends to plants indicates that miRNAs arose early in eukaryotic evolution and suggests that they have been shaping gene expression since the emergence of multicellular life. Although the evolution of the RNAi and PTGS pathways and their related proteins has been attributed to defense against viruses and transposons (Ketting and Plasterk 2000; Vance and Vaucheret 2001), the presence of miRNAs in plants suggests that Dicer and Argonaute proteins also have ancient roles in miRNA processing and function.
One difference between plant and animal miRNAs is the dsRNA precursor from which the mature miRNAs are cleaved. Based on the length of RNA that would be necessary to allow the miRNA to be incorporated into an RNA duplex suitable for Dicer cleavage, we predict that plant miRNA precursors can be more than three times as large as those of animals (Table 1). However, we have not detected plant precursor molecules during our Northern analysis of wild-type or caf RNA. Our method may not be sufficiently sensitive to detect very low levels of precursors. Perhaps precursor transcripts are more rapidly cleaved and turned over in Arabidopsis than in metazoans, or plant precursors might be too large or diffuse in size for Northern analysis techniques maximized for the resolution of the ∼21-nt mature RNAs. For instance, plant miRNAs might be processed cotranscriptionally, directly from transient primary transcripts. This would be in contrast to metazoan miRNAs, which often appear to be processed from metastable stem–loop precursors that have been preprocessed from a primary transcript (Lau et al. 2001). Although the common role of Dicer homologs in the production of plant and animal miRNAs highlights the similarities between their mechanisms of production, there might be differences in the structure and production of precursors, cellular compartmentalization, timing of precursor processing, or types of cofactors involved in processing.
The increasing number of miRNAs being identified raises the question of what their cellular functions are. Although some might regulate translation via base-pairing to target gene 3′ UTRs in a manner similar to regulation by lin-4 and let-7 RNAs, it is not clear whether all will be found to perform similar biochemical functions. One hint that miRNAs could perform other types of RNA-mediated gene regulation is our finding that miR171 could interact with the coding region of three GRAS family transcription factors through perfect complementarity rather than the limited base-pairing seen between lin-4 and let-7 and the 3′ UTRs of their targets. If these genes are regulatory targets of miR171, the miRNA could act like other ∼21-nt regulatory RNAs and direct mRNA degradation or epigenetic modification of the genomic sequence.
A role for the miRNAs in development of both plants and animals is suggested by the phenotypes of Dicer and Argonaute family mutants. In C. elegans, developmental defects resulting from reduction of function of dcr-1 (Dicer) and alg-1/alg-2 (Argonaute-like gene) have been attributed to the improper processing of miRNA precursors and a reduction in mature miRNA expression (Grishok et al. 2001). The mutant animals essentially reiterate stem-cell-like divisions and delay the switch to a later-stage developmental program. An intriguing parallel in Arabidopsis is that mutant alleles of caf/sin1 delay the meristem switch from vegetative to floral development (Ray et al. 1996a) and cause overproliferation of the floral meristem (Jacobsen et al. 1999), which suggests a distant link between the pathways affected by Dicer mutants in plants and animals. Mutations in two Arabidopsis Argonaute family genes also alter meristem development. The argonaute mutants disrupt axillary shoot meristem formation and leaf development (Bohmert et al. 1998), and ZWILLE/PINHEAD is required for shoot meristem maintenance and floral development (Moussian et al. 1998; Lynn et al. 1999). The existence of miRNAs in plants suggests that aberrant processing of miRNAs could be responsible for some if not all of the developmental defects in caf mutants, and it is possible that the same will be true for argonaute or zwille/pinhead mutants. However, ARGONAUTE is also required for PTGS (Fagard et al. 2000; Morel et al. 2002), and a related protein is required for RNAi in animals (Tabara et al. 1999; Hammond et al. 2001; Williams and Rubin 2002). In fact, the Drosophila Argonaute family member aubergine, a gene required for oogenesis (Schupbach and Wieschaus 1991), is involved in the endogenous RNAi-like silencing of Stellate by dsRNA produced from both DNA strands of the Suppressor of Stellate locus (Aravin et al. 2001), raising the possibility that the Arabidopsis argonaute or caf phenotypes reflect the role of these proteins in the production of endogenous siRNAs that control gene expression. Further investigation of the roles of small RNAs such as those from the Chromosome 2 cluster (Fig. 5) will address this possibility.
Finally, we suspect that other classes of Dicer- and Argonaute-dependent small RNAs are present in Arabidopsis. Noncoding RNAs continue to be discovered in a wide range of organisms, and the roles they play in the cell are only beginning to be understood (Eddy 2001). In many ways, the most interesting possibility is that no one class of RNAs can be responsible for the phenotypes of Dicer and Argonaute family mutations because organisms use such a rich variety of RNA-mediated gene regulation in their development.
Materials and methods
Plant growth and RNA isolation
Total RNA from wild-type Arabidopsis thaliana (Columbia accession) was isolated from 6-day-old seedlings grown on agar-based medium overlaid with filter paper and from flowers and stems of 4-week-old plants grown in soil using Trizol (GIBCO BRL). Total RNA was prepared from leaves and siliques using a modification of the method described in Nagy et al. (1988), in which the LiCl precipitation was replaced by ethanol precipitation. For isolation of RNA from carpel factory plants, progeny of CAF/cafheterozygous plants (in the Landsberg erecta accession) were grown on medium supplemented with 12 μg/mL kanamycin for 8 d, after which kanamycin-resistant individuals were transferred to soil and grown for an additional 24 d under continuous illumination. Plants were then scored as having (caf/caf) or lacking (CAF/caf) the carpel factory phenotype (Jacobsen et al. 1999), and RNA was prepared from leaves, stems, and flowers using a modification of the Nagay et al. (1988) method (see above). Wild-type plants (Landsberg erecta accession) were processed similarly, except that seeds were originally sown on medium lacking kanamycin.
RNA analysis
Endogenous 18-nt to 26-nt RNAs from seedlings and flowers were isolated from total RNA by 15% PAGE and cloned as described (Lau et al. 2001). The laboratory protocol is available at http://web.wi.mit.edu/bartel/pub/. For Northern analysis, 20 μg of total RNA per lane was separated on a 15% polyacrylamide gel, electroblotted to a nylon membrane, and hybridized to end-labeled anti-sense DNA probes (Lee et al. 1993).
Sequence analysis
Sequences of RNA clones were compared with the Arabidopsis genome downloaded from ftp://ncbi.nlm.nih.gov/genbank/genomes/A_thaliana/ (13-Aug-2001). Predicted secondary structures were generated using the Zucker folding algorithm and manually inspected for fold-backs with the RNA sequence in the stem as is characteristic of metazoan miRNAs (Lau et al. 2001). To identify Oryza sativa homologs, the miRNAs were compared with the rice genome sequence downloaded from the Beijing Genomics Institute Web site at http://btn.genomics.org.cn/rice (first draft) using the BLAST algorithm, and the adjoining sequences were analyzed for fold-back secondary structures as described above.
Acknowledgments
We thank the Arabidopsis Biological Resource Center at Ohio State University for seeds segregating for the caf mutation, Nelson Lau for reagents and advice in the cloning of endogenous Dicer products, Lee Lim for advice on bioinformatics, and Phil Zamore and members of the Bartel laboratories for comments on the manuscript. This research was supported in part by the Robert A. Welch Foundation (C-1309).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bartel@rice.edu; FAX (713) 348-5154.
E-MAIL dbartel@wi.mit.edu; FAX (617) 258-6768.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1004402.
References
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AA, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in Drosophila melanogastergermline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Bender J. A vicious cycle: RNA silencing and DNA methylation in plants. Cell. 2001;106:129–132. doi: 10.1016/s0092-8674(01)00441-x. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:295–296. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsiscontrolling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase in Arabidopsisis required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLaurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsisroot. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Leneckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Boutet S, Morel J-B, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegansdevelopmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A novel species of small antisense RNA in posttranscriptional gene silencing. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond SC, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates posttranscriptional gene silencing in Drosophilacells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. RNAi: Nature abhors a double-strand. Curr Opin Genet Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNAseIII gene in Arabidopsiscauses unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Plasterk RHA. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SEJ, Bernstein E, Sijen T, Hannon GJ, Plasterk RHA. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lai EC. MicroRNAs are complementary to 3′ UTR motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsisdevelopment and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Matzke AJ, Pruss GJ, Vance VB. RNA-based silencing strategies in plants. Curr Opin Genet Dev. 2001;11:221–227. doi: 10.1016/s0959-437x(00)00183-0. [DOI] [PubMed] [Google Scholar]
- Morel J, Mourrain P, Béclin C, Vaucheret H. DNA methylation and chromatin structure affect transcriptional and posttranscriptional transgene silencing in Arabidopsis. Curr Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. ArabidopsisSGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsisembryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Kay SA, Chua N-H. Analysis of gene expression in transgenic plants. In: Gelvin SB, Schilperoort RA, editors. Plant molecular biology manual. 1988. , Part B4, pp. 1–29. Kluwer, Dordrect. [Google Scholar]
- Nykäken A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda M, Maller B, Srinivasan A, Fishman M, Hayward D, Ball E, et al. Conservation across animal phylogeny of the sequence and temporal regulation of the 21 nucleotide let-7heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Ray A, Lang JD, Golden T, Ray S. SHORT INTEGUMENT (SIN1), a gene required for ovule development in Arabidopsis, also controls flowering time. Development. 1996a;122:2631–2638. doi: 10.1242/dev.122.9.2631. [DOI] [PubMed] [Google Scholar]
- Ray S, Golden T, Ray A. Maternal effects of the short integumentmutation on embryo development. Dev Biol. 1996b;180:365–369. doi: 10.1006/dbio.1996.0309. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Bettinger JC, Pasquinelli AE, Rougvie AE, Horvitz HR, Ruvkun G. The 21 nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild-type Arabidopsisand two female-sterile mutants. Plant Cell. 1992;4:1237–1249. doi: 10.1105/tpc.4.10.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogasterII. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Vance V, Vaucheret H. RNA silencing in plants—Defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophilaembryos. Proc Natl Acad Sci. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]