Abstract
Proteasomes are responsible for most intracellular protein degradation in eukaryotes. The 20S proteasome comprises a dyad-symmetric stack of four heptameric rings made from 14 distinct subunits. How it assembles is not understood. Most subunits in the central pair of β-subunit rings are synthesized in precursor form. Normally, the β5 (Doa3) propeptide is essential for yeast proteasome biogenesis, but overproduction of β7 (Pre4) bypasses this requirement. Bypass depends on a unique β7 extension, which contacts the opposing β ring. The resulting proteasomes appear normal but assemble inefficiently, facilitating identification of assembly intermediates. Assembly occurs stepwise into precursor dimers, and intermediates contain the Ump1 assembly factor and a novel complex, Pba1–Pba2. β7 incorporation occurs late and is closely linked to the association of two half-proteasomes. We propose that dimerization is normally driven by the β5 propeptide, an intramolecular chaperone, but β7 addition overcomes an Ump1-dependent assembly checkpoint and stabilizes the precursor dimer.
Keywords: proteasome, ubiquitin
Introduction
The ubiquitin–proteasome system affords the major mechanism for regulated intracellular protein degradation in eukaryotes (Baumeister et al, 1998). Polymers of ubiquitin are conjugated to a substrate protein, resulting in recognition and destruction of the substrate by the 26S proteasome. Proteasome substrates include many important regulatory proteins as well as abnormal proteins. The 26S proteasome is a ∼2.5 MDa complex with a 20S core particle bound at one or both ends by a 19S regulatory particle (RP). The 20S proteasome core is a cylindrical structure comprising 28 subunits arranged in four coaxially stacked rings. An interior chamber houses the protease active sites; access to this chamber is restricted by a set of narrow channels in the outer α-subunit rings.
All 20S proteasome subunits are encoded by a family of related but distinct genes (Baumeister et al, 1998). In most archaea, proteasomes have just two subunit types, α and β. Eukaryotic 20S proteasome subunits can be grouped with either the archaeal α or β subunit. Both eukaryotic and archaeal proteasomes have an α7β7β7α7-ring arrangement, except that multiple distinct α- and β-type subunits are part of the same particle in eukaryotes. In yeast, seven different α-type and seven different β-type subunits are in each 20S particle (Chen and Hochstrasser, 1995; Groll et al, 1997). Besides the basic set of 14 subunits, vertebrates encode three additional subunits that specifically replace their closest paralogs when antigen-presenting cells are stimulated with γ-interferon (Aki et al, 1994). These ‘immunoproteasomes' have altered peptide-bond cleavage specificities. How the 20S proteasome assembles into its highly regular arrangement of subunits, including correct replacements with alternative subunits, is a key unsolved issue.
The β subunits carry the proteolytic active sites (Baumeister et al, 1998; Heinemeyer et al, 2004). Only three of the seven β subunits in eukaryotes bear functional sites: β1, β2, and β5. In yeast, these subunits are named Pre3 (β1), Pup1 (β2), and Doa3/Pre2/Prg1 (β5), and their activities can be followed by cleavage of small peptides that monitor postglutamyl peptide-hydrolyzing, trypsin-like, and chymotrypsin-like activities, respectively. All of the active subunits are synthesized with N-terminal propeptides, which are removed autocatalytically near the end of proteasome assembly. The resulting N-terminal threonine residue, Thr1, carries the catalytic nucleophile.
One of the least understood areas of 26S proteasome physiology is the mechanism by which this enormous complex is assembled in vivo from its 33–35 different subunits (reviewed in Kruger et al, 2001; Heinemeyer et al, 2004). Even for the 20S proteasome, which is understood in great structural detail (Baumeister et al, 1998), little is known about how it assembles. A handful of apparent assembly intermediates have been identified, but their components are still incompletely catalogued. The most abundant of these intermediates, loosely referred to as a ‘half-proteasome', has a full set of α subunits but in mammals lacks three or four of the β subunits (Nandi et al, 1997), suggesting an ordered addition of β subunits to a preformed α ring. Propeptide processing of the β subunits occurs following apposition of two half-proteasomes, yielding the mature 20S proteasome (Chen and Hochstrasser, 1996).
Certain protease propeptides can facilitate folding or assembly of the mature domain of the linked protein and have been called ‘intramolecular chaperones'; this concept has also been applied to 20S proteasome subunit propeptides (Chen and Hochstrasser, 1996; Zühl et al, 1997). Eukaryotic β-subunit propeptides, particularly the β5-subunit propeptide, aid assembly by mechanisms that are still obscure. The bacterial Rhodococcus β-subunit propeptides appear to promote subunit folding and α-ring assembly (Zühl et al, 1997; Kwon et al, 2004). The yeast β5 subunit and its mammalian ortholog LMP7 cannot incorporate efficiently into proteasomes in vivo without their propeptides (Chen and Hochstrasser, 1996; Schmidt et al, 1999). Little beyond this is known at present.
Efficient 20S proteasome biogenesis in vivo also requires a number of assembly factors (Heinemeyer et al, 2004; Hirano et al, 2005). The best characterized is Ump1 (Ramos et al, 1998). Loss of Ump1 in yeast causes a growth defect but is not lethal. Proteasome assembly in the mutant is inefficient and is associated with aberrations in the processing of active β-subunit precursors. Recently, a novel heterodimeric assembly factor, PAC1–PAC2, has been described in human cells (Hirano et al, 2005). PAC1–PAC2 appears to associate with and help assemble the α ring. No counterpart to this heterodimer was reported in yeast.
Here, we describe a series of genetic and biochemical experiments that yield significant new insights into eukaryotic 20S proteasome assembly. Assembly occurs stepwise into precursor dimers, and we observed that the intermediates, but not mature proteasomes, are associated with several assembly factors, including Ump1 and a novel complex, Pba1–Pba2, which is the likely equivalent of the vertebrate PAC1–PAC2 complex. Notably, β7 incorporation occurs late and is closely linked to association of the two half-proteasomes. On the basis of our findings, we propose a model in which the β5 propeptide drives proteasome dimerization, but β7 addition is needed to overcome an Ump1-dependent assembly checkpoint and stabilize the precursor dimer.
Results
Proteasomes in β5Δpro β7HC cells
Yeast cells normally require the β5 (Doa3) propeptide for proteasome assembly and cell viability (Chen and Hochstrasser, 1996). In a screen for high-copy suppressors of β5 propeptide mutants, we recently found that overexpression of the proteasome β7 (Pre4) subunit strongly suppresses this assembly defect (manuscript in preparation). Remarkably, suppression occurs even when the β5 propeptide is deleted entirely (β5Δpro).
An important question is whether 20S proteasomes assembled under these conditions differ from those assembled in cells with a normal β5 subunit. We affinity-purified proteasomes from β5 (wild type (WT)) and β5Δpro strains which expressed β7 from a high-copy plasmid and had a chromosomal Flag epitope-tagged allele of β4 (Ramos et al, 1998). Purified particles were resolved by two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), and gels were stained for protein (Figure 1A). WT 20S proteasomes yielded the expected pattern of 14 subunits. Proteasomes from the β5Δpro strain were indistinguishable. When β5 and β5Δpro proteasomes were assayed for peptide cleavage at the three distinct catalytic centers, the mutant particles also showed nearly WT activities (Figure 1B). Interestingly, β5Δpro incorporates very poorly into 20S proteasomes when cells coexpress full-length β5, indicating a strong assembly disadvantage for β5Δpro (Chen and Hochstrasser, 1996). However, when such cells overproduced β7, the amount of β5Δpro found in mature particles was greatly increased (Supplementary Figure S1).
Figure 1.

20S proteasomes from β5proΔ β7HC cells are similar to WT proteasomes. (A) Two-dimensional gel analysis of affinity-purified proteasomes. Cells expressed a β4 (PRE1) allele chromosomally tagged with Flag and His6 epitopes (β4′). 20S proteasomes (30 μg) purified on an anti-Flag column from β5 β7HC (MHY2831) and β5Δpro β7HC (MHY2832) cells were subjected to isoelectric focusing in a pH range of 3–10, followed by SDS–PAGE and GelCode Blue staining. (B) Normalized peptidase activities of the three active centers (averages of two measurements). The nearly identical reduction for all sites relative to WT likely reflects a slightly lower concentration for the mutant particles.
If extracts from yeast cells frozen in liquid N2 are resolved by fast protein liquid chromatography gel filtration at 4°C, transient assembly intermediates can be detected (Ramos et al, 1998). When this was done with β5Δpro cells overexpressing β7, a buildup of the ‘15S' or ‘half-proteasome' intermediate was observed (Figure 2A). The position of the 15S peak in the fractionation was determined by detection of the Ump1 assembly factor and β2 precursor (proβ2), which both accumulate there (Ramos et al, 1998). These data suggest that proteasome assembly is less efficient in cells lacking the β5 propeptide and that the β5 propeptide is important for a step in proteasome assembly after 15S formation.
Figure 2.

Accumulation of proteasomal subparticles in β5Δpro β7HC cells. (A) Superose-6 chromatography followed by SDS–PAGE and anti-HA immunoblot analysis of whole-cell extracts from WT (MHY2498) and β5Δpro (MHY2506) cells. Alleles encoding β2-HA2 and Ump1-HA2 proteins replaced the corresponding endogenous loci. Peak positions of mature proteasomes, proteasome precursor complexes, and free subunits are indicated at the top of each panel. (B) Superose-6 column fractions resolved by nondenaturing PAGE with subsequent anti-HA immunoblotting. Proteasomes and proteasome subparticles are indicated at right (see Figure 3 legend). Ferritin (437 kDa) and thyroglobulin (669 kDa) were used as approximate size markers.
We conclude that increased cellular levels of β7 allow assembly of structurally normal 20S proteasomes in the absence of the β5 propeptide and that the β5 propeptide and β7 are likely to facilitate a common step in proteasome assembly.
Proteasomal subparticles detected by native gel analysis
The enhanced accumulation of proteasomal subparticles in the β5Δpro β7HC strain suggested that this strain might aid identification of novel assembly intermediates. However, the resolution of Superose-6 chromatography was insufficient to separate what might be multiple distinct intermediates, based on the broad distribution of the proβ2 protein (Figure 2A). We therefore selected different fractions from the gel filtration column and further resolved them by nondenaturing PAGE. Such native gels allowed the detection of distinct proteasomal species by immunoblotting against tagged Ump1 and β2 proteins (Figure 2B). Detection was enhanced in fractions from cells lacking the β5 propeptide (right side), but importantly, they were also detected in WT cells upon longer film exposure (bottom panel). Therefore, these species are not the result of aberrant proteasome assembly caused by deletion of the β5 propeptide. Ump1-HA was detected exclusively in two of the faster migrating species (labeled 15S and 15S-Blm10; Supplementary Figure S2). These data indicate that the ‘15S half-proteasome' defined by gel filtration analysis likely represents a series of proteasomal subparticles rather than a single molecular complex.
Characterization of novel proteasomal subparticles
The amount of putative proteasome assembly intermediates present in bands such as those in Figure 2B was not sufficient for identification by liquid chromatography/tandem mass spectrometry (LC-MS/MS). We therefore replaced the gel filtration separation step with an affinity purification procedure (Verma et al, 2000) and purified proteasomes from large cultures. Immunopurified proteins were resolved on native gels (Figure 3A). Protein species similar to those seen in Figure 2B were observed. The presence of proteasomal subunits in the various bands was verified by immunoblotting (not shown). Individual bands were excised, and trypsin-generated peptides were identified by LC-MS/MS (Table I). Every complex that we examined in this way contained all seven different α subunits but contained different subsets of β subunits.
Figure 3.
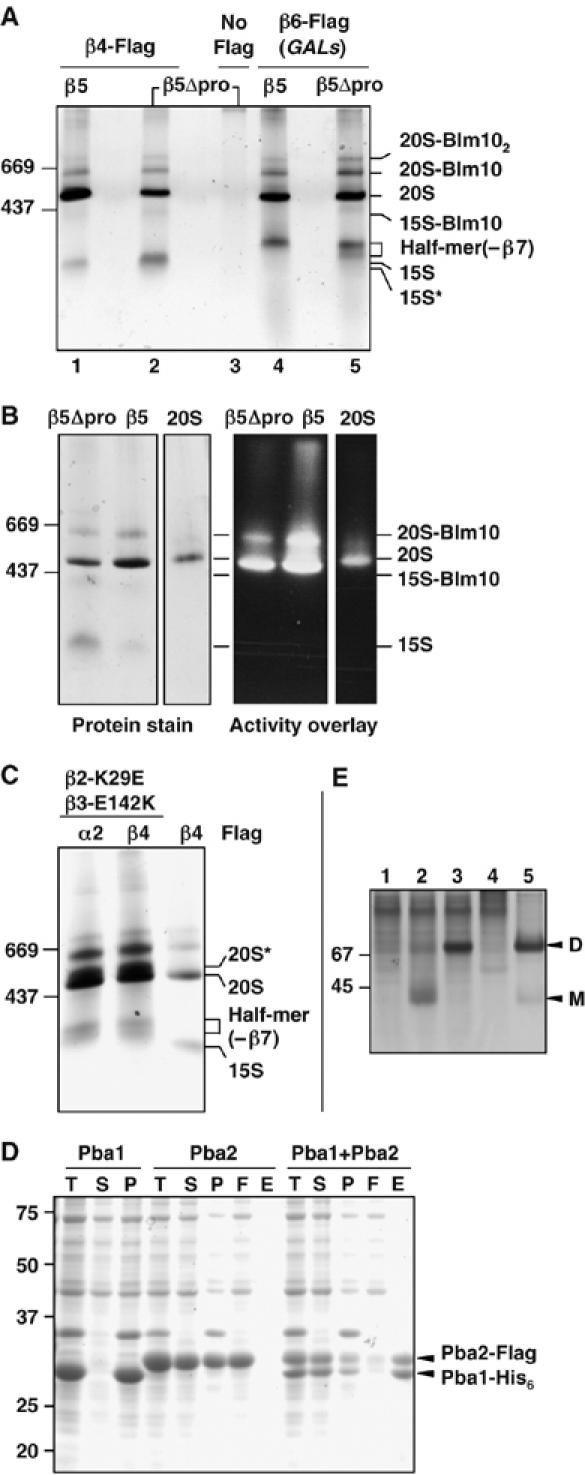
Native gel analysis of affinity-purified proteasomal particles. (A) GelCode Blue-stained native gel showing affinity-purified protein complexes. Extracts from yeast β5 or β5Δpro cells with either Flag-His6-tagged β4 (lanes 1 and 2) or β6 proteins (lanes 4 and 5) that also expressed β7 from a high-copy plasmid were purified on an anti-Flag affinity column. Control purification from untagged cells is shown in lane 3. (B) Native gel stained for protein (left) and assayed for peptidase activity by an activity overlay assay with the fluorogenic LLVY-AMC substrate (right). Conventionally purified 20S proteasomes (‘20S') were used for comparison. (C) Protein complexes affinity-purified with anti-Flag from β2-K29E β3-E142K cells and resolved on a GelCode Blue-stained native gel (first two lanes). For comparison, an anti-Flag purification from β5Δpro cells is also shown (last lane). (D) Pba1 and Pba2 bind directly to one another (GelCode Blue-stained SDS gel). T, total cell lysate; S, soluble fraction; P, pellet; F, Ni-NTA column flow-through; E, eluate from Ni-NTA. (E) Pba1 and Pba2 form a heterodimer. E. coli lysates expressing the following proteins were resolved by native PAGE (10%) and visualized with GelCode Blue: 1, Pba1-His; 2, Pba2-Flag; 3, Pba1-His+Pba2-Flag; 4, control lysate; 5, partially purified Pba1-His–Pba2-Flag complex. D, dimer; M, monomer.
Table 1.
Composition of proteasomal complexes determined by LC-MS/MS analysis (see Supplementary Table III for details)
| Particle name | Tagged subunit | Relevant genotype | α subunits | β subunits | Other |
|---|---|---|---|---|---|
| 15S* | β4 | β5Δpro β7HC | All | (β2), β3, β4 | Pba1 and Pba2 |
| 15S | β4 | β5Δpro β7HC | All | β2–β4 | Ump1, Pba1, and Pba2 |
| 15S-Blm10 | β4 | β5Δpro β7HC | All | β2–β4 | Blm10, Pba1 (Cdc48) |
| Half-mer (−β7) | β6 (GALs promoter) | β5Δpro β7HC and β5 β7HC | All | β1–β6 | Ump1, Pba1, and Pba2 |
| α2 or β4 | β2-K29E | All | β1–β6 | Ump1, Pba1, and Pba2 | |
| β3-E142K | |||||
| Precursor dimer (20S*) | α2 or β4 | β2-K29E | All | All | Ump1, Pba1, and Pba2 |
| β3-E142K | |||||
| Mature 20S | β4 | β5Δpro β7HC | All | All | None |
When proteasomes were purified via a Flag epitope tag on β4, four complexes in addition to mature 20S proteasomes were identified (Figure 3A; Table I). The most abundant species, labeled ‘15S', contained three β subunits, β2, β3, and β4, in addition to all the α subunits. The Ump1 assembly factor and two additional proteins, Pba1 (YLR199c) and Pba2 (YKL206c), were also detected. The Pba (proteasome biogenesis-associated) polypeptides are likely proteasome assembly factors (see below). Although the 15S Ump1-containing species detected by gel filtration was previously regarded as a ‘half-proteasome', we did not detect four of the β subunits in this gel band from either β5 or β5Δpro cells. The presence of Ump1 in this complex also demonstrated that the β5 propeptide is not necessary for Ump1 binding to the 15S proteasome precursor, consistent with in vitro data that indicate Ump1 can interact with β5Δpro (Heink et al, 2005). Intriguingly, the ‘13S intermediate' characterized in mammalian proteasome assembly also consists of the α ring plus β2, β3, and β4 (Nandi et al, 1997; Schmidtke et al, 1997), arguing for conservation of the 20S assembly mechanism.
The species labeled ‘15S*' appeared to be identical to 15S except that we failed to identify either Ump1 or β2 by LC-MS/MS. Because 15S* levels were very low, it was possible that these proteins were present but below the limit of detection. Anti-HA immunoblotting of cells expressing both HA-tagged Ump1 and β2 reacted with the 15S* complex (Figure 2B); however, when only Ump1 was HA-tagged, the 15S* species was not detected even with long film exposure (Supplementary Figure S2). This suggests that 15S* is identical to 15S except for the absence of Ump1. The simplest interpretation of these data is that Ump1 enters the assembly pathway after association of the β2, β3, and β4 subunits with the α ring and after Pba1 and Pba2 binding.
The band labeled ‘15S-Blm10' in Figure 2 contained the same proteasomal subunits found in the 15S complex but also included the ∼247 kDa Blm10 polypeptide (Fehlker et al, 2003; Schmidt et al, 2005). Blm10 binds one or both ends of the mature 20S proteasome cylinder. Consistent with this, we also detected Blm10 and mature 20S proteasome peptides in the two prominent bands above free 20S proteasome (Figures 2 and 3). Substrate overlay assays confirmed that the 20S-Blm10 species contains active enzyme, whereas no activity was evident in the 15S-Blm10 species (Figure 3B).
Several other yeast strains were tested for accumulation of potential proteasome assembly intermediates. First, we generated a matched pair of β5 or β5Δpro strains that expressed Flag-tagged β6 (Pre7) from a galactose-regulated promoter (GALs). This caused modest overexpression of β6 (not shown). As before, β7 was also expressed from a high-copy plasmid. Interestingly, a novel protein complex migrating slightly slower than the 15S intermediate was detected in these cells (Figure 3A, lanes 4 and 5). LC-MS/MS revealed a full set of α subunits, six of the seven β subunits, Ump1, Pba1, and Pba2. Remarkably, despite its strong overexpression and accumulation as a free subunit (Supplementary Figure S3A and unpublished data), β7 was missing from the complex, which we designated the ‘half-mer (−β7)'. Immunoblotting of protein complexes separated on native gels confirmed the absence of β7 from this complex; β7 was readily detected in the 20S-Blm10 complex, which, based on immunoblotting against β6, is less abundant than the half-mer (−β7) (Supplementary Figure S3B). These findings suggested that the increase in β6 levels could drive assembly beyond the 15S intermediate stage but that β7 integration became rate-limiting for dimerization. A proteasome intermediate of this composition has not been reported previously for any organism. We note that results with mammalian β7 are also consistent with late entry of β7 into assembling proteasomes (Thomson and Rivett, 1996).
Unexpected support for a half-mer (−β7) intermediate came from affinity purifications from a completely different strain, specifically, a β2-K29E β3-E142K mutant without overexpression of β6 or β7. This double mutant had been constructed as part of a pseudoreversion analysis to show that β2 and β3 were direct neighbors within the β ring (Arendt and Hochstrasser, 1997). The double mutant restores a salt bridge between β2 and β3 but reverses its polarity; it is still strongly defective for ubiquitin-dependent proteolysis. From the proteasome crystal structure, the β2-K29 and β3-E142 residues also make electrostatic contacts with an absolutely conserved pair of residues (E184 and R185) in the β6 subunit of the trans β ring. Given the structural constraints at this nexus of three subunits, the β2-K29E β3-E142K mutant should still be perturbed in its interactions with the trans β ring, and such a perturbation might yield increased levels of half-proteasomes or incompletely matured particles.
In affinity purifications with either Flag-tagged α2 or β4, the half-mer (−β7) was the fastest migrating species detected. This implied that the complementary β2-K29E and β3-E142K mutations restored sufficient intraring interactions between β2 and β3 to allow their incorporation into half-mers but still had a defect in dimerization. A defect in the dimer interface was also suggested by the prominent species just above the mature 20S proteasome in these purifications (20S* in Figure 3C): peptide sequencing of this complex showed that it contained all 14 proteasome subunits as well as Ump1, Pba1, and Pba2. Peptides derived from the β2 and β5 propeptides were also found, underscoring the fact that this was a precursor complex. On the basis of its migration on native gels, we infer that 20S* is the proteasomal precursor dimer (the ‘preholoproteasome'; see Baumeister et al, 1998), which had been postulated as a normal assembly intermediate (Chen and Hochstrasser, 1996; Heinemeyer et al, 2004). Presumably, the precursor dimer accumulates in this mutant as a result of a perturbed β-ring–β-ring interface that interferes with autocatalytic subunit cleavage and final proteasome maturation. Although its full composition was not analyzed, a complex of comparable size had been observed by Fehlker et al (2003), and this ‘nascent core particle' may be equivalent to the 20S* precursor dimer.
The number of MS/MS peptide spectra derived from each protein in a mixture can be compared, and such spectral counts correlate closely with relative protein amounts (Old et al, 2005). We compared spectral counts between each 20S subunit in each precursor complex relative to their counts in the mature 20S proteasome or the 20S-Blm102 complex, in which all 20S subunits are present at a 1:1 ratio (Supplementary Table IV). Using the Rsc metric (equal to the log2 ratio of abundance between two samples) of Old et al (2005), relative levels of all the subunits detected among the different complexes were statistically similar to those in the mature 20S proteasome and the 20S-Blm102 complex (Rsc<2). However, certain subunits that were not detected in some of the subcomplexes, such as β7 in the half-mer (−β7), were readily detected in the benchmark complexes, yielding Rsc values >2 in each case. Therefore, spectral counting suggests that these subunits were not missed because of undersampling by MS/MS but because they were underrepresented (absent) in the specific subcomplex. This is consistent with immunoblot analyses of these subcomplexes (Supplementary Figure S3B and data not shown).
In summary, native gel analysis of affinity-purified proteasomal complexes together with LC-MS/MS protein identification uncovered a multitude of potential 20S proteasome assembly intermediates and also identified a novel pair of potential assembly factors, Pba1 and Pba2.
A yeast assembly factor related to human PAC1–PAC2
A recent study on human 20S proteasome assembly implicated a heterodimer of the PAC1 and PAC2 proteins; this complex appears to help assemble the α ring and maintain it in a state competent for β-subunit incorporation (Hirano et al, 2005). Modest sequence similarity (19% identity) between human PAC2 and yeast YKL206c (Pba2) was noted by Hirano et al (2005), but no counterpart to PAC1 was detected. We have found weak similarity between human PAC1 and yeast Pba1 (YLR199c) (Supplementary Figure S4). Moreover, purification of recombinant Pba1 and Pba2 from Escherichia coli demonstrated interaction in what appears to be a 1:1 complex (Figure 3D); Pba1 was completely insoluble unless coexpressed with Pba2. Native gel analysis yielded an apparent size consistent with a heterodimer (Figure 3E). Therefore, Pba1 and Pba2 form a heterodimer just as the human PAC1 and PAC2 proteins do. The Pba1–Pba2 complex is found exclusively in proteasome precursor species, consistent with a role in proteasome assembly.
Loss of Pba1 and/or Pba2 caused a very mild defect in proteasome biogenesis based on the slightly increased accumulation of proβ5 (and Ump1) in assembly intermediates relative to processed β5 in mature particles (Supplementary Figure S5A). Consistent with this, substantial growth defects were not seen with pba1Δ and/or pba2Δ, but genetic interactions with the ump1Δ and pre9Δ proteasomal mutations were observed (Supplementary Figure S5B).
Importance of the β7 tail in suppressing loss of the β5 propeptide
The accumulation of half-mer (−β7) proteasomal subparticles under several different conditions, including cells in which β7 was highly overexpressed, suggested that β7 incorporation into nascent half-proteasomes is a slow step in the assembly pathway and closely coupled to half-mer dimerization. A striking feature of β7 is a unique 15-residue C-terminal tail that extends across the dyad axis and inserts into a groove between the β1 and β2 subunits in the opposing β ring (Figure 4A). The β7 tail likely facilitates half-mer dimerization (Ramos et al, 2004). If enhancement of dimerization underlies the ability of high-copy β7 to suppress the β5 propeptide deletion, then truncating the β7 tail should abolish suppression. Using a high-copy pre4-1 allele of β7, which deletes the last 15 residues of the subunit, this was indeed observed (Figure 4B). Levels of truncated β7 expression were comparable to WT (not shown). The Trp211 residue in the β7 tail is strictly conserved and is largely buried in the interface with β1 in the trans β ring (Figure 4A). Mutation of this residue to alanine also eliminated β5Δpro suppression.
Figure 4.

A β7tail trans-ring interaction with β1 required for suppression of β5Δpro lethality. (A) Structure of the central proteasomal β rings (from PDB 1RYP) viewed down the dyad axis. Figure was prepared with RasMol 2.6. Arrowheads indicate the strictly conserved Trp211 residue (white) in the β7 tail. (B) Deletion of the β7 tail (residues 211–225; pre4-1) or mutation of Trp211 to Ala abolishes β7HC suppression of β5Δpro. MHY1179 cells with the indicated YEplac181 plasmid-borne β7 alleles and pRS317 plasmid-borne β5 or β5Δpro were grown on fluoroorotic acid (FOA) medium (30°C, 5 days) to evict the WT β5 and β7 URA3-based plasmids originally present. (C) Mutation of β1 residues that disrupt the trans-interaction with the β7 tail impairs β7HC suppression of β5Δpro. MHY1179 cells cotransformed with plasmids encoding the indicated β7 and β1 alleles (and also carrying a pRS317 plasmid expressing either WT β5 or β5Δpro) were streaked on FOA and grown at 30°C for 3 days (top) or 6 days.
Alteration of residues in β1 that help form the tightly packed interface with Trp211 of the β7 tail also greatly impaired the ability of β7HC to suppress β5Δpro (Figure 4C). Both high-copy β1-G171W and β1-V172W abolished suppression, but they caused no growth defects in WT (β5) cells. Notably, the combined expression of the complementary β7-W211A and β1-V172W (but not β1-G171W) alleles weakly but reproducibly restored a small degree of β5Δpro suppressing activity (Figure 4C). These data strongly support the notion that a β7 tail interaction with the opposing β ring during proteasome precursor dimerization is central to proteasome assembly in the absence of the β5 propeptide and imply a key role of the β5 propeptide in precursor dimerization as well.
High-copy β7 also suppresses a specific β5 point mutant
If β7 overexpression stimulates a specific proteasome assembly step, it might be able to suppress the phenotypic abnormalities of other proteasome mutants if they were defective in the same assembly step. We tested for β7HC suppression of several β5 point mutants and a mutant for the neighboring β4 subunit, which are all temperature-sensitive and canavanine-hypersensitive (Figure 5A). Strikingly, a single mutant was suppressed, β5(doa3-1). High-copy mutant β7(pre4-1) did not suppress doa3-1 (not shown). Such allele-specific suppression is consistent with the possibility that the β5(doa3-1) mutant shares a defect in proteasome assembly with β5Δpro. To examine this, proteasomes from β5(doa3-1) cells (+/− β7HC) were fractionated by gel filtration, and proteasome maturation was assayed by immunoblotting against the β1 and β5 catalytic subunits (Figure 5B). A substantial fraction of proβ1 and proβ5 accumulated in the ‘15S' precursor fractions and as free subunits in β5 (doa3-1) cells that did not overexpress β7. With β7HC, a much larger portion of these precursors were processed and incorporated into mature 20S and 26S proteasomes. Therefore, the β5 (doa3-1) mutant is abnormal for proteasome assembly, and β7 overexpression can partially suppress this defect.
Figure 5.

High-copy β7HC suppresses a specific β5 point mutant. (A) Suppression analysis of various β5 and β4 alleles by β7HC. Top panel: mutant β5 (doa3/prg1) or β4 (pre1) or congenic WT strains were transformed with either empty vector or a high-copy β7 plasmid and then streaked on 1 μg/ml canavanine (30°C, 5 days). Lower panel: exponentially growing cultures were plated on YPD in 1:6 serial dilutions and incubated for 2 days at the indicated temperatures; the yeast strains were derived from MHY784 by transformation with the indicated plasmids followed by eviction of YCp50-DOA3 on FOA. (B) The β5 (doa3-1) mutant has an assembly defect that is partially rescued by β7HC. Panels show anti-β5 and anti-β1 immunoblots of Superose-6 column fractions from extracts of doa3-1 cells that had been transformed with empty vector or a high-copy β7 plasmid. Anti-β1 antibody cross-reacts with an unknown protein (asterisk).
The doa3-1 mutation changes the absolutely conserved Asp51 residue to an Asn residue. We have previously shown that β5 (doa3-1) proteasomes are globally altered in structure and tend to dissociate into half-mers when run on native gels (Chen and Hochstrasser, 1995). This suggests that a defect in the interface between half-proteasomes can impair assembly. As described above, this is suppressible by β7HC in a tail-dependent manner. Hence, these results strengthen the hypothesis that a key assembly function shared by the β5 propeptide and the β7 tail is in half-mer dimerization.
A link between β6 N-terminal sequences and Ump1
Given that β7HC can suppress β5Δpro lethality, we investigated whether the intervening β6 subunit might also affect this process. The β6 subunit has a 19-residue N-terminal propeptide that is expected to be in close proximity to β7 before β6 processing in the precursor dimer (Groll et al, 1999). Surprisingly, whereas β5Δpro is lethal in the presence of WT full-length β6, the β6Δ2-19 allele restored β5Δpro viability (Figure 6A). This suppression did not require overexpression of β7, although β7HC enhanced growth.
Figure 6.

Mutations of β6 and Ump1 have related effects on proteasome assembly. (A) The growth phenotype of β6Δ2-19. Strain MHY3040 was transformed with low-copy plasmids carrying the indicated alleles and then grown on FOA at 24°C to evaluate growth following eviction of the originally present URA3-marked WT β5 and β6 plasmids. (B) Suppression of β6Δ2-28 lethality by ump1Δ. Strains were grown on FOA at 24°C to evict the URA3/β6 plasmid originally present. (C) Native gel analysis of affinity-purified proteasomal particles from ump1 cells. Gel was stained with GelCode Blue. (D) Model for 20S assembly pathway showing new intermediates. Entry of Ump1, Pba1, and Pba2 proteins are shown but are not depicted in the complexes. Extensions on subunit ovals represent propeptides except for β7, where the extension depicts the C-terminal tail that inserts between the β1 and β2 subunits in the opposing β ring. The β5 propeptide, which is highlighted, is proposed to help align and join half-mers.
The paradoxical inhibitory effect of the β6 propeptide on β5Δpro cell viability recalled a similar effect of the Ump1 proteasome assembly factor (Ramos et al, 1998). No genetic interaction between ump1Δ and β6Δ2-19 was observed (Figure 6B, bottom). However, in the mature β6 subunit, a unique nine-residue N-terminal extension (NTE) is also present, and its deletion along with the β6 propeptide (β6Δ2-28) was lethal (Figure 6B, top). When ump1Δwas combined with β6Δ2-28, β6Δ2-28 inviability was suppressed just as ump1Δ suppressed the lethality of β5Δpro. Formally, these data imply that the β6 NTE (and the β5 propeptide) promotes proteasome assembly by overcoming an inhibitory effect of Ump1.
What step in proteasome assembly might Ump1 inhibit? One potential mechanism for Ump1 action is to limit stable half-mer dimerization until all the β subunits have inserted into the half-mer, perhaps by inhibiting β7 incorporation. Such a checkpoint would make sense if dimerization of incomplete half-mers impairs subsequent proteasome maturation. When we affinity-purified proteasomal particles from ump1Δ cells and used native gel analysis to assess levels of the 15S and half-mer (−β7) species relative to 20S particles, these intermediates were now virtually undetectable (Figure 6C). The 20S proteasomes from the ump1Δ cells in Figure 6C were analyzed by LC-MS/MS. The particles appeared to be largely mature as no propeptide sequences were identified; however, in the β4-Flag purification, we detected multiple Pba1 peptides, suggesting some perturbation to late proteasome maturation because we never detected such peptides in 20S proteasomes from WT cells.
Taken together, these results suggest that the Ump1 assembly factor functions together with the β6 propeptide in an assembly checkpoint that ensures the completion of half-proteasome assembly before the tight association of these half-mers into a precursor dimer. The β6 NTE may help overcome this checkpoint by facilitating the incorporation of β7. In the absence of Ump1, the essential function of the NTE is therefore bypassed.
Discussion
The data presented here argue that eukaryotic 20S proteasome assembly proceeds in vivo by a series of discrete intermediates and in association with at least two conserved assembly factors. Several of the intermediates have not been observed previously in studies of either yeast or mammalian cells. Specific appendages that are key to assembly are the β5 propeptide, the β6 NTE, and the β7 tail, which have related functions in the joining and assembly of half-proteasome particles. Our results allow formulation of a detailed model for the later stages of eukaryotic 20S proteasome assembly.
A model for eukaryotic proteasome assembly
In Figure 6D, we present a model for the stepwise assembly of eukaryotic 20S proteasomes in vivo. In the model, the β5 propeptide helps to bring together and align proteasome half-mers and to stabilize the precursor dimer during the β-subunit precursor cleavages and conformational rearrangements that lead to mature 20S proteasomes (Groll et al, 1999). This normally essential function of β5 is partially redundant with the β7 C-terminal tail and can be largely bypassed when β7 is present in high amounts. Ump1 is proposed to inhibit stable dimerization until the rate-limiting insertion of the β7 subunit into the half-mer. The β7 tail functions as a linchpin between half-mers to help overcome Ump1 inhibition and stabilize the precursor dimer during maturation. This tail is normally not essential if functional β5 propeptide is present. In the model, the absence of Ump1 allows premature half-mer association, thereby impeding subsequent assembly and maturation steps. This simple model can explain the otherwise surprising genetic interactions seen between various β-subunit mutants as well as the paradoxical effects of Ump1 mutations on assembly (see below).
Two general problems complicate interpretation of potential protein assembly intermediates such as those suggested in Figure 6D. First, it is possible that some of the subparticles identified were not formed in vivo but derived from larger complexes during isolation. This appears unlikely here given that we have seen most of these discrete complexes by different purification methods and from different yeast strains. Isolation procedures involved rapid lysis and purification under mild conditions. We also see a strict correlation between proteasome particles that are incomplete and have unprocessed subunit precursors and their association with proteasome-specific assembly factors. The second, thornier problem is that even if an observed complex had formed in vivo, it could represent a dead-end assembly product rather than a normal or necessary intermediate on the pathway to full complex assembly. This caveat is difficult to eliminate completely, although we have seen most of the specific complexes not only in mutants but also in WT cells. Moreover, we have obtained corroborative genetic data in several cases that support the physiological significance of the observed subcomplexes. For instance, we identified a half-mer particle lacking only the β7 subunit from several very different yeast mutants in this study. Consistent with this, mutations of the neighboring β6 subunit have specific genetic interactions with β7 and β5pro mutations. Similarly, the ability of β7HC to suppress β5Δpro in a β7 tail-dependent fashion suggests that both the β7 tail and β5 propeptide function in β–trans-β interactions that facilitate dimerization. High-copy β7 also specifically suppresses the defects association with the β5 (doa3-1) mutant, and this mutant had been shown previously to impair β-trans–β interactions (Chen and Hochstrasser, 1995).
Interpretation of proteasome assembly data
Armed with the working model in Figure 6D, we can rationalize many of the disparate biochemical and genetic data described here and in previous studies. The essential function of the β5 propeptide is to help bind and align two half-proteasomes, although additional functions, such as facilitating β5 insertion into the 15S complex, are likely. Lethality of β5Δpro is overcome with high-copy β7 because the slow step of β7 incorporation into the half-mer (−β7) is enhanced sufficiently to allow dimer association and alignment via β7 tail interactions with the trans β ring. It is unlikely that the β5 propeptide acts by enhancing β7 incorporation itself because deletion of the β5 propeptide does not increase the accumulation of the half-mer (−β7) intermediate compared to that seen with WT β5 (Figure 3A; lanes 4 and 5).
Paradoxically, the Ump1 assembly factor facilitates assembly by, in our model, inhibiting stable half-mer dimerization before β7 incorporation. Ump1 can be viewed as an assembly checkpoint protein that helps ensure the proper order of proteasome assembly events. This ultimately enhances productive proteasome assembly by reducing flux through slow or dead-end assembly pathways. Such a checkpoint role is also consistent with the ability of pba1/2 mutations to suppress ump1Δ (Supplementary Figure S5B). Previous gel filtration analysis suggested that ‘15S intermediates' containing β-subunit precursors build up to high levels in ump1Δ cells (Ramos et al, 1998). The exact composition of these intermediates is unknown. ATP was included in the gel filtration analyses but not during the affinity purifications; these species therefore might represent aberrant chaperone-associated intermediates that are insufficiently long-lived to be detected after more extensive purification. Because the β5 propeptide facilitates dimerization whereas Ump1 apparently inhibits it, viability of the β5Δpro strain can be rescued by ump1Δ. The cost to the double mutant, as in the ump1Δ single, lies in its accumulation of defective dimers or other intermediates, causing reduced functional 20S proteasomes and slow growth that cannot be overcome by β7 overproduction.
A particularly simple way by which Ump1 might work would be to limit directly β7 insertion into the half-mer until all the other β subunits have incorporated. A key step here might be entry of β6; this inference is based on the unusual genetic interactions seen between mutations in β5, β6, and β7 (Figure 6). Just like ump1Δ, deletion of the β6 propeptide (residues 2–19) suppresses the lethality of β5Δpro, and the double mutant also grows slowly. Thus, the β6 propeptide could help limit incorporation of β7, the direct neighbor of β6, until all the remaining β subunits are in place in the half-mer.
Why might β2, β3, and β4 associate with the α ring before the other β subunits and why does this intermediate accumulate even in WT cells? Consideration of the surface area buried between subunits suggests a potential solution (Supplementary Table I). Of all the β subunits, β2 buries the largest surface area against the α ring, and it has by far the largest cis-β contact; this is primarily due to the long C-terminal arm that drapes over β3. A β2–β3 heterodimer, which buries 4600 Å2 against the α ring, could serve to nucleate β-ring assembly on the α-ring template. Notably, β2, β3, and β4 have three of the four largest surface areas buried against the α ring, whereas the two subunits flanking this trio, β1 and β5, have the two lowest, and neither makes extensive cis-contacts with β2 and β4, respectively. Therefore, stable association of these two subunits might normally be limiting, causing 15S precursors to accumulate. Insomuch as the 15S precursor accumulates to higher levels in β5Δpro cells (Figures 2B and 3A), the β5 propeptide probably facilitates incorporation of β5 and possibly also β1 and β6.
Clearly, these model-based interpretations of 20S proteasome assembly, although consistent with our data, will require further experimental testing. Nevertheless, the model provides a useful framework for explaining existing findings on assembly and makes testable predictions.
A conserved proteasome assembly factor
We consistently observe two additional polypeptides, Pba1 and Pba2, in proteasomal precursors (Table I). Purified Pba1 and Pba2 form a stable heterodimer. The inference that Pba1–Pba2 participates in 20S proteasome assembly derives from the finding that the polypeptides were found with multiple distinct precursor particles but never in mature 20S proteasomes. Proteasome assembly does not absolutely require Pba1-Pba2 because the corresponding deletion mutants are fully viable, but genetic interactions with proteasome mutants are consistent with a role in proteasome biogenesis.
An elegant study of proteasome assembly in mammalian cells identified a heterodimeric complex, PAC1–PAC2, which facilitates early stages of proteasome assembly, possibly by limiting off-pathway reactions such as α-ring dimerization (Hirano et al, 2005). Modest similarity between PAC2 and yeast Pba2 was noted, and we found that PAC1 and yeast Pba1 are also related. Both yeast proteins are induced during the endoplasmic reticulum (ER) unfolded-protein response, as are other ubiquitin-system components, and pba2 (add66) mutants have a mild defect in the degradation of some ER substrates (Palmer et al, 2003). Finally, Pba1–Pba2 associates with very early proteasome precursors (Table I), similar to what has been reported for the human heterodimer (Hirano et al, 2005). These data make it likely that their mechanism of action is also conserved.
We did not consistently detect any other proteins in 20S proteasome assembly intermediates except the HEAT-repeat protein Blm10, which we found not only associated with 15S intermediates but also with mature 20S proteasomes. The exact function of Blm10 has been controversial. Of the two previous models, one suggested an inhibitory Blm10 function in late proteasome assembly (Fehlker et al, 2003) and the other a role in proteasome activation (Ustrell et al, 2002; Schmidt et al, 2005). Because Blm10 associates with the α-ring surface in mature 20S proteasomes, Blm10 can probably bind this surface whenever it is not occupied by the RP or another proteasomal regulator. This might limit nonproductive binding of RPs to inactive 20S proteasome precursors. However, Blm10 likely functions primarily with the mature 20S enzyme because native PAGE of fresh yeast lysates showed that the majority of Blm10 is part of active Blm10-20S-RP hybrid proteasomes (Schmidt et al, 2005).
Assembly of multiprotein ring complexes
Many complex molecular transactions in the cell are catalyzed by multisubunit molecular machines. Often, these machines are organized into ring-shaped structures, creating a central channel or chamber, and macromolecules are moved into or out of these chambers. How such complicated ring-shaped complexes are assembled in vivo is poorly understood at best.
The eukaryotic proteasome is among the most intricate of the ring-shaped protein complexes. Our data on 20S proteasome assembly are likely relevant to the assembly of other heteromeric ring complexes such as Sm and Lsm complexes, the MCM replication complex, and class II chaperonins (see Velichutina et al, 2004). These complexes may also arise from ordered assembly pathways orchestrated by dedicated assembly factors, and their assembly might also involve assembly checkpoints that limit formation of nonproductive complexes. An enhanced understanding of proteasome assembly mechanisms also has implications for drug development. Active site inhibitors of the proteasome are in clinical use, and small molecules that interfere with proteasome assembly might provide useful alternatives to such inhibitors.
Materials and methods
Plasmid and yeast strain constructions
E. coli strains used were MC1061, Top10, and JM101, and standard methods were employed for recombinant DNA work (Ausubel et al, 1989). Yeast strains used in this study are listed in Supplementary Table II. Most of the remaining strains were derived from crosses between previously described congenic strains or by plasmid shuffling using the appropriate deletion strains and plasmids. Details on plasmid and strain constructions are available in the Supplementary Data.
Protein gel electrophoresis and immunoblotting
SDS–PAGE and immunoblot analysis of proteins were carried out according to standard procedures (Ausubel et al, 1989). For immunoblotting, gel-separated protein samples were transferred to a polyvinylidene fluoride membrane (Millipore). Blots were incubated with antibodies to HA (monoclonal 16B12; Babco), proteasome α subunits (MCP231; BioMol), Flag (M2; Sigma), LMP7-His6 (a gift from Y Yang), or affinity-purified rabbit anti-β5 and anti-β1 antibodies. Proteins were visualized by ECL (Amersham).
Native PAGE was performed as described (Hough et al, 1987), except we used a boric acid-based 6% resolving gel with a Tris-based 4% stacking gel. Gels were either stained by GelCode Blue (Pierce) or incubated for 30 min in transfer buffer containing 0.2% SDS before immunoblotting. 2D PAGE was done as described (Chen and Hochstrasser, 1995).
Affinity purification of proteasomal particles
The 20S proteasome and its precursors were affinity-purified from yeast expressing chromosomally Flag-tagged α- or β-subunit genes by a procedure modified from Verma et al (2000). Yeast cells (2 l) were grown at 30°C in synthetic medium to an OD600 of 2.0 for mature proteasome purification or to an OD600 of 1.0 for analysis of intermediates. Cells were harvested, washed once with ice-cold water, and frozen in liquid nitrogen. Cell pellets were ground to a fine power in a mortar in the presence of liquid nitrogen. The powder was suspended in 10 ml of buffer A (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10% glycerol, and 5 mM MgCl2) supplied with protease inhibitors (2 mM phenylmethylsulfonyl fluoride, 20 μg/ml pepstatin A, and 5 μg/ml each of leupeptin, chymostatin, and aprotinin). Cell debris was removed by centrifugation for 20 min in a Sorvall RC-5B tabletop centrifuge at maximum speed. The supernatant was gently mixed with 300 μl of buffer A-equilibrated anti-Flag M2 agarose beads (Sigma), and rotated for 1–2 h at 4°C. The beads were washed with 50 volumes of buffer A containing 0.2% Triton X-100. Bound proteins were eluted with 100 μl of buffer A containing 0.2 mg/ml Flag peptide. Protein concentration was determined by the Bradford method.
Glycerol gradient centrifugation and gel filtration chromatography
Proteasomes were partially separated by glycerol gradient ultracentrifugation or by gel filtration chromatography as described (Chen and Hochstrasser, 1996; Velichutina et al, 2004). Yeast cells were lysed by grinding in a mortar in the presence of liquid N2. Cell extracts were applied to a 10–40% (w/v) glycerol gradient or a Superose-6 gel filtration column. Portions of each gradient or column fraction were tested for Suc-LLVY-AMC-hydrolyzing activity or separated by SDS–PAGE and analyzed by immunoblotting.
Mass spectrometry
Purified proteasomes and proteasome subparticles were separated by 6% native PAGE and visualized with GelCode Blue. Protein bands were excised and sent to Midwest Bio Services LLC for composition analysis by Nano-LC-MS/MS. Samples were digested in-gel with trypsin, and the resulting peptide mixture was analyzed by LC-MS/MS in a DECA-XP plus ion trap mass spectrometer equipped with a nano-LC electrospray ionization source (ThermoFinnigan). Mass spectra were searched using TURBOSEQUEST software against the NIH nr protein database. Matches were retained if they had DeltaCn scores higher than 0.08 as well as Xcor scores higher than 1.5 for +1 charged peptides, higher than 2.0 for +2 charged peptides, and higher than 2.5 for +3 charged peptides. Our earlier MS/MS analysis was done as described in Kislinger et al (2003). Identified peptides are listed in Supplementary Table III. Spectral counting and analysis were done according to Old et al (2005); see Supplementary Table IV.
Supplementary Material
Supplementary Figures
Supplementary Data and Table S1, S2
Supplementary Table S3
Supplementary Table S4
Acknowledgments
We thank Jürgen Dohmen for plasmids and Jimin Wang for calculating 20S buried surface areas. This work was supported by NIH grants GM046904 and GM053756 (MH). AE was supported by the Natural Sciences and Engineering Research Council of Canada, the Ontario Research Fund, and the Ontario Genomics Institute/Genome Canada. ARK was supported in part by a postdoctoral fellowship from the Canadian Institutes of Health Research.
References
- Aki M, Shimbara N, Takashina M, Akiyama K, Kagawa S, Tamura T, Tanahashi N, Yoshimura T, Tanaka K, Ichihara A (1994) Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J Biochem (Tokyo) 115: 257–269 [DOI] [PubMed] [Google Scholar]
- Arendt CS, Hochstrasser M (1997) Identification of the yeast 20S proteasome catalytic centers and subunit interactions required for active-site formation. Proc Natl Acad Sci USA 94: 7156–7161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) (1989) Current Protocols in Molecular Biology. New York: John Wiley and Sons [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell 92: 367–380 [DOI] [PubMed] [Google Scholar]
- Chen P, Hochstrasser M (1995) Biogenesis, structure, and function of the yeast 20S proteasome. EMBO J 14: 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Hochstrasser M (1996) Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 86: 961–972 [DOI] [PubMed] [Google Scholar]
- Fehlker M, Wendler P, Lehmann A, Enenkel C (2003) Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep 4: 959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386: 463–471 [DOI] [PubMed] [Google Scholar]
- Groll M, Heinemeyer W, Jager S, Ullrich T, Bochtler M, Wolf DH, Huber R (1999) The catalytic sites of 20S proteasomes and their role in subunit maturation: a mutational and crystallographic study. Proc Natl Acad Sci USA 96: 10976–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W, Ramos PC, Dohmen RJ (2004) The ultimate nanoscale mincer: assembly, structure and active sites of the 20S proteasome core. Cell Mol Life Sci 61: 1562–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heink S, Ludwig D, Kloetzel PM, Kruger E (2005) IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc Natl Acad Sci USA 102: 9241–9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, Natsume T, Tanaka K, Murata S (2005) A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 437: 1381–1385 [DOI] [PubMed] [Google Scholar]
- Hough R, Pratt G, Rechsteiner M (1987) Purification of two high molecular weight proteases in rabbit reticulocyte lysate. J Biol Chem 262: 8303–8313 [PubMed] [Google Scholar]
- Kislinger T, Rahman K, Radulovic D, Cox B, Rossant J, Emili A (2003) PRISM, a generic large scale proteomic investigation strategy for mammals. Mol Cell Proteomics 2: 96–106 [DOI] [PubMed] [Google Scholar]
- Kruger E, Kloetzel PM, Enenkel C (2001) 20S proteasome biogenesis. Biochimie 83: 289–293 [DOI] [PubMed] [Google Scholar]
- Kwon YD, Nagy I, Adams PD, Baumeister W, Jap BK (2004) Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly. J Mol Biol 335: 233–245 [DOI] [PubMed] [Google Scholar]
- Nandi D, Woodward E, Ginsburg DB, Monaco JJ (1997) Intermediates in the formation of mouse 20S proteasomes: implications for the assembly of precursor beta subunits. EMBO J 16: 5363–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 4: 1487–1502 [DOI] [PubMed] [Google Scholar]
- Palmer EA, Kruse KB, Fewell SW, Buchanan SM, Brodsky JL, McCracken AA (2003) Differential requirements of novel A1PiZ degradation deficient (ADD) genes in ER-associated protein degradation. J Cell Sci 116: 2361–2373 [DOI] [PubMed] [Google Scholar]
- Ramos PC, Hockendorff J, Johnson ES, Varshavsky A, Dohmen RJ (1998) Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 92: 489–499 [DOI] [PubMed] [Google Scholar]
- Ramos PC, Marques AJ, London MK, Dohmen RJ (2004) Role of C-terminal extensions of subunits beta 2 and beta 7 in assembly and activity of eukaryotic proteasomes. J Biol Chem 279: 14323–14330 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D (2005) The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol 12: 294–303 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Zantopf D, Kraft R, Kostka S, Preissner R, Kloetzel PM (1999) Sequence information within proteasomal prosequences mediates efficient integration of beta-subunits into the 20 S proteasome complex. J Mol Biol 288: 117–128 [DOI] [PubMed] [Google Scholar]
- Schmidtke G, Schmidt M, Kloetzel PM (1997) Maturation of mammalian 20 S proteasome: purification and characterization of 13 S and 16 S proteasome precursor complexes. J Mol Biol 268: 95–106 [DOI] [PubMed] [Google Scholar]
- Thomson S, Rivett AJ (1996) Processing of N3, a mammalian proteasome beta-type subunit. Biochem J 315: 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustrell V, Hoffman L, Pratt G, Rechsteiner M (2002) PA200, a nuclear proteasome activator involved in DNA repair. EMBO J 21: 3516–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichutina I, Connerly PL, Arendt CS, Li X, Hochstrasser M (2004) Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. EMBO J 23: 500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 11: 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zühl F, Seemuller E, Golbik R, Baumeister W (1997) Dissecting the assembly pathway of the 20S proteasome. FEBS Lett 418: 189–194 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Data and Table S1, S2
Supplementary Table S3
Supplementary Table S4


