Abstract
The growth factor/insulin-stimulated AGC kinases share an activation mechanism based on three phosphorylation sites. Of these, only the role of the activation loop phosphate in the kinase domain and the hydrophobic motif (HM) phosphate in a C-terminal tail region are well characterized. We investigated the role of the third, so-called turn motif phosphate, also located in the tail, in the AGC kinases PKB, S6K, RSK, MSK, PRK and PKC. We report cooperative action of the HM phosphate and the turn motif phosphate, because it binds a phosphoSer/Thr-binding site above the glycine-rich loop within the kinase domain, promoting zipper-like association of the tail with the kinase domain, serving to stabilize the HM in its kinase-activating binding site. We present a molecular model for allosteric activation of AGC kinases by the turn motif phosphate via HM-mediated stabilization of the αC helix. In S6K and MSK, the turn motif phosphate thereby also protects the HM from dephosphorylation. Our results suggest that the mechanism described is a key feature in activation of upto 26 human AGC kinases.
Keywords: AGC kinase, growth factor, PKB, RSK, S6K
Introduction
A significant portion of growth factor/insulin signalling is mediated by a functionally diverse, but structurally related group of protein kinases that belong to the AGC kinase family. The group, here called the growth factor-activated AGC kinases, includes protein kinase B (PKBα-γ or AKT1-3), p70 ribosomal S6 kinase (S6K1,2), p90 ribosomal S6 kinase (RSK1-4), mitogen- and stress-activated protein kinase (MSK1,2) and several members of the protein kinase C (PKC) family. These kinases regulate cellular division, growth, survival, metabolism, motility and differentiation and several of them are implicated in human disease. The kinases function in partly distinct signalling pathways, such as the phosphoinositide 3-kinase (PI3K) pathway (PKB and S6K) (Kozma and Thomas, 2002), mitogen-activated protein (MAP) kinase pathways (RSK and MSK) (Hauge and Frodin, 2006), in calcium/lipid signalling (PKC) (Parekh et al, 2000; Newton, 2003), or in Rho GTPase signalling (PRK2) (Parekh et al, 2000). The responsiveness to distinct upstream pathways is partly due to distinct signalling modules flanking the kinase domain in the various kinases (Supplementary Figure 1).
In addition to the divergent regulation, the growth factor-activated AGC kinases share a common core mechanism of activation, which is based on three conserved phosphorylation sites. Viewed simplistically, the flanking signalling modules serve to induce proper phosphorylation of these phosphorylation sites. The three sites are located in the activation loop in the kinase domain, in the middle of a tail/linker region C terminally to the kinase domain, and within a hydrophobic motif (HM) at the end of the tail region, respectively (Figure 1B and Supplementary Figure 1). The various kinases thought to target the three phosphorylation sites are indicated in Supplementary Figure 1.
Figure 1.
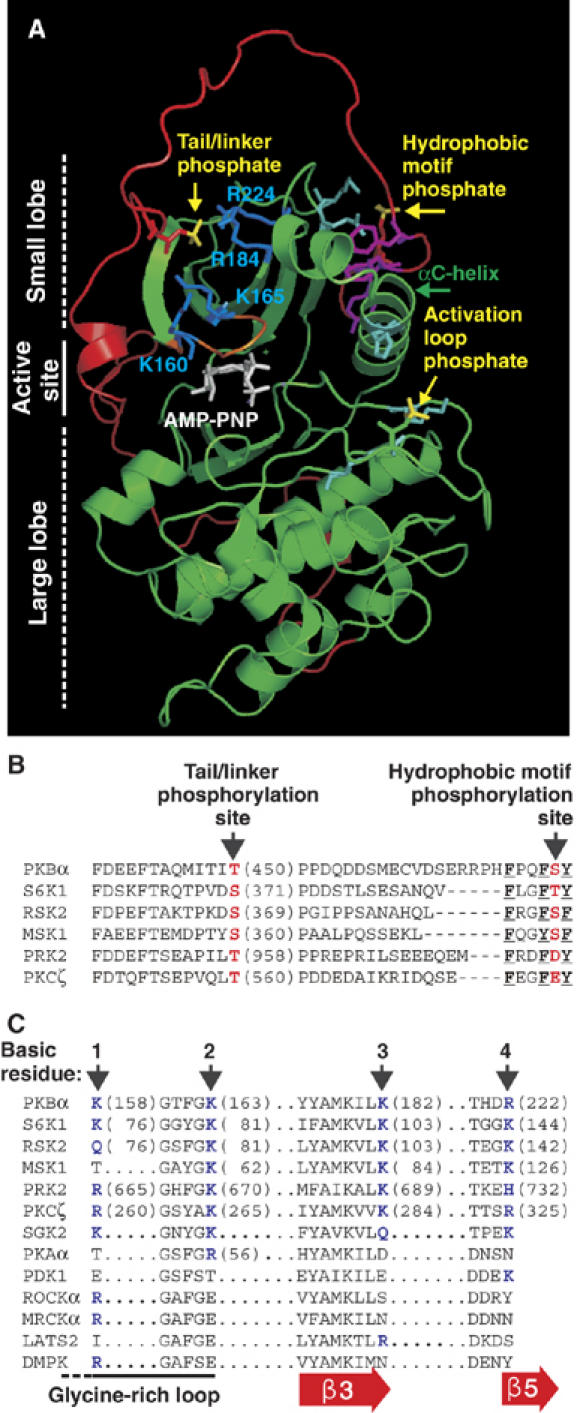
Molecular modelling suggests a widely conserved binding site for the tail/linker phosphoSer/Thr within the catalytic domain of AGC kinases. (A) Model of active PKBβ shown as a ribbon representation with side chains of selected residues. The kinase domain is shown in green and the tail/linker in red. Phosphate groups are shown in yellow. K/R residues predicted to bind the phosphate of phosphoT451 are shown in blue. Other phosphate-binding residues are in cyan. The HM, glycine-rich loop and ATP analogue are shown in magenta, orange and white, respectively. Partial sequence alignment of the tail/linker (B) and predicted binding site for the tail/linker phosphate (C) of AGC kinases. (B, C) Phosphorylation sites/phosphate-mimicking residues are shown in red. The aromatic residues that define the HM are underlined. Basic residues predicted to bind the tail/linker phosphoSer/Thr are shown in blue and labelled 1–4. Sequences are human except S6K1 (rat, p70 isoform) and RSK2 (mouse).
Intense efforts have recently established the mechanism of action of the HM and the activation loop phosphorylation sites (Biondi et al, 2000; Frodin et al, 2002; Yang et al, 2002a, 2002b; Smith et al, 2004; Engel et al, 2006). Phosphorylation of the HM triggers interaction of the phosphate with a phosphate-binding site in the small lobe of the kinase domain, providing the final binding energy required for the aromatic residues of the HM to interact with and stabilize a nearby hydrophobic pocket, which is disordered in the inactive kinase. Stabilization of the so-called αC helix in this pocket is thought to be of key importance, because this helix contains key residues for regulation of phosphotransferase activity and it may aid in stabilizing the activation loop in an optimal conformation. In RSK and S6K, the phosphorylated HM additionally functions as a phosphorylation-dependent docking site that recruits and activates the activation loop kinase PDK1 (Frodin et al, 2000; Biondi et al, 2001). The phosphate in the activation loop stimulates kinase activity by binding to basic residues in loops within the active site, which helps position catalytic residues (Knighton et al, 1991). Individual phosphorylation of the HM and the activation loop induces negligible and low-level activation, respectively. However, in combination, the two phosphorylation events synergistically stimulate kinase activity (Alessi et al, 1996; Frodin et al, 2002; Yang et al, 2002b). One mechanism for this cooperativity is thought to derive from the ability of both phosphates to promote interaction between the αC helix and the activation loop, leading to mutual stabilization of these key regulatory structures and promotion of the active, closed conformation of the kinase domain.
The phosphorylation site in the middle of the tail is the most poorly characterized of the three conserved sites, yet its mutation significantly reduces kinase activity and in some AGC kinases also HM phosphorylation (Moser et al, 1997; Bellacosa et al, 1998; Weng et al, 1998; Parekh et al, 2000; Newton, 2003; Matsuzaki et al, 2004; McCoy et al, 2004). In RSK and MSK, the site is phosphorylated by extracellular signal-regulated kinase (ERK) and p38 or ERK, respectively, during activation (Dalby et al, 1998; McCoy et al, 2004). In S6K, the site may be phosphorylated by mTOR and displays high basal phosphorylation, which is increased twofold in response to growth factor/insulin (Saitoh et al, 2002). In PKB, the site is constitutively phosphorylated by an unknown kinase (Alessi et al, 1996). In PKCs, the site is thought to be autophosphorylated during maturation to the latent catalytically competent conformation (Parekh et al, 2000; Newton, 2003).
The mechanism of action of the phosphorylation site in the middle of the tail is elusive. The AGC protein kinase A (PKA) also contains a phosphorylation site in the middle of its tail region, known as the ‘turn motif' site, because the phosphate binds nearby residues within the tail and thereby stabilizes a turn in the tail. It has been widely assumed that the tail phosphate in the growth factor-activated AGC kinases performs the same function. Consequently, the site is also known as the turn motif in these kinases and has been aligned with the turn motif of PKA (Yang et al, 2002b; Newton, 2003; Roux and Blenis, 2004).
Here, we report the mechanism(s) whereby the tail phosphorylation site activates PKBα, S6K1, RSK2, MSK1, PRK2 and PKCζ, which represent six of the seven families of growth factor-activated AGC kinases. We report that this phosphorylation site is not equivalent to the turn motif site of PKA, but rather corresponds to Glu333 of PKA. In the growth factor-activated AGC kinases, the tail phosphate binds a phosphoSer/Thr-binding site in the kinase domain near the hydrophobic pocket, serving to deliver the HM to its binding site in a zipper-like manner. Our results suggest that the tail phosphate thereby synergistically enhances kinase activation via HM-mediated stabilization of the αC helix and, in a subset of the kinases, also controls the phosphorylation state of the HM. On the basis of these findings, it could be considered referring to the tail site in growth factor-activated AGC kinases as the Z (zipper) site instead of the turn motif site.
Our results suggest that the overall mechanism described is a key feature in the activation of up to 26 human AGC kinases and has been widely conserved during evolution.
Results
A potential binding site for the tail phosphoSer/Thr is widely conserved in the catalytic domain of AGC kinases
PKBβ has been crystallized in an active conformation showing the HM bound to the hydrophobic pocket (Yang et al, 2002a). However a large portion of the tail region, including the tail phosphorylation site (T451), was not visible in the structure. The visible tail region ends with a short helix near the small lobe of the kinase domain.
Our initial modelling analysis did not support the possibility of a binding site for phosphoT451 within the tail region. We therefore analysed the small lobe of PKB for binding sites for phosphoT451 using the programs GRASP and GRID. Among various potential sites, we focused on the most interesting site, located above the ATP-binding, glycine-rich loop and formed by four basic residues, K160, K165, R184 and R224 (hereafter referred to as basic residues 1–4) of which the first two are part of the glycine-rich loop. Ab initio modelling of the noncrystallized region of the tail suggested that the phosphate of T451 might be located in the middle of this basic cluster. The location appeared energetically favourable, because the phosphate remained in the site during dynamics simulations on the model, constantly interacting with 2 or 3 of the basic residues that differed over time (Figure 1A). The four basic residues are conserved in all 23 members of the PKB, S6K, RSK, MSK, PRK and PKC families (Figure 1C and Supplementary Figure 2). They are also conserved in the three members of the SGK family of growth factor-activated AGC kinases, which have a tail phosphorylation site (Kobayashi et al, 1999), required for full kinase activity (CJ Jensen and M Frodin, unpublished observation). Finally, the tail site and the basic residues are co-conserved during evolution as illustrated by S6K orthologues from Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana and Saccharomyces pombe (Supplementary Figure 2). Modelling of S6K1 and RSK2 supported the existence of a phosphate-binding site homologous to that of PKBβ (Supplementary Figure 3). The basic residues are poorly conserved in AGC kinases not thought to contain a tail phosphorylation site (PDK1, ROCK, MRCK, LATS and DMPK, Figure 1C).
Thus, modelling and sequence conservation suggested that in the growth factor-activated AGC kinases, the tail phosphoSer/Thr interacts with a phosphate-binding site within the kinase domain, implying a different role of this phosphorylation site from that of the turn motif site in PKA. The functional characterization presented below suggests that the tail phosphate promotes zipper-like binding of the tail and HM to the kinase domain, aimed at controlling activation of the kinases by the HM.
Role of the tail site in phosphorylation and activation of AGC kinases in vivo
We first characterized further the importance of the tail site in the growth factor-activated AGC kinases. PKBα, S6K1, RSK2 and MSK1 were purified from transiently transfected COS7 cells exposed to an appropriate stimulus. PRK2, including the truncated mutant used here, PRK2Δ1−500, was active in nonstimulated COS7 cells and was therefore purified from nontreated cells.
Immunoblotting with phosphospecific antibodies showed that the tail site was constitutively phosphorylated in PKBα, PRK2 and S6K1 (sometimes a twofold induction was observed in S6K1, Supplementary Figure 5A) and strongly induced in RSK2 and MSK1 following stimulation (Figure 2). Mutation of the tail site to Ala reduced activation from ≈60% in PKBα to ≈98% in S6K1 (Figure 2). Furthermore, in S6K1, MSK1 and RSK2, mutation of the tail site reduced phosphorylation of the HM with moderate to profound effects. In PKBα, mutation of the tail site modestly enhanced phosphorylation of the HM and the activation loop. Mutation of the tail site to phosphate-mimicking Glu could substitute for phosphorylation in PKBα and RSK2, partially in MSK1, but not in PRK2 (Supplementary Figure 4), similar to findings with S6K1 (Moser et al, 1997). These results further establish the importance of the tail site in PKB, S6K, RSK and MSK and report its role in the PRK family (and in PKCζ, Figure 6) for the first time.
Figure 2.
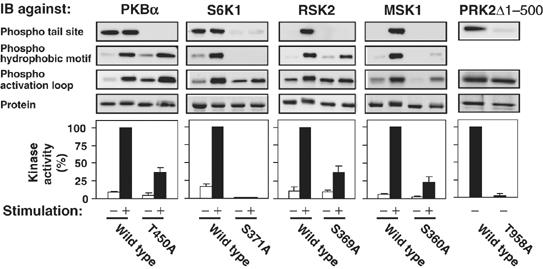
Role of the tail phosphorylation site in activation and phosphorylation of AGC kinases in vivo. COS7 cells were transfected with plasmid expressing haemagglutinin (HA)- or glutathione S-transferase (GST)-tagged wt or mutant kinase. After 16 h and a final 4 h serum-starvation period, cells were exposed to 1 μM insulin for 10 min (PKBα), to 20 nM EGF for 30 min (S6K1) or 15 min (RSK2), to 10 μg/ml anisomycin for 40 min (MSK1) or left in serum-containing medium (PRK2Δ1−500) and then lysed. The kinases were precipitated from aliquots of the cell lysates with antibody to the HA tag or with glutathione beads. The precipitates were subjected to kinase assay, to immunoblotting with the indicated phosphorylation site-specific Ab or anti-HA Ab or stained for protein. Experiments were repeated at least three times and activity data (expressed as percent) are means±s.d.
Figure 6.
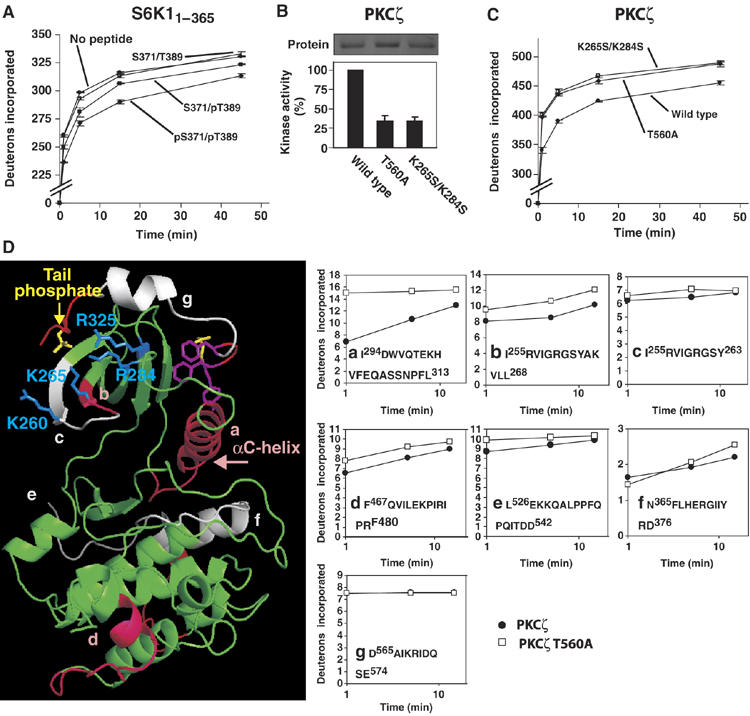
The tail phosphate promotes a compact global conformation of the AGC kinase domain and protects the tail phosphate-binding site and αC helix from solvent exposure. (A) Effect of S6K1 tail peptides, described in the legend of Figure 5, on global deuteron uptake by purified GST-S6K11−365. (B) Kinase activity of wt and mutant GST-PKCζ. (C) Global deuteron uptake by wt and mutant GST-PKCζ. (D) Local deuteron uptake by wt and mutant GST-PKCζ. In the PKCζ model, peptides showing strong and no-or-slight protection by the tail phosphate are shown in pink and grey, respectively. The panels show HX curves of the peptides. Experiments were repeated two times (A, C, D) or three times (B) and data are means ±range (A, C) or ±s.d. (B)
The predicted binding site for the tail phosphate is essential for normal activation and phosphorylation of AGC kinases in vivo
We next mutated the predicted binding site for the tail phosphate by introducing amino acids that are present at the corresponding positions in AGC kinases without a tail site, assuming that such mutations would not compromise tertiary structure. The assumption was supported by normal expression of nearly all mutants. Analysis of 76 point mutants suggested that the predicted binding site is functional in all growth factor-activated AGC kinases and identified the most important individual or combinations of basic residues in the various AGC kinase subfamilies.
In PKBα, quadruple mutation of the four basic residues (as in PKBα-K158T/K163S/K182S/R222N), reduced kinase activity by ≈40% (Fig 3A), comparable to the ≈60% reduction resulting from mutation of the tail site T450 (Figure 2). Moreover, phosphorylation of T450 was considerably reduced in PKBα-K158T/K163S/K182S/R222N, suggesting that the binding of phosphoT450 to the basic residues protects it from dephosphorylation. The finding that PKBα-K158T/K163S/K182S/R222N had somewhat higher activity than PKBα-T450A likely results from residual phosphorylation at T450 in the former mutant. The residual phosphate may interact with the introduced Thr, Ser and Asn residues and induce a small degree of activation, as these amino acids can bind phosphoSer/Thr, although with lower affinity than Lys and Arg.
Figure 3.
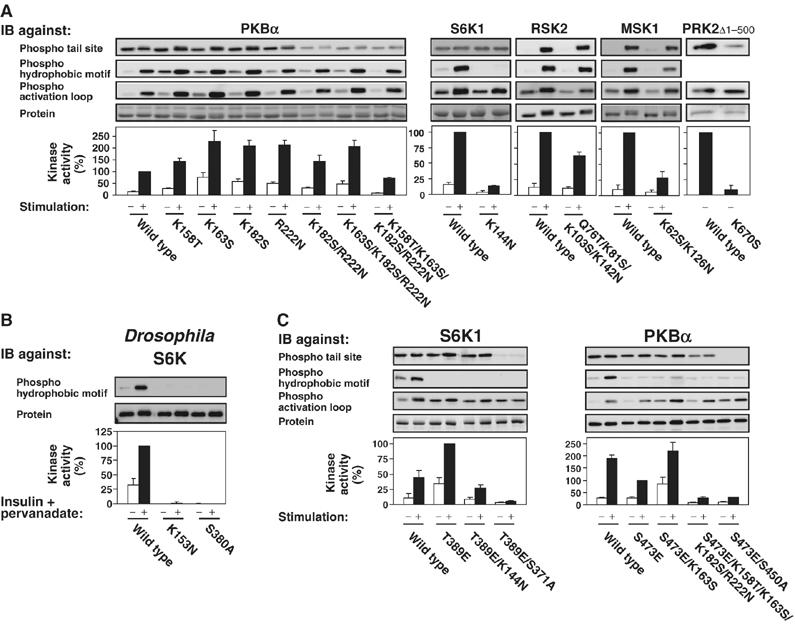
The tail phosphate-binding site is essential for normal activation and phosphorylation of AGC kinases in vivo. The activity and phosphorylation state of wt and mutant AGC kinases expressed in COS7 cells (A, C) or S2 cells (B) were analysed as described in Figure 2, except that Drosophila S6K was activated by exposure of cells to 1 μM insulin and 10 μM pervanadate for 15 min. Experiments were repeated at least three times and activity data (expressed as percentage) are means±s.d.
Surprisingly, single to triple mutation of the basic residues in PKBα resulted in significantly increased basal and insulin-stimulated kinase activity, which apparently resulted from increased phosphorylation of the HM and the activation loop (Figure 3A). These mutations also increased the biological activity of PKB, because NIH 3T3 cells virally transduced with PKBα-K163S or PKBα-K182S showed a small, but significant increase in colony formation in sparsely seeded cultures, as compared to cells transduced with wild-type (wt) PKBα (C Hauge and M Frodin, unpublished observation).
In S6K1, mutation of basic residue 4 reduced kinase activity by ≈85% (Figure 3A), comparable to the ≈98% reduction caused by mutation of the tail site S371 (Figure 2). Furthermore, mutation of basic residue 4 profoundly reduced phosphorylation in the HM and moderately in the activation loop, similar to the effects obtained by mutation of the tail site. Mutation of basic residue 1 reduced kinase activity and HM phosphorylation by ≈60% (Supplementary Figure 5A). In RSK2, individual mutation of basic residues 1–4 had negligible effect, but mutation of all four residues reduced kinase activity by ≈40%, comparable to the ≈60% reduction caused by mutation of the tail site S369, and slightly reduced HM phosphorylation. In MSK1, double mutation of basic residues 2 and 4 reduced kinase activity to almost the same low level as that obtained after mutation of the tail site S360. The double mutation also significantly reduced phosphorylation of the HM, as did mutation of the tail site. In PRK2, individual mutation of basic residue 2 (or 1 or 3, Supplementary Figure 5C), reduced kinase activity to similarly low levels as that obtained by mutation of the tail site T958. Mutation of the basic residues caused a profound reduction in phosphorylation of T958, suggesting that the binding of phosphoT958 to these residues protects it from dephosphorylation. In the various kinases, the effects of mutating the tail site and the critical basic residues were not additive (Supplementary Figure 5B and data not shown), indicating that the tail phosphate and its predicted binding site regulate kinase activity by the same mechanism.
The role of the tail phosphate appears highly conserved during evolution, because mutation of the tail site S380 and basic residue 4 (K153) abolished kinase activity and HM phosphorylation of Drosophila S6K expressed in Drosophila S2 cells (Figure 3B).
Because mutation of the tail phosphate-binding site decreased the phosphorylation of HM in S6K1 and MSK1, it could not be determined whether the site affected kinase activity by a mechanism other than regulation of HM dephosphorylation. To render the HM insensitive to phosphatases, we generated mutants with Glu in the HM phosphorylation site (S6K1-T389E and MSK1-S376E). S6K1-T389E possessed higher activity than wt S6K1 (Figure 3C), in accordance with previous findings (Weng et al, 1998). More importantly, mutation of basic residue 1 or 4 in S6K1-T389E reduced kinase activity to the same extent as did these mutations in wt S6K1 without affecting the phosphorylation state (Figure 3C and data not shown). Similar results were obtained with MSK1-S376E (data not shown). Thus, the tail phosphate-binding site can activate S6K1 and MSK1 by a mechanism distinct from the one protecting the HM from dephosphorylation.
In PKBα with Glu at the HM phosphorylation site (PKBα-S473E), the quadruple mutation (K158T/K163S/K182S/R222N) and the K163S mutation of the tail phosphate-binding site had the same effects as in wt PKBα (Figure 3C). This agrees with the results obtained with wt PKBα that severe disruption of the interaction between the tail phosphate and its binding site precludes full kinase activity and that high levels of phosphorylation in the activation loop and Glu in the HM cannot compensate for the disrupted interaction. In PKBα-S473E with deletion of the PH domain (ΔPH-PKBα-S473E), the K163S mutation did not increase phosphorylation or kinase activity of PKBα (Supplementary Figure 5D), further suggesting that this mutation stimulates PKBα activity by inducing hyperphosphorylation, which may occur at the plasma membrane. In ΔPH-PKBα-S473E, mutation of the tail site had less effect on kinase activity compared to full-length PKBα (Supplementary Figure 5D), possibly because ΔPH-PKBα-S473E was less phosphorylated at the tail site (Supplementary Figure 5E), which suggests that membrane localization of PKBα promotes phosphorylation of the tail site. However, these experiments should be interpreted with caution, because deletion of the PH domain likely alters the conformation of PKB.
We conclude that the tail phosphate interacts with a binding site widely conserved among growth factor-activated AGC kinases. We propose that the tail phosphate thereby functions as a molecular zipper that helps deliver the HM to its binding site and stabilize it there, which has two consequences. First, in all of the kinases this directly stimulates kinase activity, presumably by stabilization of the active kinase conformation. Secondly, in a subset of the kinases this controls the phosphorylation state of the HM, presumably by restricting its exposure to phosphatases and kinases. Although a functional tail phosphate-binding site is conserved in all of the kinases studied, the key basic residues involved in forming the site varies somewhat among the different AGC kinase families.
Further evidence of the tail phosphate-binding site
If the tail phosphate and the basic residues are indeed within interaction distance, it might be possible to engineer a Zn2+-binding site by replacing the tail site Ser/Thr and the basic residues with histidine residues. A Zn2+-binding site may be detected by Zn2+-dependent modulation of enzymatic function, most often inhibition due to distortion of protein structure. Zn2+ up to 3.3 μM had no effect on the activity of wt PRK2 or PRK2 with His in the tail site (PRK2-T958H) (Figure 4A). By contrast, PRK2 with His in place of basic residues 2 and 3 (PRK2-K670H/K689H) was inhibited by Zn2+ at 3.3 μM, but not at lower concentrations. Thus, the close proximity of basic residues 2–4 (wt PRK2 has His at the position of basic residue 4) allowed engineering of a Zn2+-binding site, which caused Zn2+-dependent inhibition of kinase activity, possibly due to distortion of the glycine-rich loop. More importantly, introduction of His in the tail site (T958H) of PRK2-K670H/K689H, generated a Zn2+-binding site with increased affinity, as evidenced by more profound inhibition at 3.3 μM and detectable inhibition already at 0.5 μM Zn2+. Similar results were obtained after Zn2+ site engineering in PKBα (data not shown).
Figure 4.
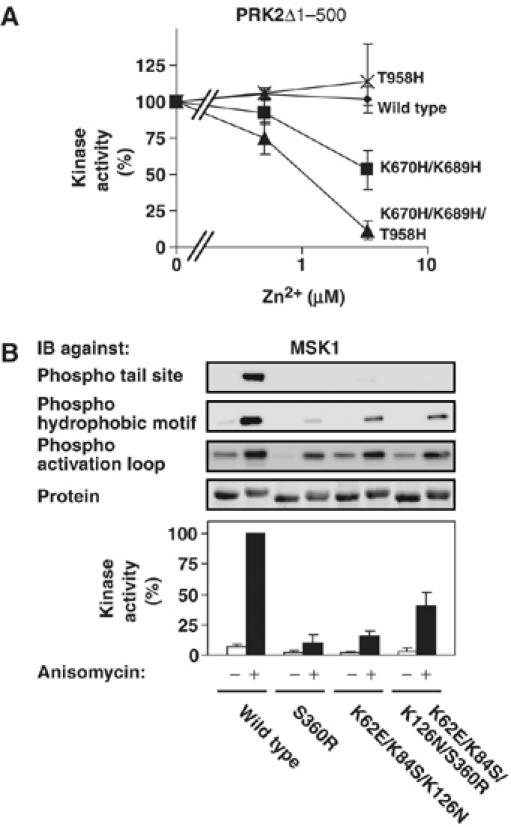
Further evidence of the tail phosphate-binding site. The activity of wt and mutant PRK2Δ1−500 (A) or MSK1 (B) expressed in COS7 cells were analysed as described in the legend of Figure 2. However, in (A) kinase activity was also determined in the presence of the indicated concentration of Zn2+ and expressed as percentage of activity in the absence of Zn2+. Experiments were repeated at least three times and activity data are means±s.d.
Furthermore, if the tail site Ser/Thr and the basic residues are within interaction distance, mutation of the tail site Ser/Thr to Arg might inhibit kinase activity to a higher extent than an Ala mutation due to electrostatic repulsion of the tail. In MSK1, an S360R mutation indeed inhibited kinase activity to a higher extent (≈90%, Figure 4B) than an S360A mutation (≈80%, Figure 2). MSK1-K62E/K84S/K126N, which has an acidic residue in place of basic residue 2 and neutral charge in place of basic residues 3 and 4 had ≈15% activity compared to wt MSK1. Strikingly, in this mutant, introduction of the S360R mutation was not inhibitory, but rather increased kinase activity to ≈40% of that of wt MSK1. In this charge-reversal mutant, R360 presumably binds E62 introduced in place of basic residue 2 and thereby partially rescues kinase activity.
In conclusion, these experiments provide evidence that the tail phosphate and the basic cluster are within interaction distance in the active AGC kinase conformation.
The tail phosphate synergistically enhances AGC kinase activation by the phosphorylated HM, dependent on the tail phosphate-binding site
We established an in vitro reconstitution assay that could test a direct activation of the AGC kinase domain by the tail phosphate and characterize its cooperation with the HM and activation loop phosphates. The deletion mutant S6K11−364, which contains the kinase domain, but lacks the region of the tail containing the tail site and the HM as well as the C-terminal autoinhibitory domain, was expressed and purified from COS7 cells, either nonphosphorylated or phosphorylated at T221 in the activation loop, achieved by co-expression with PDK1. Purified S6K11−364 was then incubated with synthetic peptides of the S6K1 tail (residues 366–395: QTPVDS371PDDSTLSESANQVFLGFT389YVAPSV), which were either nonphosphorylated (S371/T389), phosphorylated at the tail site (pS371/T389), phosphorylated in the HM (S371/pT389) or phosphorylated at both sites (pS371/pT389). Subsequently, the kinase activity of S6K11−364 was determined.
S371/T389 or pS371/T389 tail peptides did not stimulate the kinase activity of Thr221-phosphorylated S6K11−364, whereas S371/pT389 peptide induced a five- to sevenfold stimulation of kinase activity at 190 μM (Figure 5A). More importantly, pS371/pT389 peptide induced a 16- to 22-fold stimulation of kinase activity at 190 μM. These experiments revealed that the tail phosphate synergistically enhances S6K1 activation by the HM phosphate, whereas having no effect on its own. In similar experiments with a truncated PKBβ kinase domain, the tail phosphate also enhanced the ability of the HM to stimulate kinase activity (Supplementary Figure 6B).
Figure 5.
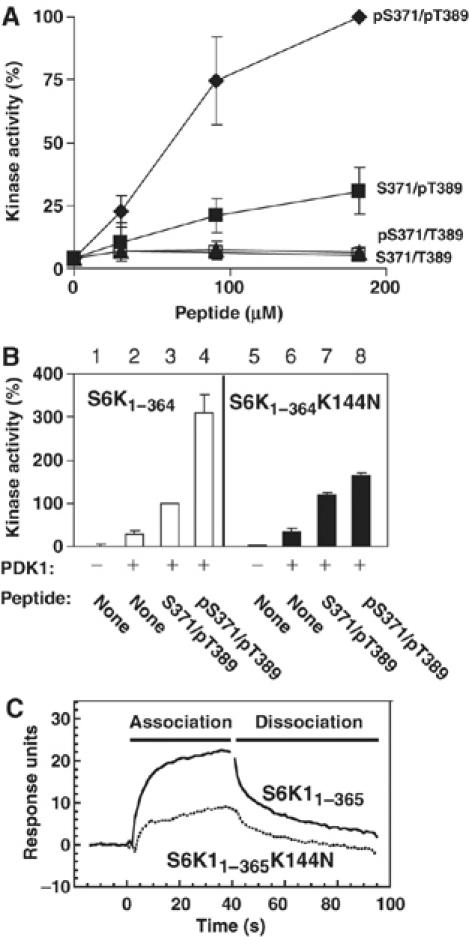
The tail phosphate synergistically enhances the ability of the HM phosphate to activate S6K, dependent on the tail phosphate-binding site. (A) The kinase activity of purified kinase domain of S6K1 (S6K11−364), pre-phosphorylated by PDK1 in the activation loop, was determined in the presence of increasing concentrations of synthetic S6K1 tail peptide (residues 366–395) that was either nonphosphorylated (S371/T389), phosphorylated at Ser371 (pS371/T389) or Thr389 (S371/pT389) or phosphorylated at both sites (pS371/pT389). (B) The kinase activity of S6K11−364 and S6K11−364 K144N, either pre-phosphorylated or not by PDK1, was determined in the absence or presence of 90 μM S371/pT389 or pS371/pT389. (C) pS371/pT389 S6K1 tail peptide was biotinylated and used to coat streptavidin Sensor Chips. The chips were thereafter analysed for binding to purified GST-S6K11−365 or GST-S6K11−365 K144N. (A–C) Experiments were repeated at least three times and activity data (expressed as percent) are means ±s.d.
We next investigated the role of the tail phosphate-binding site. The basic residue 4 mutant S6K11−364 K144N was activated normally by S371/pT389 peptide (Figure 5B, compare bars 3 and 7), but could not be hyperactivated by pS371/pT389 (compare bars 4 and 8), indicating that the tail phosphate-binding site mediates the kinase-activating effect of the tail phosphate. As a control, S6K11−364 K144N was phosphorylated and activated normally by PDK1 (compare bars 2 and 6 in Figure 5B, Supplementary Figure 6A), indicating that the K144N mutation did not compromise the tertiary structure of S6K1, but selectively disrupted the tail phosphate-binding site. As another control, PDK1, which lacks a tail phosphate-binding site (Figure 1C) but contains a pHM-binding site (Biondi et al, 2002; Frodin et al, 2002), was activated to the same extent by S371/pT389 and pS371/pT389 (Supplementary Figure 6C).
Surface plasmon resonance measurements revealed that the K144N mutation decreased the binding of S6K11−365 to pS371/pT389 peptide by ≈50% (Figure 5C). Similarly, the K144N mutation decreased the binding of S6K11−365 to pS371/T389 peptide by about 40%, whereas the K144N mutation had no effect on the binding of S6K11−365 to S371/pT389 peptide (data not shown). No specific binding of S6K11−365 or S6K11−365 K144N to S371/T389 peptide could be detected. Binding constants for these interactions could not be determined using the BiaCore instrument, because the affinities were too low for kinetic analysis, in accordance with the AC50 value of ≈60 μM for pS371/pT389 towards S6K11−365 (Figure 5A). As a control, PDK1 bound equally well to S371/pT389 and pS371/pT389 (Supplementary Figure 6D). We conclude that the binding contribution of pS371 in these experiments result from interaction with the tail phosphate-binding site.
These results support the model that the interaction between the tail phosphate and its binding site promotes the binding of the tail to the kinase domain and thereby the tail phosphate synergistically enhances kinase activation by the phosphorylated HM.
The tail phosphate promotes a compact AGC kinase conformation and protects the tail phosphate-binding site and αC helix from solvent exposure as revealed by amide hydrogen (1H/2H) exchange and mass spectrometry
Proteins analysed by hydrogen (1H/2H) exchange and mass spectrometry (HXMS) are incubated in D2O and the mass increase resulting from isotopic exchange of backbone amide protons (one per residue, except for proline) for solvent deuterons is measured by mass spectrometry. Differences in hydrogen-exchange rates between protein samples reflect differences in solvent exposure due to conformational change and/or protein–protein interaction. Sites of conformational change/interaction are revealed as regions with increased protection in analyses of peptic digests of the labelled proteins (local HXMS).
Addition of S371/pT389 and pS371/pT389 peptide to S6K11−365, resulted in ≈10 and ≈25 fewer deuterons incorporated, respectively, in S6K11−365 at early time points of analysis, as compared to no peptide added (Figure 6A). The nonphosphorylated S371/T389 peptide had no effect, demonstrating that nonspecific peptide binding to S6K11−365 did not contribute to the surface protection.
We next investigated the effect of the tail phosphate on deuteron uptake in a full-length AGC kinase. For this analysis we chose PKCζ, because only this kinase, among the ones tested, was found to be stoichiometrically phosphorylated at all the regulatory phosphorylation sites, which is critical for HXMS analysis. First, we demonstrated that the tail site T560 is essential for full PKCζ activity and identified basic residues 2 and 4 (K265 and K284) as key residues in the tail phosphate-binding site (Figure 6B). Second, we found that mutation of the tail site and its binding site greatly increased global deuteron uptake by PKCζ and to exactly the same extent (Figure 6C). At early time points of analysis, the tail phosphate protected ≈60 residues. Finally, we sought to identify specific regions in PKCζ protected by the tail phosphate by local HXMS analysis. Strikingly, peptide (a), corresponding to the regulatory αC helix was dramatically protected by the tail phosphate (Figure 6D). Moreover, protection was observed in peptide (b), but not in the overlapping peptide (c). This means that the 2 or 3 protected residues in peptide (b) are to be found in the sequence AKVLL, which encompasses a C-terminal residue of the glycine-rich loop and basic residue 2 (underlined). Protection was also observed in peptide (d), encompassing part of the αG helix, consistent with the finding that this helix is disturbed in PKB structures with a disordered αC helix. Slight protection was observed in peptide (e) corresponding to the start of the tail region. No protection was observed in peptide (f), located in a region of the large lobe not predicted to be affected by the tail phosphate or in peptide (g) located between the tail site and the HM, in agreement with the solvent exposed and flexible nature of this segment, as revealed by its near-complete deuteron uptake. For technical reasons, the sequence coverage of PKCζ was not complete and we could not therefore perform a full analysis of the effect of the tail phosphate on local HX.
The local HXMS analysis provides strong evidence that the tail phosphate interacts with basic residue 2 in the binding site and that it promotes a dramatic stabilization of the regulatory αC helix of the hydrophobic pocket. Moreover, the large extent of protection revealed by global HXMS strongly suggests that the tail phosphate promotes a significant allosteric change to a more compact conformation, which most likely corresponds to the closed, active AGC kinase conformation.
The tail site is not related to the turn motif site in PKA
We noticed that in active, but not inactive structures of PKA (1ATP/1YDR/1FMO/1L3R and 1CTP/1CMK, respectively), R56, which aligns with basic residue 2, binds E333 in the PKA tail. Moreover, E333 holds a similar position as the tail phosphate in our models (Figure 7A). Mutation of R56 or E333/E334 profoundly reduced autophosphorylation in the activation loop and catalytic activity of PKA (Figure 7B). Individual mutation of E333 had little effect, suggesting compensatory action by E332, which is in close proximity to R56 in active PKA structures. Similar compensatory action has been reported upon mutation of the tail site in PKCβII (Newton, 2001). Our results with PKA suggest that the E333/R56 interaction evolved from the tail phosphate/basic residue 1–4 interaction or vice versa, implying that the tail site should be aligned with E333, rather than with the turn motif site (Figure 7D). In such a revised alignment, the PKA turn motif site aligns well with S375 in RSK2, recently identified as a site of phosphorylation (Ballif et al, 2005). Interestingly, mutation of S375 to Ala reduced EGF-stimulated RSK2 activity by ≈20%, whereas a phosphate-mimicking Glu mutation did not (Figure 7C). The inhibitory effects of the S375A and the tail site S369A mutations were additive, indicating that the two phosphorylation sites stimulate RSK2 activity by distinct mechanisms. In agreement with this conclusion, modelling and dynamics simulations suggested that phosphoS375 does not interact with the tail phosphate-binding site but may bind neighbouring polar residues within the tail, thereby resembling the turn motif phosphate in PKA (Figure 7A).
Figure 7.
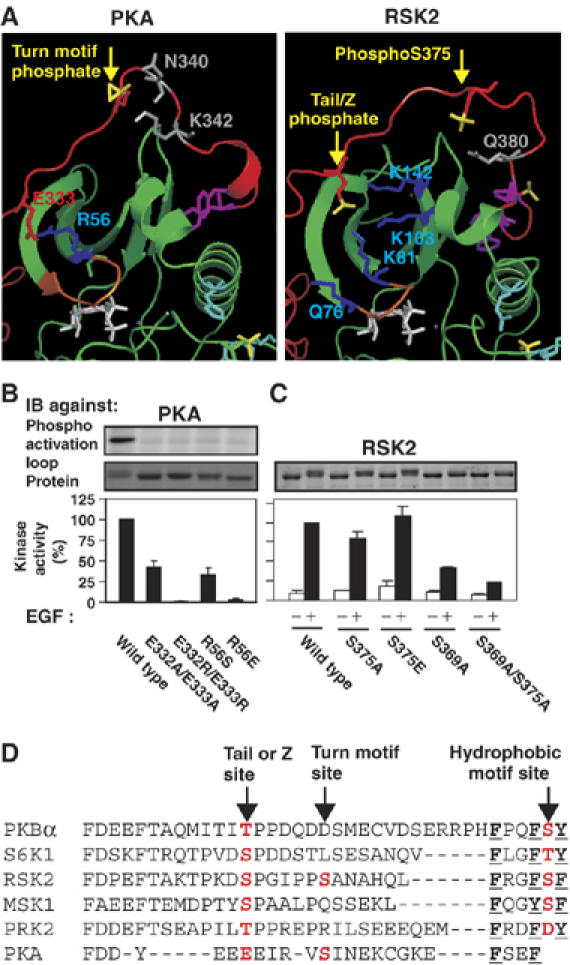
The tail phosphate may correspond to E333 of PKA. (A) Analogous positions and interactions of E333 of PKA and the tail phosphate. Left and right panels show a structure of PKA (1ATP) and our model of RSK2, respectively. Colour codes are described in the legend of Figure 1A, except that residues known/thought to interact with the turn motif phosphate of PKA and phosphoS375 of RSK2 are shown in grey. (B) Effect of mutation of E332/E333 and R56 on autophosphorylation and activity of PKAα expressed in Escherichia coli. (C) Effect of mutation of S375 in RSK2 expressed and analysed as described in the legend of Figure 2. (B, C) Data are means ±s.d. of three independent experiments. (D) Proposed alignment and naming of phosphorylation sites in the tail of AGC kinases.
We conclude that the tail site and the turn motif site are nonrelated rather than being the same site which adopts two distinct conformations in growth factor-activated AGC kinases and PKA, respectively. Thus, AGC kinases may contain either or both sites, and in PKA, the tail site consists of a phosphate-mimicking Glu. On the basis of these present findings, we propose to refer to the tail site in growth factor-activated AGC kinases as the Z (zipper) site instead of the turn motif site.
Discussion
The molecular mechanism whereby the tail phosphorylation site stimulates the activity of the growth factor-activated AGC kinases has been the last unresolved issue in their common activation mechanism. The data presented here support the following model: the tail phosphate interacts with a phosphate-binding site in the small lobe of the kinase domain, located on top of the ATP-binding, glycine-rich loop. The tail phosphate-binding site thereby provides an anchoring point for the tail, which increases the local concentration of the HM in the immediate vicinity of the binding site through which the HM stimulates kinase activity. This increase in local concentration is likely to be important, because the affinity of the phosphorylated HM for its binding site is very low, with estimated Kd values ranging from 30 to 600 μM among various AGC kinases (Frodin et al, 2002; Yang et al, 2002b; this study). By increasing the local concentration, the tail phosphate-binding site enhances the ability of the phosphorylated HM to interact with the hydrophobic pocket. Our model further proposes that the tail phosphate-binding site thereby allosterically affects the αC helix and protects the HM from dephosphorylation.
The tail phosphate-binding site thus promotes kinase activity by at least two mechanisms. In the first mechanism, which likely operates in all of the growth factor-activated AGC kinases, the tail phosphate-binding site allosterically supports the reordering of the HM-binding pocket, including the αC helix. Stabilization of the αC helix is thought to be of key importance in activation of AGC kinases. In the ordered αC helix, a conserved Glu stabilizes a conserved Lys, which positions the α- and β-phosphates of ATP, whereas other residues are thought to stabilize the activation loop, which likely constitutes a mechanism for cooperation between the HM and activation loop phosphates in stimulation of kinase activity. It is therefore a highlight of this study that local HXMS analysis showed dramatic stabilization of the αC helix by the tail phosphate. We estimate that the tail phosphate protects ≈50% of the residues in the αC helix, providing strong evidence for our model that the tail phosphate functions to aid the HM to bind and reorder the hydrophobic pocket. The results also provide the first in-solution evidence of stabilization of the αC helix during AGC kinase activation. Previously, the disorder-to-order transition of the αC helix was supported mainly by comparison of crystal structures of inactive and active PKB (Yang et al, 2002a, 2002b). Global HXMS analysis suggested that ≈60 residues were protected by the tail phosphate in PKCζ, a number exceeding the residues in the tail phosphate-binding sites and the αC helix. This suggests that the tail phosphate promotes a significant allosteric change, which we assume corresponds to stabilization also of the αB helix, another component of the HM-binding pocket and of the activation loop, leading to stabilization of the entire kinase domain in the closed, active conformation.
In the second mechanism, the tail phosphate-binding site stimulates kinase activity by increasing the phosphorylation level in the HM. This mechanism operates only in a subset of the AGC kinases such as S6K, MSK and to a slight extend RSK (this study), and likely also in several PKCs, where mutation of the tail site decreases HM phosphorylation (Parekh et al, 2000; Newton, 2003). The second mechanism is most likely linked to the first mechanism, because interaction of the phosphorylated HM with its binding site presumably renders the phosphate less accessible to phosphatases due to its binding to several charged/polar residues. The reason that the second mechanism may operate only in some AGC kinases may partly be due to the possibility that the HM in the various AGC kinases is targeted by distinct phosphatases with varying efficiency.
Basic residues 1 and 2 are located at the base of the glycine-rich loop that positions the γ-phosphate of ATP for phosphotransfer. In our HXMS analysis, the tail phosphate affected the flexibility of the segment AKVLL, which encompasses part of the glycine-rich loop and basic residue 2. It would be interesting to investigate whether the tail phosphate may promote kinase activity by modulating the position of the glycine-rich loop via basic residue 2 in addition to the two mechanisms described above.
Our data suggest that the tail phosphate-binding site is composed of four basic residues. The indispensability of individual basic residues varied among the kinases. For instance, in S6K1 and PRK2, individual mutation of basic residues 4 and 1–3, respectively, inhibited kinase activity by >85%, whereas in PKBα and RSK2, all four basic residues must be mutated to achieve substantial inhibition. In some kinases, we observed that individual mutation of a particular basic residue had negligible effect, but the same mutation enhanced the inhibitory effect of other basic residue mutations (data not shown). However, caution needs to be taken regarding conclusions on the exact contribution/importance of specific basic residues, because we introduced semiconservative substitutions like Ser, Thr or Asn with some phosphate-binding ability. Nevertheless, taken together, our results suggest that the tail phosphate may bind several distinct combinations of basic residues, which may change dynamically over time. This possibility is supported by the open appearance of the tail phosphate-binding site in the PKBβ crystal structure and by our molecular dynamics simulations. Similar dynamic interaction modes may also exist for the PKA turn motif phosphate, perhaps due to the flexible nature of the PKA tail region (Johnson et al, 2001). Thus, the turn motif phosphate binds R336, N340 and K342 in the structure 1FMO, N340 and K342 in 1ATP, and none of these residues in 1BKK. In the growth factor-activated AGC kinases, the four basic residues might perform partly distinct roles. For instance, basic residue 1 may function mainly to attract the tail phosphate, followed by docking of the tail phosphate to the more deeply positioned basic residues 2–4. Basic residues 3 and 4 are located in the nonflexible β-strands 3 and 5, respectively. Binding to these residues may fix the tail phosphate, allowing it to affect the position of the nonrigid glycine-rich loop via interaction with basic residue 2.
Recently, a crystal structure of the PKCι kinase domain was reported (Messerschmidt et al, 2005). In this structure, the tail phosphate binds basic residues 3 and 4, in agreement with our study. Unlike our model, the authors speculated that the phosphate may function (1) to stabilize the kinase domain, referring to the destabilizing effect of mutation of the turn motif site of PKA and the kinase-inactivating effect of mutation of the tail site in PKC, (2) to push the tail out of the active site or (3) to regulate the interaction of the kinase domain with the flanking signalling modules in PKC. The role of the tail phosphate and its interactions were not investigated. The side chain of the basic residue 2 Lys was not visible in the structure, but we noted that its backbone is located immediately beneath the tail phosphate, allowing binding of the side chain to the tail phosphate. We therefore mutated the tail site T555 and the basic residues 2–4 and observed a ≈25% reduction of kinase activity (Supplementary Figure 7). The mutational analysis thus revealed that the interaction is required for full PKCι activation, but also showed that the tail phosphate is less important for PKCι than for other AGC kinases that we have analysed.
This study provides the first characterization of the cooperation between all three conserved phosphates in stimulation of AGC kinase activity by using an in vitro reconstitution assay based on long tail peptides. Our data showed that the three phosphates act in a hierarchal manner: the tail phosphate has no activating effect alone or together with the activation loop phosphate. However, the tail phosphate synergistically enhances the ability of the HM phosphate to stimulate kinase activity in cooperation with the activation loop phosphate. Global HXMS analysis in this system suggested that the tail phosphate promoted a significant allosteric change, which we assume represents HM-mediated transition to the closed, active AGC kinase conformation. Our model and data thus imply that the activating conformational change induced by binding of the HM to the hydrophobic pocket is triggered not only by HM phosphorylation, as previously suggested, but is also regulated by the dual phosphorylation events in the tail site and the HM, working in a cooperative manner.
These results together with previous studies underscore the extensive cooperation of the three conserved phosphorylation sites with respect to shifting the equilibrium of the AGC kinase catalytic domain from the inactive, open conformation towards the active, closed conformation during stimulus-induced activation. This cooperativity is likely also an important determinant during inactivation. In many of the kinases, the phosphorylation sites are rapidly dephosphorylated upon cessation of the activating stimulus. In this process, dephosphorylation at one site may greatly decrease the ability of the remaining phosphates to support the active, closed and more phosphatase-resistant conformation, resulting in accelerated dephosphorylation of the remaining phosphates and complete inactivation of the kinase.
Elevated PKB activity is essential for the progression of many human cancers and may result from mutations in, for example, PI3K or phosphatase and tensin homolog deleted on chromosome 10 (PTEN). Recently, two point mutations in PKB were identified in colorectal cancer (Parsons et al, 2005). This study reports mutations in the tail phosphate-binding site, which yielded a considerable degree of activation and which stimulated cell growth (C Hauge and M Frodin, unpublished observation). Activation appeared to result from increased phosphorylation in the HM and the activation loop. Presumably, the aberrantly exposed HM of PKB in these mutants becomes hyperphosphorylated by the physiological PKB HM kinase or by some other HM kinase. Profound hyperphosphorylation of PKB was not observed after severe disruption of zipper function such as in PKBα-T450A. Conceivably, if the HM of PKB becomes excessively exposed, phosphatase action counteracts kinase action. The increased activation loop phosphorylation may be a consequence of the increased HM phosphorylation, because the phosphorylated HM is thought to promote the closed, and more phosphatase-resistant conformation of the AGC kinase domain. Alternatively, the abnormally exposed HM may increase activation loop phosphorylation by recruitment of PDK1, as HM phosphorylation may enhance phosphorylation of PKB by PDK1 under certain conditions (Scheid et al, 2002). Regardless of the mechanism, our results suggest that elevated PKB activity in cancer cells may result from mutations in the tail phosphate-binding site of PKB.
Our study suggests that the tail site and the turn motif site are two distinct phosphorylation sites that should not be aligned. Because it is problematic to use the designation ‘turn motif' for two distinct sites, we propose to refer to the tail site as the Z (zipper) site, a name that reflects one major function of this phosphorylation site according to the present findings.
In conclusion, this study provides important information on the missing pieces in the core mechanism, whereby three conserved phosphorylation sites stabilize the active conformation of up to 26 human AGC kinases, which thus represents one of the most general activation mechanisms reported in the human kinome.
Materials and methods
Procedures are described in Supplementary data.
Supplementary Material
Supplementary Methods
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Acknowledgments
We thank Simon Arthur, Dundee, for antibodies against MSK1 and Valerie Hindie, Homburg, and Francis Shaeffer, Paris, for microcalometry analysis. This work was supported by grants from the Danish Medical Research Council (22-02-0136), the Danish Cancer Society (DP01004) and the Deutsche Forshungsgeselschaft (BI 1044/2-1).
References
- Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J 15: 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- Ballif BA, Roux PP, Gerber SA, MacKeigan JP, Blenis J, Gygi SP (2005) Quantitative phosphorylation profiling of the ERK/p90 ribosomal S6 kinase-signaling cassette and its targets, the tuberous sclerosis tumor suppressors. Proc Natl Acad Sci USA 102: 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellacosa A, Chan TO, Ahmed NN, Datta K, Malstrom S, Stokoe D, McCormick F, Feng J, Tsichlis P (1998) Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene 17: 313–325 [DOI] [PubMed] [Google Scholar]
- Biondi RM, Cheung PC, Casamayor A, Deak M, Currie RA, Alessi DR (2000) Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. EMBO J 19: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi RM, Kieloch A, Currie RA, Deak M, Alessi DR (2001) The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J 20: 4380–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi RM, Komander D, Thomas CC, Lizcano JM, Deak M, Alessi DR, van Aalten DM (2002) High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. EMBO J 21: 4219–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby KN, Morrice N, Caudwell FB, Avruch J, Cohen P (1998) Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J Biol Chem 273: 1496–1505 [DOI] [PubMed] [Google Scholar]
- Engel M, Hindie V, Lopez-Garcia LA, Stroba A, Schaeffer F, Adrian I, Imig J, Idrissova L, Nastainczyk W, Zeuzem S, Alzari P, Hartmann RW, Piiper A, Biondi RM (2006) Allosteric activation of the protein kinase PDK1 with low molecular weight compounds. EMBO J 25: 5469–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodin M, Antal TL, Dummler BA, Jensen CJ, Deak M, Gammeltoft S, Biondi RM (2002) A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J 21: 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodin M, Jensen CJ, Merienne K, Gammeltoft S (2000) A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J 19: 2924–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauge C, Frodin M (2006) RSK and MSK in MAP kinase signalling. J Cell Sci 119: 3021–3023 [DOI] [PubMed] [Google Scholar]
- Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS (2001) Dynamics of cAMP-dependent protein kinase. Chem Rev 101: 2243–2270 [DOI] [PubMed] [Google Scholar]
- Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253: 407–414 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Deak M, Morrice N, Cohen P (1999) Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem J 344 (Part 1): 189–197 [PMC free article] [PubMed] [Google Scholar]
- Kozma SC, Thomas G (2002) Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays 24: 65–71 [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Yamamoto T, Kikkawa U (2004) Distinct activation mechanisms of protein kinase B by growth-factor stimulation and heat-shock treatment. Biochemistry 43: 4284–4293 [DOI] [PubMed] [Google Scholar]
- McCoy CE, Campbell DG, Deak M, Bloomberg GB, Arthur JS (2004) MSK1 activity is controlled by multiple phosphorylation sites. Biochem J 387: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt A, Macieira S, Velarde M, Badeker M, Benda C, Jestel A, Brandstetter H, Neuefeind T, Blaesse M (2005) Crystal structure of the catalytic domain of human atypical protein kinase C-iota reveals interaction mode of phosphorylation site in turn motif. J Mol Biol 352: 918–931 [DOI] [PubMed] [Google Scholar]
- Moser B, Dennis P, Pullen N, Pearson R, Williamson N, Wettenhall R, Kozma S, Thomas G (1997) Dual requirement for a newly identified phosphorylation site in p70s6k. Mol Cell Biol 17: 5648–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC (2001) Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev 101: 2353–2364 [DOI] [PubMed] [Google Scholar]
- Newton AC (2003) Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J 370: 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh DB, Ziegler W, Parker PJ (2000) Multiple pathways control protein kinase C phosphorylation. EMBO J 19: 496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, Silliman N, Ptak J, Szabo S, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Lengauer C, Velculescu VE (2005) Colorectal cancer: mutations in a signalling pathway. Nature 436: 792. [DOI] [PubMed] [Google Scholar]
- Roux PP, Blenis J (2004) ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev 68: 320–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh M, Pullen N, Brennan P, Cantrell D, Dennis PB, Thomas G (2002) Regulation of an activated S6 kinase 1 variant reveals a novel mammalian target of rapamycin phosphorylation site. J Biol Chem 277: 20104–20112 [DOI] [PubMed] [Google Scholar]
- Scheid MP, Marignani PA, Woodgett JR (2002) Multiple phosphoinositide 3-kinase-dependent steps in activation of protein kinase B. Mol Cell Biol 22: 6247–6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KJ, Carter PS, Bridges A, Horrocks P, Lewis C, Pettman G, Clarke A, Brown M, Hughes J, Wilkinson M, Bax B, Reith A (2004) The structure of MSK1 reveals a novel autoinhibitory conformation for a dual kinase protein. Structure (Camb) 12: 1067–1077 [DOI] [PubMed] [Google Scholar]
- Weng Q-P, Kozlowski M, Belham C, Zhang A, Comb MJ, Avruch J (1998) Regulation of the p70 S6 kinase by phosphorylation in vivo. Analysis using site-specific anti-phosphopeptide antibodies. J Biol Chem 273: 16621–16629 [DOI] [PubMed] [Google Scholar]
- Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D (2002a) Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat Struct Biol 9: 940–944 [DOI] [PubMed] [Google Scholar]
- Yang J, Cron P, Thompson V, Good VM, Hess D, Hemmings BA, Barford D (2002b) Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol Cell 9: 1227–1240 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8


